Abstract
Formin homology proteins are a highly conserved family of cytoskeletal remodeling proteins best known for their ability to induce the formation of long unbranched actin filaments. They accomplish this by nucleating the de novo polymerization of F-actin and also by acting as F-actin barbed end “leaky cappers” that allow filament elongation while antagonizing the function of capping proteins. More recently, it has been reported that the FH2 domains of FRL1 and mDia2 and the plant formin AFH1 are able to bind and bundle actin filaments via distinct mechanisms. We find that like FRL1, FRL2 and FRL3 are also able to bind and bundle actin filaments. In the case of FRL3, this activity is dependent upon a proximal DAD/WH2-like domain that is found C-terminal to the FH2 domain. In addition, we show that, like other Diaphanous-related formins, FRL3 activity is subject to autoregulation mediated by the interaction between its N-terminal DID and C-terminal DAD. In contrast, the DID and DAD of FRL2 also interact in vivo and in vitro but without inhibiting FRL2 activity. These data suggest that current models describing DID/DAD autoregulation via steric hindrance of FH2 activity must be revised. Finally, unlike other formins, we find that the FH2 and N-terminal dimerization domains of FRL2 and FRL3 are able to form hetero-oligomers.
Formin homology proteins (formins) are a highly conserved family of cytoskeletal regulatory proteins. Over 30 formins have been described to date, with more than 15 family members found in vertebrates (1). Formin activity is required in vivo for a diverse array of cellular functions, such as stress fiber formation, endosome motility, cell motility, cytokinetic ring formation, cell-cell junction assembly, filopodia formation, induction of cell polarity, and activation of the MAL/serum response factor signaling pathway (2-13). It is thought that at the core of all of these activities is the ability of formins to regulate actin cytoskeletal dynamics. This is achieved through the activity of two conserved formin homology domains, FH1 and FH2. FH1 consists of proline-rich repeats of varying sizes that can serve as ligands for Src homology 3 and WW domains as well as the small actin-binding protein profilin (14, 15). FH2 domains form a dimer that induces the polymerization of long, unbranched filaments. It does this by nucleating de novo actin polymerization and by associating with the F-actin barbed end as a “leaky capper,” allowing filament elongation while antagonizing the function of capping proteins (16). However, not all formins nucleate or elongate F-actin with equal potency. For example, mDia1 is an extremely efficient nucleator of polymerization, and its association with the barbed end of actin filaments accelerates elongation (16, 17). In contrast, the isolated FH2 domain of FHOD1 does not induce actin polymerization in vivo (18). In addition, FH2 activity may also be influenced by the activity of adjacent domains. For example, in INF2, a WH2 G-actin binding motif antagonizes the nucleation activity of its associated FH2 domain (19). More recently, it has been recognized that the FH2 domains of FRL1, mDia2, and AFH1 are able to bind and bundle actin filaments using at least two distinct mechanisms (20, 21).
The Diaphanous-related formins contain three dimerization interfaces: DID/DAD interactions mediate Diaphanous-related formin autoinhibition, whereas the N-terminal dimerization domain and the C-terminal FH2 each are able to form homodimers. The presence of multiple dimerization domains has called into question previous models that suggest that Diaphanous-related formins in the autoinhibited conformation are monomeric (15, 22-24). We reported previously that the dimerization and FH2 domains of mDia1 and mDia2 do not form hetero-oligomeric complexes. We wished to determine the general applicability of this observation to other formins. To this end, we performed an initial structure-function analysis of the highly similar Diaphanous-related formins FRL2 and FRL3 to determine the basic organization of these proteins and test their ability to form hetero-oligomers. As part of this study, we identified two C-terminal WH2/DAD-like motifs in FRL2 and FRL3. The more distal motif is required for FRL3 autoregulation, whereas the more proximal sequence is required for efficient F-actin bundling by both proteins. Moreover, we found that although FRL3 is autoregulated, the highly similar FRL2 is not. Indeed, we find that full-length FRL2 is constitutively active, although its DID is able to bind to its DAD both in vivo and in vitro. Finally, we find that, unlike mDia1 and mDia2, both the FH2 and N-terminal dimerization domains of FRL2 and FRL3 proteins are able to form hetero-oligomers in vivo.
MATERIALS AND METHODS
Plasmids—The constructs pMLV-LacZ, p3D.A-Luc, and pEF-SRFVP16 were described previously (25). The constructs f.l.FRL3 (codons 1-1094), FRL3N (codons 1-503), FRL3. F1F2 (codons 504-1094), FRL3.FH2 (codons 599-1094), FRL3.F1F2.1045 (codons 504-1045), FRL3.F1F2.1023 (codons 504-1023), FRL3.FH2.1045 (codons 599-1045), FRL3.FH2 1023 (codons 599-1023), f.l.FRL2 (codons 1-1028), FRL2N (codons 1-498), FRL2.F1F2 (codons 491-1028), FRL2.FH2 (codons 544-1028), FRL2.F1F2.986 (codons 491-986), FRL2.F1F2.965 (codons 491-965), and FRL2.F1F2.2+3 (codons 491-986 of FRL2 with codons 1045-1094 of FRL3) were generated by PCR using standard techniques and subcloned as indicated into the pEF-FLAG and pEFNBRSS vectors (11) with FLAG and Myc epitope tags, respectively, as well as pET-30a for expression in bacteria.
