Abstract
Acyl carrier protein (ACP) is an essential co-factor protein in fatty acid biosynthesis that shuttles covalently bound fatty acyl intermediates in its hydrophobic pocket to various enzyme partners. To characterize acyl chain-ACP interactions and their influence on enzyme interactions, we performed 19 molecular dynamics (MD) simulations of Escherichia coli apo-, holo-, and acyl-ACPs. The simulations were started with the acyl chain in either a solvent-exposed or a buried conformation. All four short-chain (≤C10) and one long-chain (C16) unbiased acyl-ACP MD simulation show the transition of the solvent-exposed acyl chain into the hydrophobic pocket of ACP, revealing its pathway of acyl chain binding. Although the acyl chain resides inside the pocket, Thr-39 and Glu-60 at the entrance stabilize the phosphopantetheine linker through hydrogen bonding. Comparisons of the different ACP forms indicate that the loop region between helices II and III and the prosthetic linker may aid in substrate recognition by enzymes of fatty acid synthase systems. The MD simulations consistently show that the hydrophobic binding pocket of ACP is best suited to accommodate an octanoyl group and is capable of adjusting in size to accommodate chain lengths as long as decanoic acid. The simulations also reveal a second, novel binding mode of the acyl chains inside the hydrophobic binding pocket directed toward helix I. This study provides a detailed dynamic picture of acyl-ACPs that is in excellent agreement with available experimental data and, thereby, provides a new understanding of enzyme-ACP interactions.
Fatty acid (FA)4 synthesis occurs in a well structured and characterized cycle of condensation and reduction reactions that elongate acyl chains before they reach maturity and are cleaved off for utilization in various anabolic pathways (1). There are two main types of machineries that perform this process, known as the type I fatty acid synthase (FAS) systems found in mammals and fungi and the type II systems existing in bacteria and eukaryotic plastids (2). Type I systems are composed of a single long polypeptide chain that contains all the enzymes and other proteins necessary to accomplish fatty acid synthesis. These mega-complexes fold into large machineries, yielding reaction chambers that are readily accessible to the enzymes of FA synthesis for rapid catalysis (3-5). In sharp contrast, type II or dissociated FAS systems express each of the enzymes necessary for fatty acid synthesis as individual proteins (6). Given how vastly different these two systems are between humans and bacteria, there has been a conscious effort to target FA synthesis in bacteria through novel antibiotics (7). A number of antibacterial agents acting on FAS components have already been discovered, demonstrating that these systems may be successfully used as a target for new types of antibiotics (8-10). For example, the pantothenamide class of antibiotics has been shown to incapacitate the FAS system by creating inactive forms of acyl carrier protein (ACP) (9).
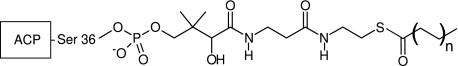
As a central player in both systems, ACP shuttles the acyl chain intermediates to various enzymes of the FAS systems. In type II systems, ACPs are small, acidic proteins of about 9 kDa. The structure of ACP is well conserved in different organisms, consisting of three major helices (I, II, IV) that run largely parallel to each other, with helix III markedly shorter and almost perpendicular to the bundle (6). The acyl chain intermediates in FA synthesis are bound covalently by means of a prosthetic linker on ACP that is derived from coenzyme A in an enzymatic reaction catalyzed by ACP synthase (ACPS) (Fig. 1). This reaction converts apo-ACP to holo-ACP, which has the phosphopantetheine arm attached to a conserved serine residue (residue 36 in Escherichia coli). The prosthetic linker contains a terminal sulfhydryl group that is utilized to covalently bind the acyl chain intermediates in a thioester linkage (Fig. 1). The hydrophobic acyl chain attached to ACP is positioned in between the four-helix bundle in a pocket that is made up of a core of hydrophobic residues (11-13). During substrate presentation, the acyl chain must somehow overcome stabilizing hydrophobic interactions with the interior of ACP and inject it into the active site of a partner enzyme. The highly dynamic nature of ACP is thought to be a crucial aspect of its ability to interact with a large number of enzymes as well as the plethora of acyl chains and their intermediates (2). These adducts include acyl chains ranging from 3 to 18 carbons in length and their β-keto-, β-hydroxy-, as well as enoyl intermediates and unsaturated fatty acyl groups. A number of structures of apo-, holo-, and acyl-ACP are available as well as structures of ACP-enzyme complexes, highlighting important aspects to ACP function such as helix II, which is known as the recognition helix (11-15). In addition, NMR studies have provided insights into the backbone dynamics of apo- and holo-ACP (16). Nevertheless, a dynamic picture of acyl-ACP has been difficult to obtain and a detailed understanding of acyl-chain:ACP interactions remains elusive. It is also unclear how the enzymes of the FAS pathway recognize the appropriate acyl-ACP. Considering that ACP makes up ∼0.25% of all soluble protein in E. coli suggests that there are mechanisms for enzymes to specifically recognize what type of acyl-ACP is bound (17). To understand this process, it is essential that a dynamic picture of the substrate behavior attached to ACP is obtained. Furthermore, if the pathway of substrate binding by ACP is revealed, potential drug target sites that are not apparent from static structures alone may be identified.
FIGURE 1.
Chemical structure diagram of the phosphopantetheine linker and the attached acyl chain via the thioester linkage. The opposite end of the prosthetic group is attached to the conserved Ser-36 in ACP. The acyl chains in our studies were extended by two carbon units ranging from n = 1 for butyryl groups to n = 8 for octadecanoyl groups.
To evaluate these questions we performed extensive molecular dynamics (MD) simulations of apo-, holo-, and saturated acyl-ACPs ranging from 4 to 18 carbon groups in length. The holo- and acyl-ACP simulations were conducted both with the prosthetic group in a solvent-exposed state as well as in a solvent-shielded conformation inside the protein. The simulations provide novel information on the manner of substrate protection by ACP as well as differentiating motions between the apo-, holo-, and acylated forms of the protein. Our findings are substantiated by excellent agreement with experimental data in the literature.
EXPERIMENTAL PROCEDURES
Simulation Setup—The MD simulations on E. coli ACP were based on the high resolution (1.2 Å) crystal structure of butyryl-ACP (Protein Data Bank code 1L0I), which contains the acyl chain buried inside the hydrophobic pocket of ACP (11). The Protein Data Bank file was first checked by submitting it to the Biotech Validation Suite for Protein Structures server, and consequently, the side chain of Gln-19 was flipped. Multiple protein conformations for certain atoms were removed based on the best structural agreement in comparison to another crystal structure of Escherichia coli ACP (11). 1L0I also contained the mutation I62M, which was used to incorporate a selenomethionine residue into the protein to address the crystallographic phase problem (11). For our studies we reversed this mutation through alignments to 1L0H in the program MOLMOL (18) and transferring of coordinates. The resulting structure was used as the starting point for all the simulations described here. The simulations conducted can be categorized into two main groups based on the initial orientation of the acyl chain in the starting structures. The first group of simulations contains ACP with its prosthetic linker in a completely solvent-exposed orientation, whereas the second group is composed of ACP with the prosthetic group in a solvent-inaccessible conformation as is seen in 1L0I. Each of these groups is composed of various ACP members, differing in the length of the prosthetic group attachment. Apo- as well as holo-ACP simulations were set up in addition to saturated acyl chain simulations ranging from butyryl-ACP (4 carbon chain) to octadecanoyl-ACP (18 carbon chain) (Table 1). For the solvent-exposed simulations, the prosthetic group was pointed vertically up from the protein, extending along the axis of helix II. This conformation was chosen as an unbiased orientation such that the prosthetic group can move in as many directions as possible. The starting structures for simulations of ACP with solvent-shielded acyl groups were derived from the butyryl-ACP crystal structure 1L0I by extending the butyryl group incrementally by two carbon groups.
