Abstract
Induced pluripotent stem cell technology, also termed iPS, is an emerging approach to reprogram cells into an embryonic stem cell-like state by viral transduction with defined combinations of factors. iPS cells share most characteristics of embryonic stem cells, counting pluripotency and self-renewal, and have so far been obtained from mouse and humans, including patients with genetic diseases. Remarkably, autologous transplantation of cell lineages derived from iPS cells will eliminate the possibility of immunological rejection, as well as current ethical issues surrounding human embryonic stem cell research. However, before iPS can be used for clinical purposes, technical problems must be overcome. Among other considerations, full and homogeneous iPS reprogramming is an important prerequisite. However, despite the fact that cells from several mouse tissues can be successfully induced to iPS, the overall efficiency of chimera formation of these clones remains low even if selection for Oct4 or Nanog expression is applied. In this report, we demonstrate that cells from the mouse meningeal membranes express elevated levels of the embryonic master regulator Sox2 and are highly amenable to iPS. Meningeal iPS clones, generated without selection, are fully and homogeneously reprogrammed based on DNA methylation analysis and 100% chimera competent. Our results define a population of somatic cells that are ready to undergo iPS, thus highlighting a very attractive cell type for iPS research and application.
Embryonic stem (ES)2 cells hold huge promise in regenerative medicine. They possess the ability to differentiate into all cell types of the human body (pluripotency) and can be cultured indefinitely in vitro without losing their characteristics (self renewal) (1, 2). One ideal way to bypass technical and ethical considerations regarding human ES cells is to produce patient specific ES-like pluripotent cells from their own terminally differentiated somatic cells. A series of major breakthroughs were achieved when Yamanaka and co-workers and other groups (3-9) demonstrated that viral transduction of a set of exogenous factors can reset the epigenetic and transcriptional state of mouse and human fibroblasts into that of pluripotent ES cells. iPS cells were shown to be molecularly and functionally similar to ES cells, including their ability to form teratomas in immunodepressed mice and to contribute to all tissues in chimeric mice. The therapeutic potential of iPS was demonstrated using gene therapy and transplantation in animal models of sickle cell anemia and Parkinson disease (10, 11). The generation of human iPS cells eliminates the previously omnipresent problem of immunological rejection and also provides a method for studying genetic diseases in culture. However, despite the huge potential implications, a number of important questions remain to be solved. The low efficiency of reprogramming initially suggested that a rare cell type, such as an adult stem cell, might be the cell of origin for iPS and that differentiated cells are instead refractory to reprogramming. To solve this, three recent studies produced iPS cells from stomach and liver cells, B lymphocytes, and also pancreatic beta cells (12-14). More important issues that have raised concerns regarding clinical application in humans are: (a) the delivery of iPS-inducing factors with viral vectors that insert into the genome and can potentially be reactivated, (b) the low efficiency generation of colonies that are competent for formation of chimeric mice, and (c) related to the latter, the lack of appropriate standard that can be used to discern a fully reprogrammed iPS clone from an incomplete one.
Herein, we report that a terminally differentiated somatic cell, the mouse meningiocyte, expresses high levels of Sox2 and is highly amenable to iPS. Our meningeal iPS clones homogeneously displayed high levels of endogenous ES marker genes, low DNA methylation profile of the promoters of ES core factors, and formed embryonic bodies. Moreover, all resulting iPS clones were competent to produce chimeras when injected into blastocysts. Taken together, our experiments show that generating iPS cells from meningeal cells has significant advantages over cells from other sources.