Cell Culture—NIH3T3 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Wisent) and antibiotics (50 units/ml penicillin and 50 μg/ml streptomycin). Raw 264.7 cells were cultured in Dulbecco's modified Eagle's medium (Wisent) supplemented as above. Cells were maintained at 37 °C in a humidified atmosphere with 10% CO2 for NIH3T3 cells and 5% CO2 for Raw 264.7 cells.
Immunofluorescence—Immunocytochemistry was performed as previously described (26, 27). Briefly, NIH3T3 cells were transfected with polyethyleneimine (1 mg/ml; 4:1 ratio with plasmid DNA) and maintained in 0.5% fetal bovine serum in Dulbecco's modified Eagle's medium overnight. Raw 264.7 cells were transfected using Lipofectamine 2000 according to the supplied protocol. Transfected cells were fixed the next day in 3.7% para-formaldehyde and permeabilized with 0.3% Triton X-100. DNA was stained with 4′,6-diamidino-2-phenylindole; F-actin was detected using fluorescein-phalloidin (catalog number F432; Molecular Probes) at a dilution of 1:40; FLAG-tagged proteins were detected using mouse anti-FLAG antibodies (Sigma F3165) at a dilution of 1:500; and Myc-tagged proteins were detected using rabbit anti-c-Myc antibodies (catalog number sc-789; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at a dilution of 1:100. Secondary antibodies used were donkey Alexa594 anti-rabbit (Molecular Probes), donkey anti-rabbit Cy3 (Jackson ImmunoResearch) at a dilution of 1:100, and donkey anti-mouse Cy5 (Jackson ImmunoResearch) at a dilution of 1:100. All optical sections were captured with a ×63 oil immersion objective (numerical aperture = 1.4) using the apotome on a Zeiss Axioimager.Z1.
Immunoprecipitation—Co-immunoprecipitation of FLAG- and Myc-tagged proteins expressed by transient transfection in NIH 3T3 cells were performed as previously described (26, 27).
Reporter Gene Assay—The serum response factor (SRF)2 reporter gene assays were performed as previously described (26, 27). Activation was standardized to an SRF-VP16 internal control included in all experiments. For inhibition and dominant negative studies, activation in absence of the inhibitor is set to 100% to allow better comparison of the relative levels of inhibition.
Actin Bundling Assay—The in vitro actin bundling assays were performed as previously described (20, 26-28). Briefly, FRL2 and FRL3 FH2 and N-terminal deletion derivatives were expressed in Escherichia coli as His tag fusion proteins and purified on Ni2+ resin. F-actin was generated from pyrene-labeled rabbit muscle actin (Cytoskeleton Inc.) according to the supplier's instructions. For actin bundling experiments, FH2 derivatives (0, 0.15, 0.3, and 0.6 μm) were incubated with 2 μm F-actin for 10 min at room temperature. Samples were centrifuged at 16,000 × g for 10 min at 4 °C. The top 80% of supernatant was transferred to a new tube, precipitated with 5 volumes of acetone, and resuspended in 20 μl of 1× SDS gel loading buffer. The remaining supernatant was removed from the pellets, which were then resuspended in 25 μl of 1× SDS gel loading buffer. Equal volumes of supernatant and pellet samples were subjected to SDS-PAGE, and proteins were visualized by Coomassie staining. For the inhibition experiments, FRL2N was incubated with the FH2 derivatives for 10 min at room temperature prior to the addition of 2 μm F-actin.
Pyrene Actin Polymerization Assay—The in vitro actin bundling assays were performed as previously described (26, 27). Briefly, FRL2 FH2, FRL2+3 FH2, and FRL2N were purified as above. FH2 (60 nm) derivatives were pre-incubated with the indicated concentrations of FRL2N or buffer for 10 min at room temperature and aliquoted into a black quartz 96-well plate. Polymerization was initiated by the addition of 5% pyrene-labeled actin for a final G-actin concentration of 2 μm. Fluorescence (excitation = 365 nm; emission = 407 nm) was read every 15 s in a Spectromax M2 fluorimeter (Molecular Devices).
RESULTS
Previous work from our group has suggested that, despite extensive sequence homology, neither the FH2 domains nor N-terminal dimerization motifs of the closely related formins mDia1 and mDia2 are able to form heterodimers (26, 27). To test the generality of this observation, we wanted to assess the ability of the related proteins FRL2 (also known as FMNL3b) and FRL3 (also known as FMNL2b) to form hetero-oligomeric complexes. These proteins share extensive homology at the protein level across their entire sequence (74% identical, 87% similar). We first generated epitope-tagged derivatives of FRL2 and FRL3 comprising the full-length protein, the isolated N terminus, and C-terminal derivatives containing FH1/FH2 (F1F2) and FH2 domains. These proteins were expressed by transient transfection in NIH 3T3 cells, and their subcellular localization and effects on actin stress fiber formation were assessed by immunofluorescence (Figs. 1 and 2).