TABLE 1.
ACP MD simulations solvent shielded acyl-chain properties
|
Acyl chain
|
Carbon groups
|
SASA
|
Acyl chain
volumec
|
Average cavity volume
|
||
|---|---|---|---|---|---|---|
| Acyl chain | Shieldeda | Phosphopantetheineb | ||||
| Å2 | % | Å2 | Å3 | Å3 | ||
| Apo | 78.5 | 25.9 | ||||
| Holo | Linker only | 202.1 | 86.7 | |||
| Butyryl | 4 | 2.1 | 98.8 | 249.1 | 82 | 140.7 |
| Hexanoyl | 6 | 3.6 | 98.4 | 257.4 | 116 | 170.9 |
| Octanoyl | 8 | 3.4 | 98.8 | 240.3 | 149 | 200.4 |
| Decanoyl | 10 | 13.4 | 95.2 | 258.9 | 182 | 231.9 |
| Dodecanoyl | 12 | 20.4 | 94.6 | 275.3 | 214 | 185.8 |
| Tetradecanoyl | 14 | 33.4 | 92.2 | 284.0 | 247 | 186.2 |
| Hexadecanoyl | 16 | 63.5 | 86.6 | 309.9 | 280 | 163.5 |
| Octadecanoyl | 18 | 67.4 | 86.9 | 315.1 | 314 | 181.2 |
Based on the total surface area of the acyl chain.
Refers to Ser-36 side chain only for apo and Ser-36 as well as the phosphopantetheine group for all others.
Calculated using the program voidoo (28).
The starting structures were initially solvated with explicit simple point charge (SPC) water in a rhombic dodecahedron box. The distance between the protein and edge of the box was ∼1.5 nm, to ensure that the minimum distance between periodic images would not be too low, even with the prosthetic group in an extended conformation directed away from ACP into the solvent. All simulations were performed using the GROMACS 3.2 MD package (19, 20) and the GROMOS96 43a2 force field containing improved alkane dihedrals parameters (21). Virtual sites were used for hydrogens to remove the highest frequency hydrogen vibrations, permitting the use of a 5-fs time-step (22). The neighbor search was conducted according to the grid method, updating the neighbor list every 4 steps and a neighbor list cut-off of 0.9 nm. All the MD simulations were carried out using periodic boundary conditions. Constant temperature (300 K, τT = 0.1ps) and pressure were maintained by coupling to a bath using the Berendsen algorithm (23). Pressure was kept at 1 bar with isotropic temperature coupling, tau τP of 1.0 ps, and compressibility of 4.5 × 10-5 bar-1. Bond lengths were restrained using the Linear Constraint Solver (LINCS) method (24). Long range electrostatic interactions were calculated according to Fast Particle-Mesh Ewald (PME) electrostatics (25) with a cutoff of 0.9 nm. Van der Waal energies were collected with a twin range cutoff of 0.9/1.4 nm. The bonds and angles of the simple point charge (SPC) water molecules were constrained using the SETTLE algorithm (26). After equilibration of the water while restraining the protein, the overall charge of the system was neutralized with 15 sodium counterions (14 ions for apo ACP). Subsequently, the protein was gradually released by decreasing the force constant on the protein in 5 distinct steps (1000, 100, 10, 1, and 0 kJ/mol nm2) of 50 ps simulations each. The simulations of ACP with the acyl chain inside the binding pocket as well as the apo- and holo-ACP simulations were conducted for 20 ns each. The simulations starting off with the prosthetic groups in solvent-exposed conformations were conducted for 50 ns each. Analysis of the trajectories was performed using various GROMACS analysis programs as well as SURFNET (27) for the ACP cavity size analysis and VOIDOO (28) for acyl volume calculations. Averaged values for the solvent-shielded simulations were calculated for the last 14 ns of the trajectory, which displayed steady r.m.s.d. values.
Phosphopantetheine Topology Building—The phosphopantetheine linker that is attached to ACP represents a new topology non existent in the GROMOS96 library of building blocks (Fig. 1). Most of the parameters such as bond lengths, charges, angles, dihedrals, and improper dihedrals were obtained from subcomponents that have been previously characterized in the GROMOS96 43a2 forcefield. Therefore, most parts of the prosthetic group could be modeled based on these previously established groups. The missing partial charges for the thioester group were calculated using quantum mechanics. The quantum mechanics calculations were performed using GAUSSIAN 03 (29) and have been previously described in detail (30). The partial charges used were 0.13 for the sulfur atom, 0.25 for the carbonyl carbon, and -0.38 for the carbonyl oxygen.
RESULTS
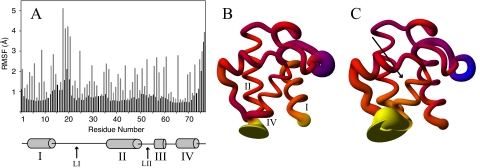
ACP Protein Dynamics—The r.m.s.d. of the backbone atoms of the trajectories remained low compared with their starting structures, stabilizing between 2 and 2.5 Å. In all the simulations ACP shows no significant degree of unfolding beyond a slight unwinding of helix IV in the extreme C-terminal part of the protein. Throughout the apo-, holo-, and acyl-ACP simulations similar regions of elevated root mean square fluctuations (RMSF) values were identified. Loop I between helices I and II shows highly flexible regions immediately after helix I and just before helix II (Fig. 2). The central region of this loop is remarkably stable, displaying backbone RMSF values around 0.6 Å, which is similar to the least flexible regions of ACP. This is because of hydrogen bonds formed between the backbone atoms of residues Asn-24, Ala-26, and Phe-28 with the backbone atoms of Gln-66, Val-65, and Thr-63, respectively, immediately preceding helix IV. Another noticeably flexible region spans residues 47 to 57, which connects helices II and III. Although the magnitude of the fluctuations in most regions of ACP is consistent between apo-, holo-, and the acyl-ACP simulations, there is a clear difference in this region. ACP displays higher RMSF values from residues 47 to 57 when harboring an acyl chain in its hydrophobic binding pocket in each of the simulations conducted. Averaged over all acyl-ACP simulations with an acyl chain occupying the binding cavity, the RMSF is 0.95 ± 0.12 Å, whereas the RMSF is only 0.79 ± 0.05 Å in apo- and holo-ACP. Side-chain fluctuations display the same pattern, with higher RMSF values in acyl-ACP compared with apo- and holo-ACP in this region. The central portions of the helices in ACP are the most rigid segments of the protein, although the ends of the helices show pronounced flexibility as well (Fig. 2).