EXPERIMENTAL PROCEDURES
Cell Culture—Meningeal membranes were obtained by surgical incision in the skull of newborn C57BL/6J mice. Membranes were digested with 0.25% trypsin-EDTA (Invitrogen) for 8-10 min at 37 °C followed by gentle dissociation with a fire-polished pipette. Samples were left to stand for 5 min to allow the tissue debris to settle down, and the resulting cell suspension was seeded at a density of 4 × 105 cells/ml (∼105 cells were obtained per newborn mouse) in 6-well dishes culture dishes pretreated with poly-d-lysine (100 μg/ml). The resulting cells were grown in high glucose Dulbecco's modified Eagle's medium (4.5 g/liter glucose, HyClone) containing 10% fetal bovine serum (HyClone), 2 mm sodium pyruvate (Invitrogen), 2 mm l-glutamine (Invitrogen), penicillin (100 units/ml), and streptomycin (100 μg/ml). Only primary cultures were used for infection. Mouse embryonic fibroblasts (MEF) and mouse skin fibroblasts (MSF) cells were prepared as described previously (15).
ES and iPS cells were cultivated on mitomycin-treated MEF (as feeder cells) in high glucose Dulbecco's modified Eagle's medium (HyClone) containing 15% fetal bovine serum (Invitrogen), leukemia inhibitory factor (LIF), penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and nonessential amino acids (Invitrogen). Before RNA or DNA purification, iPS cells were depleted of feeder cells for two passages on 0.2% gelatin. For embryonic body formation, iPS cells were harvested by trypsinization, plated on nonadherent bacterial culture dishes, and incubated in medium without LIF.
Viral Infections—4 × 104 meningeal cells in 6-well dishes were infected overnight with viral supernatants generated by transfection of Plat-E cells (Lipofectamine 2000, Invitrogen) with retroviral pMX vectors containing the cDNAs of mouse Oct4, Sox2, Klf4, and c-Myc. Two rounds of infection were performed successively (24 h each). Polybrene (Sigma) was added to increase infection efficiency. Vectors were purchased from Adgene and have been used by us before (9).
Alkaline Phosphatase (AP) Staining—Medium was gently aspirated, and cells were rinsed with 0.25 ml of PBS. Cells were then fixed with 4% paraformaldehyde in PBS, incubated at room temperature for 2 min, and rinsed twice with 0.5 ml of TBST (Tris-buffered saline + 0.05% Tween-20). Freshly prepared AP staining solution (4.5 μl 50 mg/ml nitro blue tetrazolium, 3.5 μl 50 mg/ml 5-bromo-4-chloro-3-indolyl phosphate in 100 mm Tris-HCl, pH 9.5, 100 mm NaCl, 50 mm MgCl2) was added, and plates were incubated in the dark at room temperature for 15 min. The staining solution was aspirated, and plates were rinsed with PBS.
Semiquantitative RT-PCR—RNA extraction was performed using TRIzol (Takara). Reverse transcription was done with M-MLV (Takara), and PCR was done using the rTaq kit from Takara. Genomic DNA was extracted with genomic DNA extraction kit (Takara), and PCR was performed with rTaq. The list of primers is included in supplemental Table 1.
Southern Blot—10 μg of genomic DNA from mouse iPS cell lines and R1 ES cells were digested with the appropriate BglII, EcoRI, and NcoI restriction enzymes overnight, and samples were run on a 0.8% agarose gel. After alkali denaturation, the digested genomic DNA was transferred overnight to a positive charged nylon membrane (Amersham Biosciences) and cross-linked with ultraviolet irradiation. The membrane was then incubated at 42 °C overnight with a digoxigenin-labeled DNA probe in digoxigenin Easy Hyb buffer (Roche Applied Science) with constant rotation. After stringency washing, alkaline phosphatase-conjugated anti-digoxigenin antibody (1:10,000, Roche Applied Science) was added to the membrane. Incubation with CDP-star (Roche Applied Science) was performed before exposure to an x-ray film. The probe sequence is provided in supplemental Table 1.
Immunofluorescence—Briefly, cells were fixed in 4% paraformaldehyde for 30 min at room temperature, washed with PBS, and blocked for 20 min at room temperature with 5% fetal bovine serum in PBS containing 0.1% Triton-X100. After a 1-h incubation with primary antibodies against Sox2, Rex1, Nanog (prepared in our laboratory), and SSEA1 (Chemicon) diluted in 1% fetal bovine serum in PBS, cells were washed with PBS, incubated with fluorophore-labeled appropriate secondary antibodies (Santa Cruz Biotechnology), washed, and mounted on 80% glycerol. A conventional (Olympus BX51) or a confocal immunofluorescence microscope (Leica TCS SP2 AOBS) was used for visualization.