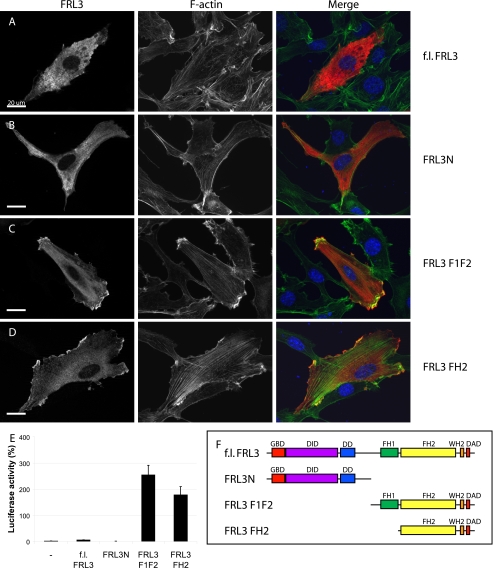
FIGURE 1.
FRL3 is an autoregulated formin. Epitope-tagged derivatives of FRL3 corresponding to the full-length protein, N terminus, FH1 + FH2, and FH2 were expressed by transient transfection in NIH 3T3 cells (0.3 μg of DNA). Protein expression was detected with 9E10 anti-Myc monoclonal (red); F-actin was visualized with fluorescein phalloidin (green). A, full-length FRL3 is distributed diffusely throughout the cytoplasm of transfected cells (100% of cells) and fails to induce stress fiber formation (94%, n = 105). B, FRL3N distributes throughout the cytoplasm and shows apparent membrane targeting in NIH 3T3 cells (99%). FRL3N-expressing cells have reduced levels of F-actin (97%, n = 113). C, expression of FRL3.F1F2 induces robust stress fiber accumulation (93%) and dense accumulation of F-actin at the cell periphery that tightly associates with the overexpressed protein (89%, n = 102). D, FRL3.FH2 induces stress fibers (73%), dense patches of F-actin in lamellopodia-like structures at the periphery of the cell and granular patches of actin throughout the cytoplasm (86%, n = 109). E, expression of FRL3.F1F2 and FH2 (1 μg of DNA transfected) induces robust activation of an SRF reporter gene. Reporter gene activity was standardized to activation induced by expression of an SRF-VP16 control fusion protein. Error bars, S.E., n = 3. F, schematic of FRL3 derivatives.
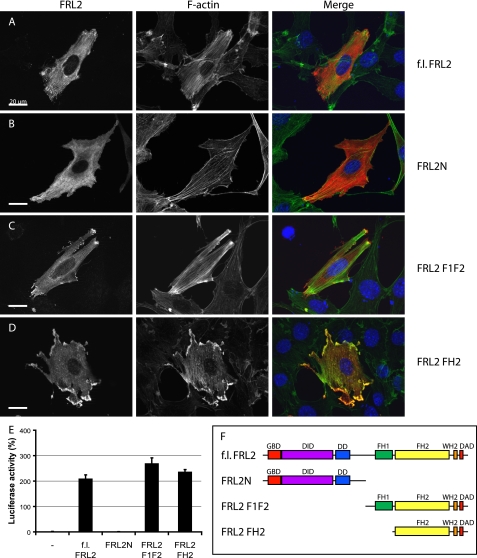
FIGURE 2.
FRL2 is not autoregulated. Epitope-tagged derivatives of FRL2 corresponding to the full-length protein, N terminus, FH1 + FH2, and FH2 were expressed by transient transfection in NIH 3T3 cells. Protein expression was detected with 9E10 anti-Myc monoclonal (red); F-actin was visualized with fluorescein phalloidin (green). A, full-length FRL2 accumulates at the cell periphery, strongly induces stress fiber formation, and associates with F-actin in transfected cells (97%, n = 101). B, FRL2N is distributed diffusely throughout the cytoplasm (96%). FRL2N-expressing cells have reduced levels of F-actin (99%, n = 103). C, expression of FRL2.F1F2 induces robust stress fiber accumulation (92%) and dense accumulation of F-actin at the cell periphery that tightly associates with the overexpressed protein (96%, n = 105). D, FRL2.FH2 induces stress fibers, dense patches of F-actin in lamellopodia-like structures at the periphery of the cell, and granular patches of actin throughout the cytoplasm (98%, n = 1-2). E, expression of full-length F1F2 and FH2 derivatives of FRL2 induces robust activation of an SRF reporter gene. Reporter gene activity was standardized to activation induced by expression of an SRF-VP16 control fusion protein. Error bars, S.E., n = 3. F, schematic of FRL2 derivatives.