FIGURE 2.
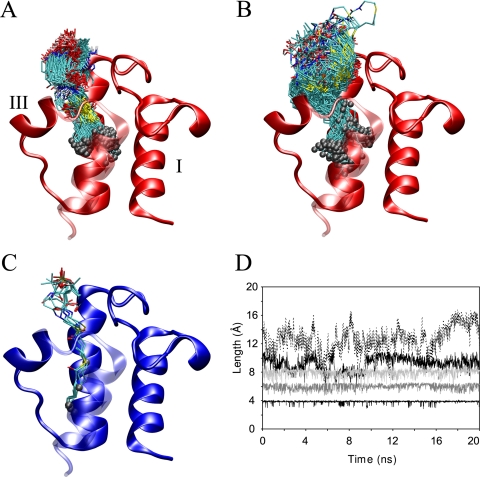
A, RMSF of apo-ACP backbone (black) and side-chain (gray) atoms, shown for each residue. B, holo-ACP in sausage representation, where the thickness of the ribbon is proportional to the backbone fluctuations seen during the simulation. The diagram is colored according to the difference in fluctuations compared with apo-ACP, ranging from blue (less flexible than apo-ACP) to yellow (more flexible than apo-ACP) and red being similarly flexible to apo-ACP. C, sausage representation of octanoyl-ACP colored analogous to B. The arrow indicates the position of the bound acyl chain.
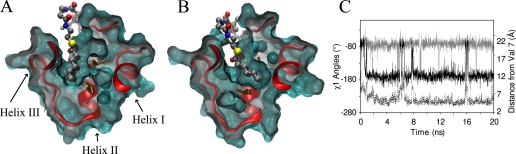
Buried Acyl-ACP Simulations—Acyl-ACP MD simulations with solvent-shielded acyl groups ranging from 4 to 18 carbons in length were performed based on the butyryl-ACP crystal structure (11). All of the simulations retained the acyl chain inside the binding pocket, consistent with what has been described in acyl ACP structures from E. coli and spinach (11-13). The binding pocket is located between the three major helices, with the entrance close to the N-terminal end of helix II and additionally bordered by loop I and helix III. The binding cavity is split up into two different binding pockets that are observed to house the tip of the acyl chains (Fig. 3B). The first pocket is similar to the starting structure, analogous to the conformation found in the crystal and NMR acyl-ACP structures between helices II, III, and IV. The second subpocket of the cavity is located more toward helix I, using the same entrance but now residing between helices I, II, and IV (Fig. 3B, Table 2). Whether the tip of the acyl chain binds the first or the second pocket of the binding cavity is determined through a quasi switch that is mediated by the side-chain orientation of Leu-42 and Leu-46. The χ1 angle of Leu-46 measures ∼68° in the starting conformation but switches to ∼-180° almost always when the acyl chain occupies the second sub-pocket (Fig. 3C). The switch in the dihedral angle of Leu-46 opens up a path into the alternate binding cavity of ACP. As a second requirement for the acyl chain to occupy this side of the binding pocket, the χ1 angle of Leu-42 must switch to the energy minimum around -68°. In the starting structures the χ1 angle of Leu-42 measures ∼180° but during the simulation it commonly switches to about -68°. Although this shift does not necessitate acyl chain entry into sub-pocket II, the Leu-42 χ1 angle always measures -68° when sub-pocket II is occupied (Fig. 3C). Similarly to Leu-46, the movement of Leu-42 is required to open up the pathway into the second binding cavity, such that the tip of the acyl chain may enter. Overall, the majority of the simulation time is spent with the tip of the acyl chain oriented into sub-pocket II. The decanoyl chain is the exception to this pattern, as its group resides in sub-pocket I for most of the simulation. In sub-pocket I, the decanoyl chain wraps around helix III at times rather than stick straight into the cavity. The octadecanoyl group adopts a similar conformation and at times penetrates into the solvent between helices III and IV. This behavior was not seen in any of the other simulations.
FIGURE 3.
Clipped surface representation of hexanoyl-ACP highlighting sub-pocket I (A) and sub-pocket II (B). The protein is oriented such that helix IV is directed toward the viewer, similarly to e.g. Figs. 2 and 4A. The side chains of the crucial residues Leu-42 and Leu-46 (brown) are shown in both a closed (A) and an open (B) conformation toward sub-pocket II, demonstrating their role as gating residues. C, χ1 angles of Leu-42 (gray) and Leu-46 (black) of dodecanoyl-ACP. The conformation of these residues is linked to the opening and closing of sub-pocket II and its occupancy by the acyl chain, which is indicated by the distance to Val-7 that resides at the bottom of sub-pocket II (dotted line). When the χ1 of Leu-46 measures -180°, the distance to Val-7 is low (∼5 Å), indicating sub-pocket II is filled by the acyl chain.
TABLE 2.
ACP residues contacted in the acyl-chain binding pocket
|
MD
simulationsa
|
Protein Data Bank structures
|
||||
|---|---|---|---|---|---|
| Common residues | Sub-pocket 1 only | Sub-pocket 2 only | NOEs C10b,c | NOEs C18b,c | X-ray, C10d |
| Val-7, Ile-10, Ile-11 | Glu-4 | ||||
| Phe-28, Val-29 | Val-29 | Phe-28, Val-29 | |||
| Thr-39 | Glu-30, Thr-39 | Thr-39 | |||
| Leu-42, Val-43, Leu-46 | Leu-42, Leu-46 | L46, Glu-47 | Leu-42, Leu-46, Glu-47 | ||
| Ala-59 | Ile-54 | Pro-55, Ala-59 | Thr-52, Ile-54, Ala-59 | ||
| Glu-60, Ile-62, Ala-68 | Glu-60, Ile-62, Ala-68 | Ile-62, Ala-67 | Glu-60, Ile-62, Thr-63, Ala-68 | ||
| Ile-72 | Tyr-71 | Ile-72 | Tyr-71 | Tyr-71, Ile-72 | |
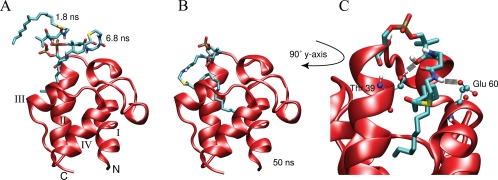
Solvent-exposed Acyl-ACP Simulations—Five of the eight 50-ns acyl-ACP MD simulations with the acyl chain oriented into the solvent in the starting structure displayed a transition into the hydrophobic binding pocket. The five forms of ACP are butyryl, hexanoyl, octanoyl, decanoyl, and hexadecanoyl. The structures converged very well with the simulations of solvent-shielded acyl-ACPs, yielding very low r.m.s.d. values (supplemental Fig. 1). In the other three trajectories the acyl chain never traversed into the hydrophobic pocket, and usually the prosthetic linker was engaged in hydrogen bonding with the protein which prevented the arm from sampling a broader region of the conformational space. Notably, the longer acyl-ACP simulations did not immerse into the pocket as frequently, as only one of the four longer (>C10) acyl chains was able to find the binding pocket. In those cases additional simulations were conducted with new sets of randomly generated velocities, yet a transition into the binding pocket was not observed. Typically, the acyl chain starts off in an entirely solvent-exposed state and rapidly fluctuates due to the energetic penalty of having the extended hydrophobic acyl chain in contact with the aqueous solvent (Fig. 4A). Using the solvent-accessible surface areas of the acyl chains, the difference in solvation free energy of the acyl chain going from the entirely solvent-exposed state to the buried conformation was calculated (31). The differences measure between 7 and 25 kJ/mol, depending on the acyl chain length, with longer chains resulting in larger energy differences. The acyl chain then usually folds onto itself and associates with the protein. During this interplay, the prosthetic linker is stabilized via hydrogen bonding with the protein. The acyl chain by contrast continues to flop around in the solvent because it does not form favorable interactions with the solvent or the protein surface, and this appears to be the source of disorder for the phosphopantetheine group. In the butyryl, hexanoyl, octanoyl, and hexadecanoyl simulations, the acyl chain then finds the entrance to the hydrophobic cavity and, once found, moves inside rapidly (Fig. 4B). The decanoyl-ACP simulation is an exception to this, as it entered into the hydrophobic pocket by slowly pushing through the side of the protein between helix II and loop II. The prosthetic linker is always actively involved in hydrogen bonding with water molecules or the protein itself. Throughout the 50 ns of simulation time, the prosthetic group averages about 10 hydrogen bonds with the solvent at any given time. These interactions with the solvent are observed independently of the prosthetic group orientation; in both the solvent-exposed and buried acyl-chain states. Hydrogen bonds are formed consistently between the prosthetic linker and two residues lining the entrance to the hydrophobic cavity. The hydroxyl group of Thr-39 in helix II and also the side-chain carboxyl group of Glu-60 form hydrogen bonds with virtually all potential donors/acceptors on the prosthetic arm, independent of which acyl chain is attached (Fig. 4C).