Bisulfite Genomic Sequencing—Bisulfite treatment was performed using the CpGenome modification kit (Chemicon) according to the manufacturer's recommendations. PCR primers for Oct4 and Nanog promoters are included in supplemental Table 1. PCR products were cloned into pMD18-T (Takara), and 10 randomly selected clones for each gene were sequenced with M13 forward and M13 reverse primers.
Blastocyst Injection—Diploid or tetraploid blastocysts (94-98 h after human chorionic gonadotropin injection) derived from ICR mice were placed in a drop of potassium simplex optimization medium with 15% FCS under mineral oil. A flat tip microinjection pipette with an internal diameter of 12-15 μm was used for iPS cell injection. After injection, blastocysts were returned to potassium simplex optimization medium and placed at 37 °C until transferred to recipient females.
DNA Microarray—Total RNA was isolated from cells using RNeasy mini kit (Qiagen, Valencia, CA). 3 μg of total RNA were used for cDNA synthesis, primed with T7-oligo(dT) promoter (Affymetrix, Santa Clara, CA) and using SuperScript II polymerase (Invitrogen). Biotin-labeled cRNA was synthesized by in vitro transcription using an RNA transcript labeling kit (Affymetrix). After being fragmented, cRNA was hybridized to a mouse GeneChip 430.2 (Affymetrix). Arrays were scanned with a GeneArray scanner 7G (Affymetrix). Data were background-subtracted and normalized with the Robust Multichip Average method using the software ArrayAssist 5.5.1 (Stratagene Corp. and Strand Life Sciences Pvt. Ltd.). Comparison between meningeal cells, MEF and MSF, is provided in supplemental Table 2. Probes that were not involved in any Kyoto Encyclopedia of Genes and Genomes (Kegg) pathway or corresponded to any Gene Ontology (GO) term were eliminated; when there were two or more probes corresponding to a single gene, only the one with the highest value was kept.
RESULTS AND DISCUSSION
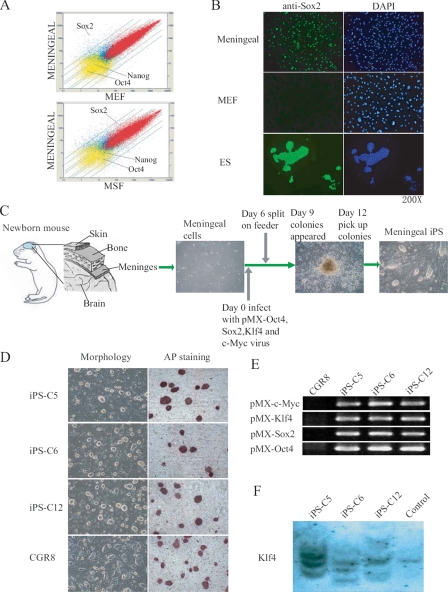
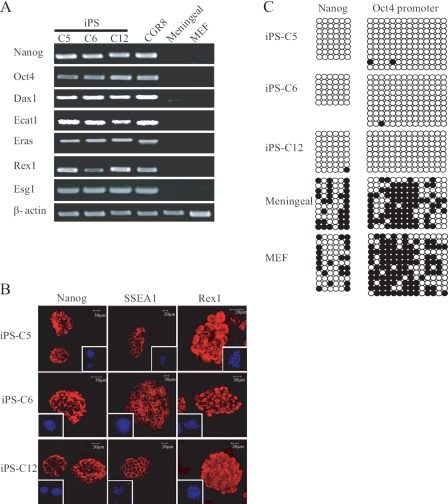
Mouse meninges, like in humans, contain and protect the brain from injuries and are composed of three layers: dura mater (external), arachnoid (attached to the dura mater), and pia mater (internal). Meningeal cells in mammals are derived from the neural crest and can differentiate into several lineages and are thus multipotent, including osteoblasts and neuronal cells, both in vivo and in vitro (16). The whole genome gene expression profile showed that meningeal cells significantly differ from MEF and MSF (Fig. 1A and supplemental Table 2). Red dots indicate genes that are expressed in both types of cells, blue dots indicate genes that are expressed in only one of them, and yellow indicate genes that are expressed in none. Interestingly, meningeal cells expressed a much higher mRNA level of Sox2 than MEF (48 times higher) and MSF (10 times higher) (Fig. 1A, arrows, and supplemental Table 2). Sox2 is a master regulator of ES cell pluripotency needed to generate iPS cell lines from fibroblasts and most other cell types (3). Immunofluorescence microscopy demonstrated correlation between Sox2 mRNA and protein expression (Fig. 1B). We therefore postulated that meningeal cells may be a better source for iPS generation than other somatic cell types used so far. To this aim, 40 × 103 meningeal cells (only primary cultures were used) were retrovirally transduced in 6-well dishes using the standard Oct4, Sox2, Klf4, and c-Myc combination of factors (Fig. 1C). Control incubation with a vector encoding