As expected, expression of full-length FRL3 did not induce F-actin accumulation, consistent with the presence of well conserved autoregulatory N-terminal DID and C-terminal DAD domains (Fig. 1A). The full-length protein exhibited a diffuse cytoplasmic distribution similar to that seen with full-length mDia1 (27). Expression of the FRL3 N terminus (FRL3N) inhibited stress fiber formation (Fig. 1B). The FH1/FH2 derivative of FRL3 (FRL3.F1F2) induced robust accumulation of stress fibers and a dense band of F-actin at the periphery of the cell. The protein was mostly distributed diffusely in the cytoplasm with some concentrated at the periphery and apparently co-localized with F-actin (Fig. 1C). FRL3.FH2 also strongly induced stress fiber formation and F-actin accumulation in lamellopodial-like projections at the periphery of the cell; the epitope-tagged protein showed apparent tight association with the F-actin in these lamellae (Fig. 1D). The immunofluorescence results were confirmed using an SRF reporter gene assay (Fig. 1E). This reporter gene is activated by the actin/MAL/SRF pathway in response to depletion of the cellular pool of G-actin and serves as a reliable quantitative measure of the activity of factors that induce actin polymerization (11-13, 25, 26, 29, 30). As expected, expression of FRL3.F1F2 or FRL3.FH2 strongly activated the reporter gene, whereas expression of full-length FRL3 and FRL3N had no effect.
Expression of N-terminal (FRL2N), FH1/FH2 (FRL2.F1F2), and FH2 (FRL2.FH2) deletion derivatives of FRL2 yielded similar results as the corresponding derivatives of FRL3 with the notable exception that expression of full-length FRL2 also resulted in the robust accumulation of stress fibers in transfected cells (Fig. 2, A-D). These results were confirmed in the SRF reporter gene assay (Fig. 2E). This suggests that, despite the presence of highly conserved DID and DAD regulatory motifs, FRL2 is not subject to autoregulation.
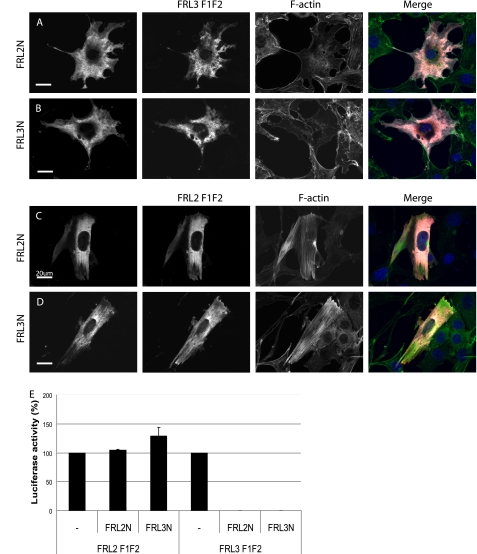
To investigate the basis for the apparent constitutive activity of full-length FRL2, we took advantage of the ability of the DID/DAD autoregulatory interaction to be recapitulated in trans using isolated N- and C-terminal deletion derivatives. Notably, in the case of mDia1 and mDia2, the isolated N terminus of mDia1 is able to inhibit the isolated C terminus of mDia2 and vice versa. Using a similar strategy, we wanted to determine if the failure in FRL2 autoinhibition lies with the DID or DAD of FRL2 (Fig. 3). FRL3.F1F2 was co-expressed with FRL2N or FRL3N in NIH 3T3 cells, and the effects on actin polymerization were assessed by immunofluorescence. In both cases, expression of either FRL2N or FRL3N was sufficient to completely inhibit the ability of FRL3.F1F2 to induce stress fiber formation (Fig. 3, A and B). Consistent with this being a direct effect, FRL3.F1F2 was extensively co-localized with FRL2N and FRL3N. FRL3.F1F2-induced activation of SRF was also completely inhibited by co-expression of either FRL2N or FRL3N (Fig. 3E). In contrast, expression of either FRL2N or FRL3N had no effect on FRL2.F1F2 activity as assessed by immunofluorescence and the SRF reporter gene assay (Fig. 3, C-E). Thus, FRL2 contains a functional DID motif that is capable of inhibiting FRL3 activity in trans but does not contain a functional DAD.
FIGURE 3.
FRL2 contains a functional DID but not a functional DAD. A and B, FRL3.F1F2 (red) was coexpressed with FRL2N or FRL3N (white) by transient transfection. Co-expression of either FRL2N or FRL3N (1.2 μg of DNA transfected) inhibits FRL3.F1F2 (0.3 μg of DNA)-induced stress fiber formation (green, 98%, n = 103) (compare cells in A and B with Fig. 1C). C and D, FRL2.F1F2 (red) was co-expressed with FRL2N or FRL3N (white) by transient transfection. Co-expression of either FRL2N or FRL3N did not inhibit FRL2.F1F2-induced stress fiber formation (green) (97%, n = 100). E, expression of FRL2N or FRL3N (1 μg of DNA transfected) inhibits FRL3.F1F2 (0.1 μg of DNA)-induced activation of an SRF reporter gene but has no effect on FRL2.F1F2-induced SRF activation. Reporter gene activation in the absence of inhibitor was standardized to 100%. Error bars, S.E., n = 3.