FIGURE 4.
Snapshots of the hexadecanoyl-ACP simulation started with a solvent-exposed acyl chain. The prosthetic group was initially extended away from the protein entirely, extending upwards from helix II. Soon after the start of the simulation, the acyl chain folds onto itself and moves toward the protein as is seen in two snapshots at 1.8 and 6.8 ns (A). When the tip of the acyl chain finds the entrance to the cavity it moves into the pocket, drawing the linker away from transiently formed hydrogen bonds with the protein (B). C, shows a close up view of the prosthetic group inside the hydrophobic cavity along with the two major hydrogen bonding partners, Thr-39 and Glu-60.
Acyl Chain-Binding Pocket Interactions—Although none of the acyl groups escape from the hydrophobic cavity of ACP, the acyl chains do undergo significant movement inside the cavity. The fluctuations of the acyl chains are dependent on the length of the acyl chain bound in the hydrophobic pocket. The shortest acyl chains with four and six carbon groups move similarly, as they can penetrate to the bottom of the cavity, but do not remain there consistently. The longer hexanoyl group remains in the cavity more steadily, because its acyl group can reach the bottom of the pocket more easily. The octanoyl group is more adept at this, filling the cavity with its acyl chain throughout the MD trajectory and fluctuating only very little. The longer acyl groups temporarily reach the bottom of they hydrophobic pocket as well; however, unlike the octanoyl group, they are unable to remain there for any extended period of time. The extent to which the longer (>C8) acyl chain remains buried at the bottom of the cavity appears to be related to the hydrogen bonding interactions formed between the prosthetic linker and the rest of ACP as well as the length of the acyl chain present. In general, a longer carbon tail destabilizes the acyl chain due to the size of the hydrophobic portion that does not fit into the pocket, but its mobility is also affected by the hydrogen bonding contacts made between the phosphopantetheine linker and the rest of the protein.
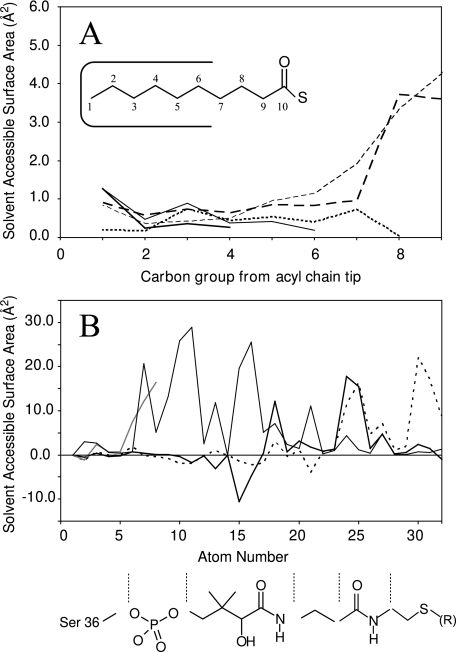
This pattern is also reflected by the solvent-accessible surface area (SASA) of the various acyl groups. The tip of the acyl groups are similarly protected independent of the length of the carbon chain. In each case the seven last carbon groups are well protected as they reside inside the hydrophobic binding pocket with SASAs of <2 Å2 (Fig. 5A). The butyryl-, hexanoyl-, and octanoyl-ACPs display the lowest SASA for their acyl chain during the simulations, averaging to no more than 3.6 Å2 of the fatty acyl group exposed to the solvent. The SASA increases dramatically with acyl chains longer than eight carbon groups in length (Table 1). For example, decanoyl-ACP has almost three times as much of its acyl chain exposed (Table 1). Increases in SASA of the acyl chain are seen consistently with increasing fatty acyl length, up to 67 Å2 for octadecanoyl-ACP. Although only seven carbon groups fit into the binding pocket, in octanoyl-ACP the eighth carbon group is also shielded from the solvent (Fig. 5A). This is because the eighth carbon is part of the thioester link and thereby is additionally protected by the oxygen atom attached to it, which is positioned to face the solvent. The phosphopantetheine linker is also least solvent-accessible when shorter acyl groups are attached. The majority of the linker is similarly solvent-accessible in all the simulations except for the sulfur atom and the two methylene groups preceding the thioester linkage (Fig. 5B). In holo-, butyryl-, hexanoyl-, octanoyl-, and decanoyl-ACP, these atoms are also highly shielded from the solvent, whereas they are vastly more solvent-exposed in the presence of longer acyl groups (Fig. 5B).
FIGURE 5.
A, averaged solvent-accessible surface area (SASA) of the acyl chains in butyryl- (thick solid), hexanoyl- (thin solid), octanoyl- (thin dotted), decanoyl- (thick dashes), and octadecanoyl-ACP (medium dashed). The first seven carbon groups are well protected from the solvent, with SASAs measuring less than 2 Å2 in each case. For octanoyl-ACP, the eighth carbon group is additionally protected behind the carbonyl oxygen. The inset describes the numbering as presented. B, the SASA of the phosphopantetheine linker plotted for each atom of apo- (gray) and holo-ACP (thin). Acyl-ACP SASAs are shown as the averaged difference to holo-ACP values (shorter chain: C4-C10, thick black; longer chain: C12-C18, dotted). This highlights the difference in SASA of the sulfur group and the two preceding carbon atoms (atoms 30-32). The prosthetic group diagram is provided as a reference, aligned to the corresponding atom numbers.