green fluorescent protein demonstrated close to 100% infection efficiency (data not shown). Mouse ES culture medium with LIF was added at day 2 after infection and renewed daily, and at day 6, 105 cells were transferred to 10-cm dish plates coated with mouse fibroblast feeder cells. Colonies (around 1000 colonies out of 105 transduced meningeal) typically arose 12 days after incubation with the four factors, which is similar to previous reports using fibroblasts (9). These had the classical compact morphology of ES colonies (Fig. 1C). Moreover, at day 14, 80% of them proved positive for AP staining (data not shown), a marker for dedifferentiated ES cells. 12 colonies from a different culture dish were picked for further characterization, of which 10 were capable of sustained proliferation on feeder cells. The overall efficiency of our approach (∼0.8%) was therefore similar to fibroblasts and four factors without selection (0.5%-0.8%) (9, 17), and moderately lower than using neural stem cells in the same culture conditions (3.6%) (18). Of the 10 meningeal iPS colonies, three were selected, and AP staining of the derived colonies proved like-wise positive (Fig. 1D). Stable integration of the respective retroviruses in the genome of the three iPS clones was verified by semiquantitative RT-PCR using primers that amplify the transgene and the vector (Fig. 1E). Southern blot for Klf4 also demonstrated genomic integration of multiple copies of this transgene (Fig. 1F). Integration happened in different locus in the three iPS cell lines, in agreement with previous reports demonstrating that iPS does not require integration of exogenous factors near specific genes (12). iPS cells expressed high levels of endogenous ES markers, including Nanog, Oct4, and SSEA1, as assessed by semiquantitative RT-PCR (Fig. 2A) and immunofluorescence microscopy (Fig. 2B). A well known characteristic of ES cells is the low degree of DNA methylation in the promoters of key regulators including Oct4, Nanog, and many others (4, 5). DNA methylation of ES master genes has also been reported to closely correlate with the efficiency of iPS reprogramming (19). Bisulfite sequencing demonstrated low or absent methylation of the endogenous Oct4 and Nanog promoters in our iPS clones and high methylation in MEF and uninfected meningeal cells (Fig. 2C).
FIGURE 1.
Generation of iPS cells from mouse meningeal cells. A, DNA microarray comparison between meningeal cells and the indicated cell types. A scatter-plot representation of the expression values for all probe sets is shown. Positions of Oct4, Sox2, and Nanog are marked with arrows. The parallel lines indicate 2-, 3-, 5-, and 10-fold changes in gene expression (up or down). B, microscope immunofluorescence for Sox2 (green) of meningeal cells and MEF. 4′,6-Diamidino-2-phenylindole (DAPI) staining is shown in blue. Bars indicate the magnification. C, scheme showing the extraction of meningeal membranes from newborn mice and subsequent transduction of meningeal cells using a mixture of retroviruses. D, cells derived from three selected iPS clones formed ES-like colonies and showed positive for AP staining. E, semiquantitative RT-PCR demonstrated integration of the corresponding transgenes in the genome of the three meningeal iPS clones. Sets of primers that amplify both the vector and the transduced factor were used. F, Southern blot also demonstrated integration of the Klf4 transgene in the genome of the three iPS clones. R1 mouse ES cells were used as a control.