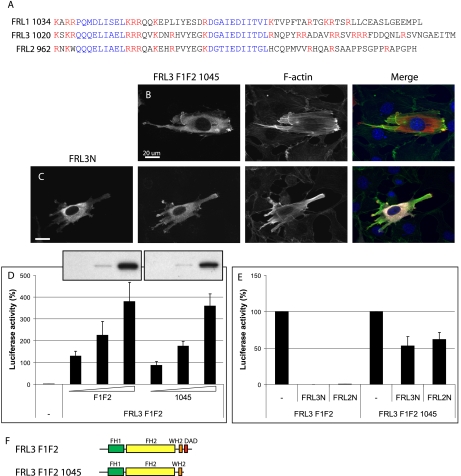
Further examination of the FRL2 and FRL3 C-terminal sequences revealed the presence of two WH2/DAD-like motifs in both proteins (Fig. 4A). To identify which of these motifs is required for the inhibitory DID interaction, we generated a C-terminal deletion derivative of FRL3.F1F2 that removed the more distal DAD motif (FRL3.F1F2.1045). The ability of FRL3N to inhibit the activity of this C-terminal truncation mutant was assessed by immunofluorescence. FRL3N was unable to inhibit FRL3.F1F2.1045-induced stress fiber formation (Fig. 4, B and C). Similar results were obtained with these derivatives in the SRF reporter gene assay (Fig. 4E). Deletion of the distal DAD motif of FRL3 had only a moderate effect on activity as assessed by immunofluorescence or in the SRF reporter gene assay. Based on these results, we refer to the more distal motif as DAD and the more proximal motif as WH2 (see below).
FIGURE 4.
The most distal DAD-like motif in FRL3 is required for autoregulation. A, alignment of the FRL1, -2, and -3 C-terminal domains. Two WH2/DAD-like motifs (blue) are present in the C termini of FRL2 and FRL3. Basic residues are highlighted in red. B and C, FRL3.F1F2.1045 (red)-induced stress fiber (green) formation is largely unaffected by co-expression of FRL3N (white) (94%, n = 102 versus 72%, n = 101). D, deletion of the FRL3 DAD domain does not significantly affect FRL3.F1F2 activity. Inset, relative levels of expression of FRL3.F1F2 and FRL3.F1F2.1045, as determined by immunoblotting using the same exposure of the same gel. Reporter gene activity was standardized to activation induced by expression of an SRF-VP16 control fusion protein. Error bars, S.E., n = 3. E, SRF reporter gene activation by FRL2.FH2.1045 is not significantly inhibited by FRL2N or FRL3N. Reporter gene activation in the absence of inhibitor was standardized to 100%. Error bars, S.E., n = 3. F, schematic of FRL3 derivatives.
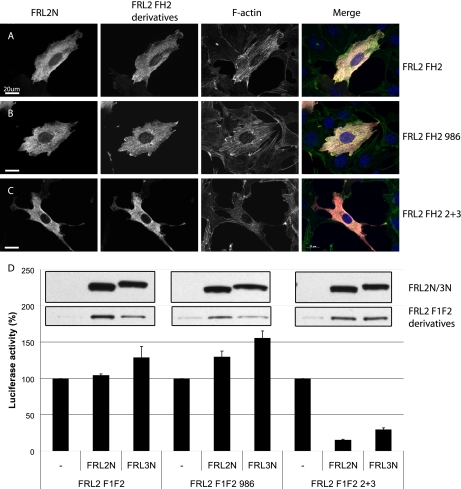
Having mapped the DAD domain in FRL3, we then wanted to determine if it is the DAD domain and sequences C-terminal to it in FRL2 that are insensitive to DID inhibition or if it is a general property of the FH2 domain of FRL2. We constructed chimeric FRL2 derivatives containing the FRL3 DAD and sequences C-terminal to it (FRL2.F1F2.2+3 and FRL2.FH2.2+3) and tested the ability of FRL2N to inhibit their activity. As expected, FRL2N was unable to inhibit FRL2.F1F2 or FH2 activity (Fig. 5, A and D); nor did it inhibit the activity of FRL2 derivatives that lack the DAD domain (FRL2.F1F2.986 or FH2.986) (Fig. 5, B and D). However, unlike the other FRL2 derivatives, the chimeric protein was inhibited by co-expression of either FRL2N or FRL3N (Fig. 5, C and D). Similar results were obtained with chimeric versions of the full-length FRL2 and FRL3 proteins (supplemental Fig. 1) Thus, it is the FRL2 DAD or sequences C-terminal to it that fail to support DID-dependent inhibition of FH2 activity.
FIGURE 5.