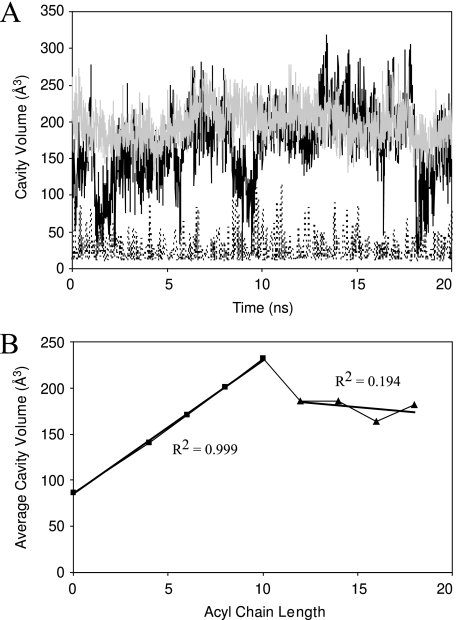
The size of the hydrophobic binding pocket in ACP is highly dynamic. In apo-ACP, the pocket is closed ∼50% of the time but opens up and gets as large as ∼115 Å3 (Fig. 6). With the prosthetic linker inside the binding pocket in holo-ACP, the size is slightly larger and expands further when housing the butyryl, hexanoyl, octanoyl, and decanoyl acyl groups (Table 1). Increasing the size of the acyl chain beyond 10 carbon groups does not result in a larger binding cavity. Rather, the cavity sizes average between 163 and 186 Å3 with no correlation to acyl chain length beyond 10 carbons, whereas for shorter acyl chains the correlation is very high. From holo- to decanoyl-ACP, the correlation coefficient (R2) between acyl chain length and cavity size measures 0.999, whereas for longer acyl chain ACPs, R2 equals 0.194 (Fig. 6B). Fluctuations observed in the presence of longer acyl groups depress the average volume such that they end up more similar, whereas the maximum cavity sizes observed are still larger in simulations of longer acyl chains. For example, housing the octadecanoyl chain in the binding pocket leads to predominantly large cavity sizes (>200 Å3), except when the acyl chain almost completely exits the binding pocket. During these periods the size of the cavity shrinks dramatically (<50 Å3), thereby depressing the average pocket size (Fig. 6A). This also explains the large cavity observed in octanoyl-ACP; because it consistently fills the cavity, the average is closer to the maximum volume observed during the simulation.
FIGURE 6.
A, cavity volumes as a function of time for various representative ACP forms. ApoACP (dotted curve) displays the smallest cavity size, with substantial fluctuations, yielding cavity sizes as large as ∼115 Å3. Note that cavity sizes of 0 Å3 are not shown since they cannot be detected. Octanoyl- (gray) and octadecanoyl-ACP (black) have much higher cavity volumes due to the acyl chain occupying the cavity. The fluctuations in cavity volume are noticeably larger in octadecanoyl-ACP. B, the average cavity volume as a function of acyl chain length. The linear best fit lines are shown for short chain (0-10 carbon groups) and long chain (12-18 carbon groups) averages along with the R2 values for the two segments.
DISCUSSION
Comparison to Protein Data Bank Structures—At the commencement of this study, a structure for E. coli butyryl-ACP had recently been published which provided the template for the simulations conducted (11). Since then additional acyl-ACP structures have been published from both E. coli and spinach, which provide good subjects for cross-referencing our results (12, 13). The E. coli crystal structures provide the most direct point for comparison with our MD simulations because there are no potential differences introduced by the primary structure. The spinach ACP sequence is only 39% identical, which could be responsible for some of the differences seen between our simulations and these NMR structures. All of the trajectories with solvent-shielded acyl chains display low backbone r.m.s.d. to the E. coli crystal structures, measuring between 1.3 and 1.6 Å (residues 6-75) (Fig. 7). Spinach ACPs differ quite substantially from the E. coli structures, both compared with the x-ray structures and the simulations. Aligning the simulation trajectories and the E. coli crystal structures to spinach ACP, r.m.s.d. values ranging from 5.6 to 6.2 Å for backbone atoms (residues 6-75) are obtained. Furthermore, in the spinach octadecanoyl-ACP structure, part of loop I is disordered, which was attributed to dynamics induced by the long attached acyl chain (13). Such increased flexibility is not observed in our long-chain ACP simulations. This suggests that there may be slight differences between E. coli and spinach ACPs in their mechanism of accommodating acyl chains. This has also been hypothesized for mammalian ACPs, which may not bind acyl chains in a hydrophobic pocket at all (32). Direct comparisons are possible between the E. coli butyryl-, hexanoyl-, and decanoyl-ACP structures (1L0I, 2FAC, 2FAE, respectively) and our simulations. The acyl group in decanoyl-ACP agrees very well with the published structure, whereas the butyryl and hexanoyl chains have slightly higher average r.m.s.d. for the whole arm (2.5, 4.0, and 3.9 Å acyl-chain r.m.s.d., respectively). These differences are mainly seen because the butyryl group's prosthetic linker is highly mobile and because the hexanoyl chain occupies sub-pocket II for large portions of the simulation. Some of the motions of the butyryl group involve the penetration deeper into the hydrophobic binding pocket. The phosphopantetheine linker in that conformation corresponds to an alternative binding mode identified in the hexanoyl- and heptanoyl-ACP crystal structures of E. coli and forms the same hydrogen bonding pattern (12). This behavior was not observed in any of the other simulations and, therefore, suggests that this represents a minor binding conformation and additionally demonstrates the excellent agreement between the simulations and x-ray structures.
FIGURE 7.
Snapshots of E. coli hexanoyl- and octadecanoyl-ACPs shown in A and B, respectively, taken every 50 ps over the full 20 ns of MD trajectory. C, crystal structures ranging from butyryl- to decanoyl-ACP by Roujeinikova et al. (11, 12). Panels A-C show ACP with helix IV in a transparent representation oriented toward the viewer, and gray spheres highlight the tip of the acyl chains in each structure. D, the distance of the acyl chain from the first to the last carbon group in the acyl chain of butyryl- (lower black line), hexanoyl- (dark gray), octanoyl- (light gray), decanoyl- (black), and hexadecanoyl-ACP (dotted). Notice that up to octanoyl-ACP the chain is largely extended inside the cavity, whereas beginning with decanoyl-ACP the chain is increasingly distorted, which is also reflected in panels A and B.