FIGURE 2.
Characterization of meningeal iPS clones. A, semiquantitative RT-PCR demonstrated that the three iPS clones express key ES markers; CGR8 mouse ES cells, uninfected meningeal cells, and MEF were used as positive control and negative controls, respectively. B, confocal immunofluorescence microscopy showed homogeneous expression of Nanog, Rex1, and SSEA1 in the selected iPS clones. Bars indicate the magnification. The square on the margin shows 4′,6-diamidino-2-phenylindole staining in lower magnification. C, analysis of the methylation state of the Oct4 and Nanog promoters using bisulfate sequencing. Open circles indicate unmethylated, and filled circles indicate methylated CpG dinucleotides.
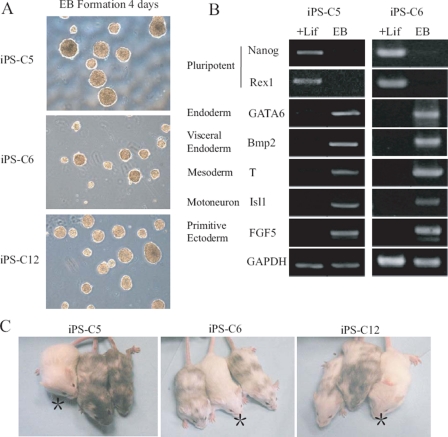
To demonstrate pluripotency, meningeal iPS clones were cultured in nonadherent culture dishes without LIF and allowed to form embryoid bodies (Fig. 3A). Expression of differentiation markers was further analyzed in two of the three colonies at day 9 by semiquantitative RT-PCR and showed efficient transformation into cells of all three germ layers: increased expression of GATA6 and Bmp2 (endoderm), Isl1 and T (mesoderm), and FGF5 (ectoderm) (Fig. 3B). Conversely, expression of Nanog and Rex1 went down with embryonic body differentiation. Remarkably, when injected into blastocysts derived from ICR mice, all three iPS clones gave rise to neonatal animals that were highly chimeric on the basis of their mixed coat color (Fig. 3C), thus showing highly homogeneous and complete reprogramming. For details about the number of injected blastocysts and of chimeric animals produced, see Table 1.
FIGURE 3.
In vitro reprogrammed meningeal iPS cells are pluripotent. A, cells from the three selected iPS clones were grown as embryonic bodies to assess their differentiation capability. Phase contrast captures after 4 days of embryonic body formation are shown. B, gene expression of embryoid bodies (at day 9) derived from two meningeal iPS colonies was analyzed by semiquantitative RT-PCR for markers of the three germ layers. iPS cells cultured in ES medium with LIF were used as a control. C, meningeal iPS cells injected into blastocysts contribute to tissue formation in mice, as denoted by the mixed coat-skin color. Asterisks indicate control mice.
TABLE 1.