A chimeric FRL2 protein containing the FRL3 DAD is inhibited by DID in trans. A, FRL2.FH2 (red)-induced stress fiber formation (green) is not inhibited by co-expression of FRL2N (white) (97% alone, n = 100 versus 99% with FRL2N, n = 100). B, FRL2.FH2.986-induced stress fiber formation is not inhibited by co-expression of FRL2N (98%, n = 100). C, FRL2.FH2.2+3-induced stress fiber formation is inhibited by co-expression of FRL2N (25%, n = 102). D, FRL2.F1F2- or FRL2.F1F2.986-induced SRF reporter gene activation is not affected by co-expression of FRL2N orFRL3N. FRL2.F1F2.2+3 SRF activation is strongly inhibited by FRL2N and FRL3N. Inset, top panel, relative levels of expression of FRL2N and FRL3N as determined by immunoblotting using the same exposure of the same gel; bottom panel, relative levels of expression of FRL2.F1F2, FRL2.F1F2.986, and FRL2.F1F2.2+3, as determined by immunoblotting using the same exposure of the same gel. Reporter gene activation by FRL2.F1F2, FRL2.F1F2.986, or FRL2.F1F2.2+3 in the absence of inhibitor was standardized to 100% for each set. Error bars, S.E., n = 3.
The inability of FRL2 C-terminal derivatives to be inhibited by FRL2N or FRL3N implies two possible models; either the FRL2 DID and DAD domains are unable to form a stable complex, or they form a complex that is not competent to repress activation of the adjacent FH2 domain. To distinguish between these two models, we used co-immunoprecipitation assays to determine if the DID and DAD domains of FRL2 are able to interact in vivo. In these assays, we tested the ability of FLAG-tagged FRL2N or FRL3N to co-immunoprecipitate FRL2 and FRL3 C-terminal derivatives either containing or lacking the DAD motif. As expected, we found that FRL2N and FRL3N were able to efficiently co-immunoprecipitate FRL3.FH2 in a DAD-dependent manner (Fig. 6, A and B). Unexpectedly, we found that FRL2N and FRL3N were also able to co-immunoprecipitate FRL2.F1F2 with a similar efficiency as with FRL3.FH2 or FRL2.F1F2.2+3 (Fig. 6, A and B). The interaction between FRL2N and FRL2.F1F2 was also DAD-dependent (Fig. 6, A and B). Thus, the DID and DAD domains of FRL2 are able to interact in vivo, but in the case of FRL2.F1F2 or FRL2.FH2, this interaction is not inhibitory.
FIGURE 6.
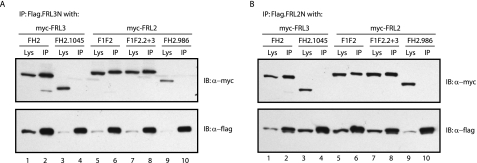
C-terminal derivatives of FRL2 and FRL3 bind to N-terminal FRL2 and FRL3 derivatives in a DAD-dependent manner. A, FLAG-tagged FRL3N (1.5 μg of DNA) was co-expressed in NIH 3T3 cells with Myc-tagged FRL3.FH2 or FH2.1045 and FRL2.F1F2, F1F2.2+3, or F1F2.986 (1.5 μg of DNA). The FLAG-tagged protein efficiently co-immunoprecipitated (IP) FRL3.FH2, FRL2.F1F2, and FRL2.F1F2.2+3 but not FRL3.FH2.1045 or FRL2.F1F2.986 (compare lanes 1 and 2 with lanes 3 and 4 and lanes 5-8 with lanes 9 and 10). B, FLAG-tagged FRL2N was co-expressed in NIH 3T3 cells with Myc-tagged FRL3.FH2 or FH2.1045 and FRL2.F1F2, F1F2.2+3, or F1F2.986. The FLAG-tagged protein efficiently co-immunoprecipitated FRL3.FH2, FRL2.F1F2, and FRL2.F1F2.2+3 but not FRL3.FH2.1045 or FRL2.F1F2.986, which lack the most distal DAD motif (compare lanes 1 and 2 with lanes 3 and 4 and lanes 5-8 with lanes 9 and 10).
Having shown that the DID/DAD interaction mediates heterodimerization between the N and C termini of FRL2 and FRL3, we used similar co-immunoprecipitation assays to assess the ability of the isolated N termini, FH2, and full-length proteins to form heterodimers. In contrast to our previous findings with mDia1 and mDia2, we found that we were able to efficiently co-immunoprecipitate hetero-oligomeric complexes of both the N-terminal dimerization domains and FH2 domains of FRL2 with FRL3 (Fig. 7, A and B). Consistent with these results, we were also able to efficiently co-immunoprecipitate hetero-oligomeric complexes of full-length FRL2 with full-length FRL3 (Fig. 7C).
FIGURE 7.