Interestingly, the second sub-pocket (oriented toward helix I) identified in our MD simulations is not seen in the acyl-ACP x-ray and NMR structures previously published (11-13). Both sub-pockets in ACP are capable of binding the same size acyl chains, as the SASAs of the acyl groups do not change depending on which sub-pocket is occupied. The changes that occur between the two pockets are very subtle and only require the reorientation of Leu-42 and Leu-46 as well as a slight tilt of Phe-28 to open the binding pocket. It is possible that in the NMR structure such small changes could not be differentiated based on the NOE pattern. Comparing the NOEs formed by the prosthetic group observed by Zornetzer et al. (13) and the residues contacted in each of the binding pockets, one cannot rule out the second sub-pocket as an acyl chain binding site in the spinach structures (Table 2). In the crystal structures the lattice provided in the crystal environment may shift the equilibrium between the two pockets such that only one binding orientation is observed (12). Inspection of other published ACP structures reveals that residues equivalent to Leu-46 also adopt χ1 angles similar to the ones seen in our simulations, making this switch a realistic possibility (33, 34). Furthermore, a number of ACP structures also indicate the presence of a second sub-pocket analogous to what we observe in our simulations. ACPs from Bacillus subtilis (1HY8), Mycobacterium tuberculosis (1KLP), and from the Streptomyces coeli-color actinorhodin polyketide synthase system (2AF8) all display cavities where the second sub-pocket is observed (34-36). Another ACP structure from B. subtilis (1F80) bound to ACPS displays a continuous tunnel from its prosthetic linker into the second sub-cavity, exactly as is seen in our simulations (14). These observations further support the idea that ACP can harbor acyl chains in its hydrophobic pocket in two ways, either pointing toward helix III or helix I.
Acyl-chain Properties—Recent structures of acyl-ACPs have confirmed the hypothesis that the acyl chain is harbored inside the hydrophobic binding pocket of ACP (11-13). The simulations here provide a dynamic picture of ACP associated with a variety of saturated acyl chains spanning from 4 to 18 carbon groups in length. The acyl chain in each simulation remains in the hydrophobic core of ACP, in agreement with NMR and x-ray data (12, 13). Furthermore, we observed the transition from a solvent-exposed acyl chain into the hydrophobic pocket of ACP in a number of simulations, and the two types of simulations converge very well (supplemental Fig. 1). Inside the binding pocket the acyl chains are highly mobile, which is not readily obvious from the previously published structures (Fig. 7). The amount of movement observed in the acyl chain is dependent on the size of the acyl chain attached. The octanoyl acyl chain represents the ideal length for binding in the hydrophobic pocket. Its acyl chain is just long enough to reach the bottom of the cavity while shielding all of its hydrophobic carbon groups from the solvent. At the same time its prosthetic linker is not drawn into the hydrophobic cavity but remains available for hydrogen bonding with the solvent and the surface of the protein. The short chain acyl groups are unable to fill out the hydrophobic cavity completely, creating a small gap at the bottom of the cavity. This results in instability of butyryl and hexanoyl groups, which is reflected in their elevated motions. Although the tip of the acyl chain does reach the bottom of the cavity at times, the linker arm is not positioned optimally in those cases. The linker is then pulled into the hydrophobic pocket as well, which is unfavorable due to the polar nature of the phosphopantetheine arm and prevents the acyl chain from reaching the bottom of the cavity consistently. Long chain acyl groups are highly dynamic for different reasons. Acyl chains greater than eight carbon groups in length are too long to fit into the pocket completely and consequently must expose parts of their aliphatic chain to the polar solvent. Decanoyl-ACP represents an intermediate to these behaviors, as its chain is only slightly larger than the octanoyl chain. Because of its size there is increased strain on the acyl chain that forces it to compact to fit into the pocket entirely and shield all of its acyl chain from the solvent. This is seen in the distance between the first and tenth carbon groups of the decanoyl chain (Fig. 7D). Unlike the shorter chains (less than eight carbon groups), its acyl group is not extended very frequently but fluctuates heavily in length. That is, because the decanoyl chain is too long to fit into the cavity in an entirely extended conformation, the chain must bend and adopt less favorable dihedral angles to fit into the cavity and shield all the aliphatic groups from the solvent. Because this is not the most favorable conformation for the acyl group, interplay between pushing the acyl group inside and straightening out the acyl chain is observed, which results in only partial burying of the decanoyl group. This pattern is not apparent by comparing the acyl chain and binding pocket volumes (Table 1). The measured cavity volumes would indicate that acyl chains up to 10 or 12 carbons in length could be accommodated. This is because the cavity is wide and has a number of grooves but is not very long, and as such its volume is larger than the acyl chain volume it can possibly hold.
Which acyl chain is optimally bound by ACP is further demonstrated by the SASA of the acyl group. The acyl chain is well protected from the solvent up to the first eight carbon groups (Fig. 5A). This is in very good agreement with experimental data, which has suggested that the binding pocket can optimally house acyl chains up to eight carbon groups in length (37). It has been shown that the hydrophobic character of ACP increases from dodecanoyl- to hexadecanoyl-ACP based on binding to octyl-Sepharose (37, 38). This is seen in the simulations as well, as the hydrophobic SASA of the acyl chain increases continuously with an increase in acyl chain length beyond octanoyl-ACP and, therefore, strengthens the validity of our findings. To further characterize the strength of binding by various acyl chains, isothermal titration calorimetry experiments or free energy perturbation calculations could be conducted to obtain more quantitative data on the factors governing acyl chain-ACP interactions.
Comparison of Apo-, Holo-, and Acyl-ACP—The dynamic nature of ACP has long been considered one of its vital attributes for the successful interaction with different enzymatic partners. The simulations of the various forms of ACP are in good agreement with dynamic data obtained from NMR studies (16, 39). The regions of elevated flexibility consist of the N and C termini, the loop regions, and the ends of the helices independent of the type of ACP simulated (Fig. 2). Most notably, the end of helix II leading into loop II represents the only region with consistently higher fluctuations in the acyl-ACP simulations compared with the apo and holo proteins. This region contains a high proportion of acidic residues (seven acidic residues from 47-57) and no basic residues and is well known for its importance in ACP-enzyme interactions (15, 40, 41). Especially the negatively charged residues on helix II, the recognition helix, are crucial for protein-protein interactions (15, 40). A high density of charged residues and few hydrophobic residues is commonly associated with natively unfolded proteins and with that flexibility, which may be important for maintaining the motions needed in this region of ACP (42). FoldIndex, a tool for the prediction of intrinsically unfolded proteins, indicates that the C-terminal regions of helix II and loop II are the most likely regions of ACP to be intrinsically unfolded (43). Therefore, the high density of negatively charged residues in this area appears important for maintaining the flexibility needed in this region of the protein. The presence of a basic residue might lead to the formation of salt bridges that would alter the manner in which ACP accommodates acyl chains and how it interacts with enzymatic partners. The portion of helix II that shows increased fluctuations in acyl-ACPs consists of residues 47-50, which includes three consecutive Glu residues. Changes in the dynamic behavior of ACP in this segment could, therefore, be reflected in its ability to interact with different enzymes.