Summary of colonies used for blastocyst injections and the corresponding yields
| iPS cell lines | Injected blastocysts | Pups | Chimeras |
|---|---|---|---|
| C57iPS-C5 | 52 | 16 | 2 |
| C57iPS-C6 | 55 | 28 | 9 |
| C57iPS-C12 | 79 | 18 | 6 |
Two relevant aspects should be noted in our study. First, our data reinforce the idea that susceptibility to iPS generation may be universal and not restricted to given cell types. This implies that the low efficiency of iPS cell derivation unlikely reflects the reprogramming of rare adult stem cells included in the starting cell population. It does not exclude, however, that adult stem cells, and also fully differentiated cells with inherent plasticity, may be more amenable to iPS than other cell types, need less factors, or be more sensitive to alternative cocktails. This would in fact explain why iPS efficiency from stomach, liver epithelial cells, and pancreatic beta cells is significantly lower than in fibroblasts (12, 14). Also supporting this idea, pro B lymphocytes are more easily reprogrammed to iPS than mature B cells, which require additional genetic manipulation of the B cell-specific transcription factor Pax5 (13). Genetic or pharmacologic manipulation of chromatin modifiers may as well increase reprogramming efficiencies in a cellular context-specific manner. Second and more importantly, we provide a cell model to study iPS that has several advantages when compared with others. Meningeal cells express high levels of Sox2, and its extraction at least in animals is simple and only requires an incision in the skull. Remarkably, meningeal iPS clones are highly homogeneous in their characteristics and yield 100% efficiency of chimeras. Among other things, recent work on the field has concentrated on both improving iPS efficiency and reducing the number of transduced factors (18, 20-22). Raising the efficiency of iPS colony generation is important to produce colonies even when the starting number of cells is scarce. It may also facilitate substitution of one or more factors by alternative approaches including chemicals or siRNA. On the other hand, reducing the number of factors will decrease the risk of viral reactivation but will not eliminate it unless a different methodology is developed. A previous report has demonstrated that in the course of iPS, most transduced cells become trapped in an intermediate stage, suggesting that stochastic events are needed to proceed to complete reprogramming (19). iPS remains also a highly inefficient method not just regarding the number of cells that progress to the clonal stage but more crucially in the ability of those cells to contribute to formation of chimeras. The latter is the ultimate test for pluripotency but logically cannot be applied in humans. In the future, scrupulous profiling of human iPS cells at both the RNA and the microRNA level may thus be needed to ascertain “purity.” Otherwise, besides additional problems, inadequate reprogramming is likely to result in incomplete differentiation into cells of specific lineages for therapeutic purposes. For these reasons, at this moment of time and while the research progresses, it is fundamental to develop alternative models that increase the quality of iPS. High basal levels of Sox2 and the inherent plasticity of mouse meningeal cells may underlie their high susceptibility to undergo full iPS; the number of colonies produced with meningeal cells is, however, not significantly higher than with fibroblasts. Neural stem cells also express high levels of Sox2 and are amenable to iPS, albeit with notable less efficiency, using a two-factor combination (18). Neural stem cells in humans could only be made available after brain surgery of patients and subsequent tissue removal, whereas in mouse, they require a more delicate procedure than meningeal cells. The authors (18) hypothesized that other cell types expressing adequate endogenous levels of complementary factors may as well be reprogrammed using a reduced number of exogenous factors. It is conceivable that meningeal cells will be prone to reprogramming using two or three factors (apart from the Sox2, Klf4, and Oct4 combination), a possibility that is currently being tested in our laboratory. In addition, we are performing studies that attempt to delineate whether other characteristics of meningeal cells, besides Sox2, contribute to their highly flexible phenotype, as this may provide clues for further work. In this regard, meningeal cells may as well be more susceptible to a fully chemical/small interfering RNA iPS or to alternative delivery systems such as adenoviruses. In summary, we propose that our model will help accelerate the ongoing research and future clinical application of iPS. Given the amazing progress achieved in hardly 2 years of existence, the overcome of current problems is contemplated with optimism by researchers worldwide.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 30725012 and 30630039; Chinese Academy of Sciences Grant KSCX2-YW-R-48; Guangzhou Science and Technology Grant 2006A50104002; Ministry of Science and Technology 973 Grants 2006CB701504, 2006CB943600, 2007CB948002, and 2007CB947804; and National High Technology Project 863 Grant 2005AA210930. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental tables.
Footnotes
The abbreviations used are: ES, embryonic stem; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast; MSF, mouse skin fibroblasts; LIF, leukemia inhibitory factor; AP, alkaline phosphatase; PBS, phosphate-buffered saline; RT-PCR, reverse transcription-PCR.