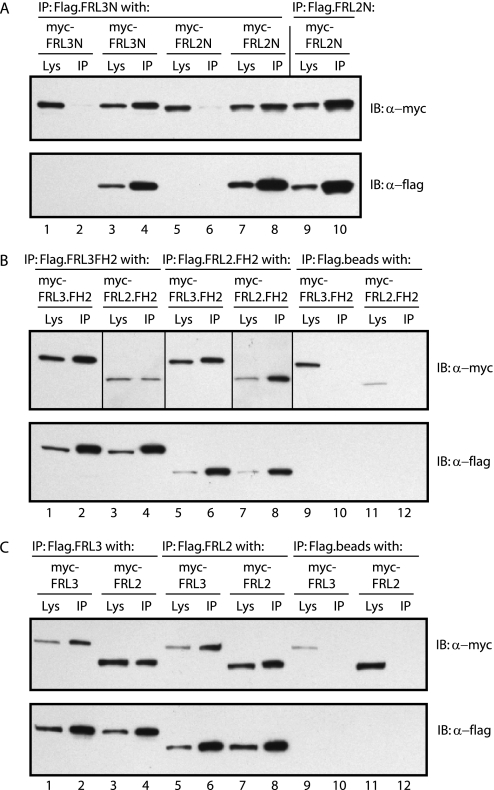
FRL2 and FRL3 are able to form hetero-oligomers. A, FRL2N and FRL3N form hetero-oligomers in vivo. FLAG-tagged FRL3N, or FRL2N, was co-expressed in NIH 3T3 cells with Myc-tagged FRL3N or FRL2N. FLAG-tagged FRL3N efficiently co-immunoprecipitated (IP) FRL3N and FRL2N (compare lanes 1 and 2 with lanes 3 and 4 and lanes 5 and 6 with lanes 7 and 8). FLAG-tagged FRL2N efficiently co-immunoprecipitated FRL2N. FLAG beads alone served as a negative control (lanes 1 and 2 and lanes 5 and 6). B, FRL2.FH2 and FRL3.FH2 form hetero-oligomers in vivo. FLAG-tagged FRL3.FH2 or FRL2.FH2 was co-expressed in NIH 3T3 cells with Myc-tagged FRL2.FH2 and FRL3.FH2. FLAG-tagged FRL3.FH2 efficiently co-immunoprecipitated FRL2.FH2 and FRL3.FH2 (compare lanes 1 and 2 with lanes 3 and 4 and lanes 9 and 10). FLAG-tagged FRL2.FH2 also efficiently co-immunoprecipitated FRL2.FH2 and FRL3.FH2 (compare lanes 5 and 6 with lanes 7 and 8 and lanes 11 and 12). FLAG beads alone served as a negative control (lanes 9-12). Vertical lines separate different exposures of the same immunoblot (lanes 1, 2, 5, 6, and 9-12 versus lanes 3, 4, 7, and 8). C, full-length FRL2 and FRL3 form hetero-oligomers in vivo. FLAG-tagged FRL3 or FRL2 was co-expressed in NIH 3T3 cells with Myc-tagged FRL2 and FRL3. FLAG-tagged FRL3 efficiently co-immunoprecipitated FRL3 and FRL2 (compare lanes 1 and 2 with lanes 3 and 4 and lanes 9 and 10). FLAG-tagged FRL2 also efficiently co-immunoprecipitated FRL2 and FRL3 (compare lanes 5 and 6 with lanes 7 and 8 and lanes 11 and 12). FLAG beads alone served as a negative control (lanes 9-12).
Both FRL2 and FRL3 contain two C-terminal WH2/DAD-like motifs (Fig. 4A). Previous reports have implicated similar motifs in modifying FH2 activity (19). Therefore, we generated additional FH2 C-terminal truncation mutants with each of these motifs deleted in turn and assayed the effect on FH2 activity in vivo. Expression of FRL3.FH2 induces the formation of thick actin stress fibers and punctate actin aggregates and the accumulation of F-actin in lamellopodia-like projections (Fig. 8A). Deletion of the distal DAD motif (FRL3.FH2.1045) had little effect on FH2 activity in this assay (Fig. 8B). Further deletion of both distal and proximal WH2/DAD-like motifs (FRL3.FH2.1023) caused an apparent loss of the punctate F-actin aggregates and reduction in peripheral F-actin accumulation (Fig. 8C). We also assessed the function of the FRL3.FH2 deletion derivatives using the SRF reporter gene assay, where a progressive loss of activity was observed as each WH2/DAD-like motif was deleted. The effect of removing these motifs from FRL2.FH2 was less pronounced, as assessed by immunofluorescence, and had no effect on FRL2.FH2 activity in the SRF reporter gene assay (Fig. 9).
FIGURE 8.