Biological Considerations—ACP has a widespread and profound role in bacteria, representing a major hub of protein interactions in E. coli (44). In addition, the bacterial and mammalian FAS systems involving ACP are considerably different, making this interaction network a promising target for the development of new antibiotics (7, 8). As such, the structural and dynamic characterization of ACP and its acyl chains are crucial in the advancement of this field. Our simulations indicate that there is a hierarchy of preferred acyl chain binding partners in terms of their ability to bind ACP and rest in the hydrophobic pocket. All our evidence suggests that octanoyl-ACP provides the best fit for the hydrophobic binding pocket, whereas both shorter and longer acyl groups represent sub-optimal binding. This is important for ACP-enzyme interactions, during which the acyl chain has to be released from the binding pocket before becoming available to any enzymes. In the case of octanoyl-ACP, the hydrophobic interactions will be stronger compared with short-chain and long-chain acyl groups. Long-chain acyl groups are large enough to fill the binding cavity equally well as octanoyl-ACP but partially exit periodically. This essentially would make it easier for enzymes to draw the longer (>C8) acyl chains out of the pocket and utilize them as substrates. In addition, in long-chain acyl-ACPs some of the hydrophobic acyl group is partially exposed at all times, which would enable hydrophobic interactions between the enzyme and acyl chain, whereas the tip of the acyl chain still rests inside the pocket. Similarly, holo- and short-chain acyl-ACPs are capable of shielding the terminal two carbon groups and the sulfhydryl group of the phosphopantetheine linker from the solvent by keeping them close to the helical turn in loop I and helix III. In acyl-ACP containing greater than 10 carbon groups the hydrophobic tail is too long, and as such, the region between loop I and helix III is instead occupied by methylene groups of the acyl chain, leaving the end of the prosthetic linker solvent-exposed. This explains what is observed in thioester cleavage studies, which showed that the shorter acyl-ACP forms were well protected from base hydrolysis (37). Furthermore, this suggests a mechanism for enzymes to specifically recognize long chain ACPs rather than incompatible shorter versions. These principles can also be applied to the partner enzyme perspective; depending on the structures of the active and substrate binding sites, interactions analogous to those in ACP may occur where binding of certain acyl chain lengths is done more effectively, which will affect the rate of substrate binding and release.
Knowledge of the optimal binding conformation may also prove important in the continued effort to develop novel antibiotics targeting FAS systems. Understanding how ACP accommodates acyl chains in the binding pocket may provide clues as to how compounds may be modified to exhibit stronger binding. For example, a pantothenate analog that is capable of occupying sub-pocket II in ACP might exhibit stronger binding, which would further decrease its turnover rate and thereby increases its potency (9). Such potential applications could be investigated using ACP mutants to help pinpoint residues of importance in sub-pocket II. Furthermore, it would be interesting to determine the role of the hydrogen bonding residues Thr-39 and Glu-60 at the portal to the hydrophobic pocket. Compounds attenuating these interactions may provide a good target for potential therapeutics.
Conclusions—ACP and its role in fatty acid synthesis have been intensively studied for many years and continue to grow in importance as a potential target for novel antibiotics. This is chiefly because of the significant differences between the dissociated microbial type II FAS systems and the large mega-complexes of the human type I FAS systems (8). Notwithstanding the interest in these systems, it has been difficult to elucidate the interactions between ACP and its associated acyl chains. The acyl-ACP NMR and crystal structures provide informative snapshots of the acyl chains as they are buried inside ACP, but a dynamic picture of the interactions between ACP and its acyl chains has yet to be described (11-13). Here we have used MD simulations to address this problem. Our simulations depict the transition of a fully solvent exposed acyl chain into the hydrophobic binding pocket of ACP yielding final structures that match the published high resolution structures. This suggests that the simulations accurately depict the energetic interactions between the acyl chain and the protein, as it is able to find the perceived energy minimum in five of eight trajectories. The three trajectories that did not enter the binding pocket may have as well, with more exhaustive sampling. Inside the binding pocket the acyl chains exhibit properties in excellent agreement with experimentally derived observations. As such, these simulations provide a detailed and dynamic view of the interactions of ACP and its acyl chains.
Supplementary Material
Acknowledgments
This work was stimulated by numerous discussions with Dr. David Byers from Dalhousie University in Halifax, Canada. The authors thank Drs. L. Monticelli, C. Kandt, and J. MacCallum for help with the MD analysis. We further thank Dr. Z. Xu for the calculation of partial charges on the prosthetic linker.
This work was supported by operating grants from the Canadian Institute of Health Research (to H. J. V. and D. P. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: FA, fatty acid; FAS, FA synthase; ACP, acyl carrier protein; ACPS, ACP synthase; MD, molecular dynamics; RMSF, root mean square fluctuations; SASA, solvent-accessible surface area; NOE, nuclear Overhauser effect.
References
- 1.Zhang, Y. M., and Rock, C. O. (2008) Nat. Rev. Microbiol. 6 222-233 [DOI] [PubMed] [Google Scholar]
- 2.Byers, D. M., and Gong, H. (2007) Biochem. Cell Biol. 85 649-662 [DOI] [PubMed] [Google Scholar]
- 3.Leibundgut, M., Jenni, S., Frick, C., and Ban, N. (2007) Science 316 288-290 [DOI] [PubMed] [Google Scholar]
- 4.Jenni, S., Leibundgut, M., Boehringer, D., Frick, C., Mikolasek, B., and Ban, N. (2007) Science 316 254-261 [DOI] [PubMed] [Google Scholar]
- 5.Maier, T., Jenni, S., and Ban, N. (2006) Science 311 1258-1262 [DOI] [PubMed] [Google Scholar]
- 6.White, S. W., Zheng, J., Zhang, Y. M., and Rock, C. O. (2005) Annu. Rev. Biochem. 74 791-831 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Y. M., White, S. W., and Rock, C. O. (2006) J. Biol. Chem. 281 17541-17544 [DOI] [PubMed] [Google Scholar]