References
- 1.Evans, M. J., and Kaufman, M. H. (1981) Nature 292 154-156 [DOI] [PubMed] [Google Scholar]
- 2.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. (1998) Science 282 1145-1147 [DOI] [PubMed] [Google Scholar]
- 3.Takahashi, K., and Yamanaka, S. (2006) Cell 126 663-676 [DOI] [PubMed] [Google Scholar]
- 4.Okita, K., Ichisaka, T., and Yamanaka, S. (2007) Nature 448 313-317 [DOI] [PubMed] [Google Scholar]
- 5.Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., Bernstein, B. E., and Jaenisch, R. (2007) Nature 448 318-324 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007) Cell 131 861-872 [DOI] [PubMed] [Google Scholar]
- 7.Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, J. A. (2007) Science 318 1917-1920 [DOI] [PubMed] [Google Scholar]
- 8.Park, I. H., Zhao, R., West, J. A., Yabuuchi, A., Huo, H., Ince, T. A., Lerou, P. H., Lensch, M. W., and Daley, G. Q. (2008) Nature 45 141-146 [DOI] [PubMed] [Google Scholar]
- 9.Qin, D., Li, W., Zhang, J., and Pei, D. (2007) Cell Res. 17 959-962 [DOI] [PubMed] [Google Scholar]
- 10.Hanna, J., Wernig, M., Markoulaki, S., Sun, C. W., Meissner, A., Cassady, J. P., Beard, C., Brambrink, T., Wu, L. C., Townes, T. M., and Jaenisch, R. (2007) Science 318 1920-1923 [DOI] [PubMed] [Google Scholar]
- 11.Wernig, M., Zhao, J. P., Pruszak, J., Hedlund, E., Fu, D., Soldner, F., Broccoli, V., Constantine-Paton, M., Isacson, O., and Jaenisch, R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5856-5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoi, T., Yae, K., Nakagawa, M., Ichisaka, T., Okita, K., Takahashi, K., Chiba, T., and Yamanaka, S. (2008) Science 321 699-702 [DOI] [PubMed] [Google Scholar]
- 13.Hanna, J., Markoulaki, S., Schorderet, P., Carey, B. W., Beard, C., Wernig, M., Creyghton, M., Steine, E. J., Cassady, J. P., Foreman, R., Lengner, C. J., Dausman, J., and Jaenisch, R. (2008) Cell 133 250-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadtfeld, M., Brennand, K., and Hochedlinger, K. (2008) Curr. Biol. 18 890-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi, K., Okita, K., Nakagawa, M., and Yamanaka, S. (2007) Nat. Protoc. 2 3081-3089 [DOI] [PubMed] [Google Scholar]
- 16.Gagan, J. R., Tholpady, S. S., and Ogle, R. C. (2007) Birth Defects Res. C Embryo Today 81 297-304 [DOI] [PubMed] [Google Scholar]
- 17.Blelloch, R., Venere, M., Yen, J., and Ramalho-Santos, M. (2007) Cell Stem Cell Rev. 1 245-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. B., Zaehres, H., Wu, G., Gentile, L., Ko, K., Sebastiano, V., Arauzo-Bravo, M. J., Ruau, D., Han, D. W., Zenke, M., and Scholer, H. R. (2008) Nature 454 646-650 [DOI] [PubMed] [Google Scholar]
- 19.Mikkelsen, T. S., Hanna, J., Zhang, X., Ku, M., Wernig, M., Schorderet, P., Bernstein, B. E., Jaenisch, R., Lander, E. S., and Meissner, A. (2008) Nature 454 49-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi, Y., Tae Do, J., Desponts, C., Hahm, H. S., Scher, H. R., and Ding, S. (2008) Cell Stem Cell 2 525-528 [DOI] [PubMed] [Google Scholar]
- 21.Mali, P., Ye, Z., Hommond, H. H., Yu, X., Lin, J., Chen, G., Zou, J., and Cheng, L. (2008) Stem Cells (Durham) 26 1998-2005 [DOI] [PubMed] [Google Scholar]
- 22.Brambrink, T., Foreman, R., Welstead, G. G., Lengner, C. J., Wernig, M., Suh, H., and Jaenisch, R. (2008) Cell Stem Cell 2 151-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.