- 8.Wright, H. T., and Reynolds, K. A. (2007) Curr. Opin. Microbiol. 10 447-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, Y. M., Frank, M. W., Virga, K. G., Lee, R. E., Rock, C. O., and Jackowski, S. (2004) J. Biol. Chem. 279 50969-50975 [DOI] [PubMed] [Google Scholar]
- 10.Wang, J., Soisson, S. M., Young, K., Shoop, W., Kodali, S., Galgoci, A., Painter, R., Parthasarathy, G., Tang, Y. S., Cummings, R., Ha, S., Dorso, K., Motyl, M., Jayasuriya, H., Ondeyka, J., Herath, K., Zhang, C. W., Hernandez, L., Allocco, J., Basilio, A., Tormo, J. R., Genilloud, O., Vicente, F., Pelaez, F., Colwell, L., Lee, S. H., Michael, B., Felcetto, T., Gill, C., Silver, L. L., Hermes, J. D., Bartizal, K., Barrett, J., Schmatz, D., Becker, J. W., Cully, D., and Singh, S. B. (2006) Nature 441 358-361 [DOI] [PubMed] [Google Scholar]
- 11.Roujeinikova, A., Baldock, C., Simon, W. J., Gilroy, J., Baker, P. J., Stuitje, A. R., Rice, D. W., Slabas, A. R., and Rafferty, J. B. (2002) Structure 10 825-835 [DOI] [PubMed] [Google Scholar]
- 12.Roujeinikova, A., Simon, W. J., Gilroy, J., Rice, D. W., Rafferty, J. B., and Slabas, A. R. (2007) J. Mol. Biol. 365 135-145 [DOI] [PubMed] [Google Scholar]
- 13.Zornetzer, G. A., Fox, B. G., and Markley, J. L. (2006) Biochemistry 45 5217-5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parris, K. D., Lin, L., Tam, A., Mathew, R., Hixon, J., Stahl, M., Fritz, C. C., Seehra, J., and Somers, W. S. (2000) Structure 8 883-895 [DOI] [PubMed] [Google Scholar]
- 15.Rafi, S., Novichenok, P., Kolappan, S., Zhang, X. J., Stratton, C. F., Rawat, R., Kisker, C., Simmerling, C., and Tonge, P. J. (2006) J. Biol. Chem. 281 39285-39293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, Y., Kovrigin, E. L., and Eletr, Z. (2006) Biochem. Biophys. Res. Commun. 341 776-783 [DOI] [PubMed] [Google Scholar]
- 17.Rock, C. O., and Jackowski, S. (1982) J. Biol. Chem. 257 10759-10765 [PubMed] [Google Scholar]
- 18.Koradi, R., Billeter, M., and Wüthrich, K. (1996) J. Mol. Graph. 14 51-55 [DOI] [PubMed] [Google Scholar]
- 19.Berendsen, H. J. C., van der Spoel, D., and van Drunen, R. (1995) Comput. Phys. Commun. 91 43-56 [Google Scholar]
- 20.Lindahl, E., Hess, B., and van der Spoel, D. (2001) J. Mol. Model 7 306-317 [Google Scholar]
- 21.Scott, W. R. P., Hunenberger, P. H., Tironi, I. G., Mark, A. E., Billeter, S. R., Fennen, J., Torda, A. E., Huber, T., Krüger, P., and van Gunsteren, W. F. (1999) J. Phys. Chem. A 103 3596-3607 [Google Scholar]
- 22.Feenstra, K. A., Hess, B., and Berendsen, H. J. C. (1999) J. Comput. Chem. 20 786-798 [DOI] [PubMed] [Google Scholar]
- 23.Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., Dinola, A., and Haak, J. R. (1984) J. Chem. Phys. 81 3684-3690 [Google Scholar]
- 24.Hess, B., Bekker, H., Berendsen, H. J. C., and Fraaije, J. G. E. M. (1997) J. Comput. Chem. 18 1463-1472 [Google Scholar]
- 25.Darden, T., York, D., and Pedersen, L. (1993) J. Chem. Phys. 98 10089-10092 [Google Scholar]
- 26.Miyamoto, S., and Kollman, P. A. (1992) J. Comput. Chem. 13 952-962 [Google Scholar]
- 27.Laskowski, R. A. (1995) J. Mol. Graph. 13 323-330 [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt, G. J., and Jones, T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50 178-185 [DOI] [PubMed] [Google Scholar]
- 29.Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Montgomery, J. A., Jr., Vreven, T., Kudin, K. N., Burant, J. C., Millam, J. M., Iyengar, S. S., Tomasi, J., Barone, V., Mennucci, B., Cossi, M., Scalmani, G., Rega, N., Petersson, G. A., Nakatsuji, H., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Klene, M., Li, X., Knox, J. E., Hratchian, H. P., Cross, J. B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R. E., Yazyev, O., Austin, A. J., Cammi, R., Pomelli, C., Ochterski, J. W., Ayala, P. Y., Morokuma, K., Voth, G. A., Salvador, P., Dannenberg, J. J., Zakrzewski, V. G., Dapprich, S., Daniels, A. D., Strain, M. C., Farkas, O., Malick, D. K., Rabuck, A. D., Raghavachari, K., Foresman, J. B., Ortiz, J. V., Cui, Q., Baboul, A. G., Clifford, S., Cioslowski, J., Stefanov, B. B., Liu, G., Liashenko, A., Piskorz, P., Komaromi, I., Martin, R. L., Fox, D. J., Keith, T., Al-Laham, M. A., Peng, C. Y., Nanayakkara, A., Challacombe, M., Gill, P. M. W., Johnson, B., Chen, W., Wong, M. W., Gonzalez, C., and Pople, J. A. (2004) Gaussian 03, Revision C. 02, Gaussian, Inc., Wallingford, CT
- 30.Kandt, C., Xu, Z. T., and Tieleman, D. P. (2006) Biochemistry 45 13284-13292 [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg, D., and Mclachlan, A. D. (1986) Nature 319 199-203 [DOI] [PubMed] [Google Scholar]
- 32.Ploskon, E., Arthur, C. J., Evans, S. E., Williams, C., Crosby, J., Simpson, T. J., and Crump, M. P. (2008) J. Biol. Chem. 283 518-528 [DOI] [PubMed] [Google Scholar]
- 33.Sharma, A. K., Sharma, S. K., Surolia, A., Surolia, N., and Sarma, S. P. (2006) Biochemistry 45 6904-6916 [DOI] [PubMed] [Google Scholar]
- 34.Xu, G. Y., Tam, A., Lin, L., Hixon, J., Fritz, C. C., and Powers, R. (2001) Structure 9 277-287 [DOI] [PubMed] [Google Scholar]
- 35.Wong, H. C., Liu, G., Zhang, Y. M., Rock, C. O., and Zheng, J. (2002) J. Biol. Chem. 277 15874-15880 [DOI] [PubMed] [Google Scholar]
- 36.Crump, M. P., Crosby, J., Dempsey, C. E., Parkinson, J. A., Murray, M., Hopwood, D. A., and Simpson, T. J. (1997) Biochemistry 36 6000-6008 [DOI] [PubMed] [Google Scholar]
- 37.Cronan, J. E., Jr. (1982) J. Biol. Chem. 257 5013-5017 [PubMed] [Google Scholar]
- 38.Rock, C. O., and Garwin, J. L. (1979) J. Biol. Chem. 254 7123-7128 [PubMed] [Google Scholar]
- 39.Andrec, M., Hill, R. B., and Prestegard, J. H. (1995) Protein Sci. 4 983-993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong, H. S., Murphy, A., McMaster, C. R., and Byers, D. M. (2007) J. Biol. Chem. 282 4494-4503 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y. M., Wu, B., Zheng, J., and Rock, C. O. (2003) J. Biol. Chem. 278 52935-52943 [DOI] [PubMed] [Google Scholar]
- 42.Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000) Proteins 41 415-427 [DOI] [PubMed] [Google Scholar]
- 43.Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., Silman, I., and Sussman, J. L. (2005) Bioinformatics 21 3435-3438 [DOI] [PubMed] [Google Scholar]
- 44.Butland, G., Peregrin-Alvarez, J. M., Li, J., Yang, W. H., Yang, X. C., Canadien, V., Starostine, A., Richards, D., Beattie, B., Krogan, N., Davey, M., Parkinson, J., Greenblatt, J., and Emili, A. (2005) Nature 433 531-537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.