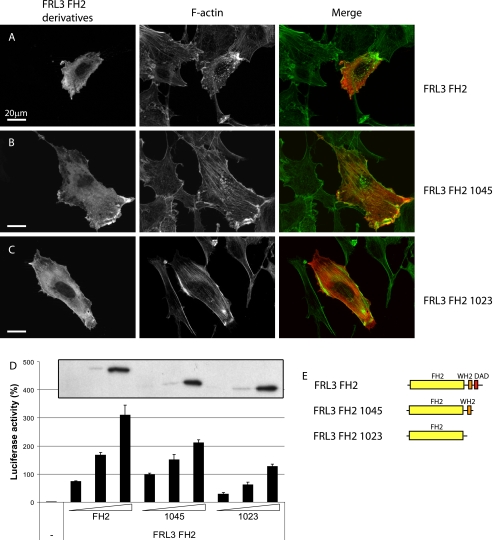
Deletion of the C-terminal DAD motifs reduces FRL3.FH2 activity in vivo. A, as in Fig. 1, expression of FRL3.FH2 induces thick stress fibers, actin aggregates, and peripheral F-actin accumulation (98%, n = 101). B, FRL3.FH2.1045 behaves similarly to FH2 (83%, n = 100). C, FRL3.FH2.1023 induces thick stress fibers (75%, n = 102) but no actin aggregates or peripheral F-actin. D, deletion of the distal and proximal DAD motifs reduces the ability of FRL3.FH2 to induce activation of an SRF reporter gene. Inset, relative levels of expression of FRL3.FH2, FRL3.FH2.1045, and FRL3.FH2.1023 as determined by immunoblotting. Reporter gene activity was standardized to activation induced by expression of an SRF-VP16 control fusion protein. Error bars, S.E., n = 3. E, schematic of FRL3.FH2 derivatives.
FIGURE 9.
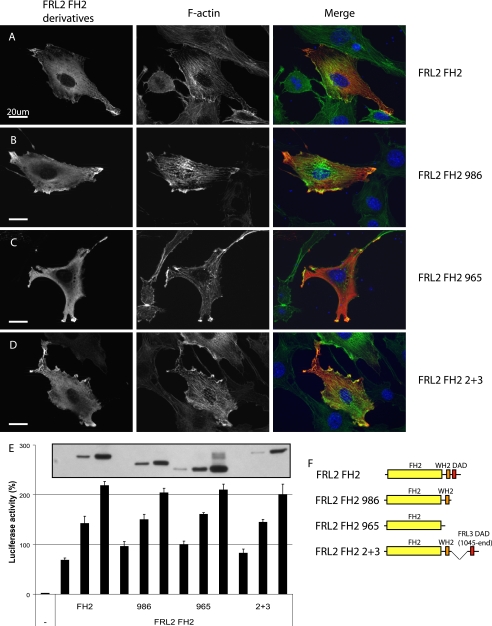
Deletion of the C-terminal DAD modifies FRL2.FH2 activity in vivo. A, as in Fig. 2, expression of FRL2.FH2 induces thick stress fibers, actin aggregates, and peripheral F-actin accumulation (97%, n = 100). B, FRL2.FH2.986 behaves similarly to FH2 (99%, n = 100). C, FRL2.FH2.965 induces fewer actin aggregates and peripheral F-actin (88%, n=100). D, FRL2.FH2.2+3 behaves similarly to FRL2.FH2(98%, n = 100). E, deletion of both distal and proximal DAD motifs does not affect FRL2.FH2-induced SRF reporter gene activation. Inset, relative levels of expression of FRL2.FH2, FRL2.FH2.986, FRL2.FH2.965, and FRL2.FH2.2+3, as determined by immunoblotting. Reporter gene activity was standardized to activation induced by expression of an SRF-VP16 control fusion protein. Error bars, S.E., n = 3. F, schematic of FRL2.FH2 derivatives.
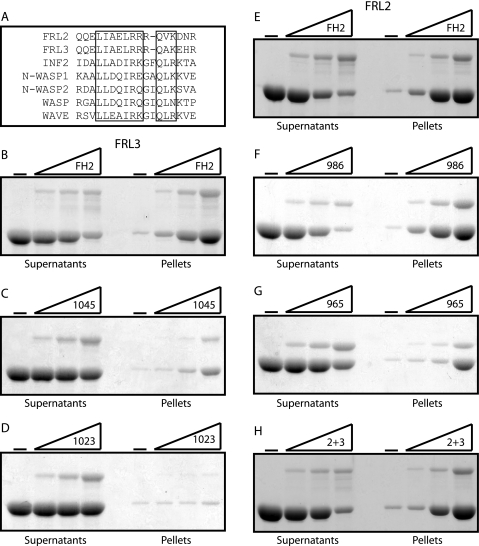
A notable effect of deleting the two WH2/DAD-like motifs of FRL3 was a decrease in FH2-induced punctate F-actin staining (Fig. 8C), which was also diminished in cells expressing the corresponding FRL2 construct (FRL2.FH2.965) (Fig. 9C). The related protein FRL1 has been shown to bind and bundle F-actin (20, 28), and actin bundling by the actin-binding protein espin is dependent on its WH2 domain (31). The proximal WH2/DAD-like motif shares extensive homology with other WH2 domains (Fig. 10A). Therefore, we tested the ability of FRL2 and FRL3 FH2 deletion derivatives to bundle actin filaments in vitro. We found that purified FRL2.FH2 and FRL3.FH2 proteins are able to bundle actin filaments in vitro (Fig. 10, B and D). In the case of FRL3, removal of the distal DAD reduces actin bundling (Fig. 10C), and further truncation to remove the proximal WH2/DAD-like motif completely eliminates actin bundling by this protein (Fig. 10D). As we observed in vivo, the effects of deletion of the WH2/DAD-like motifs on FRL2.FH2 activity were less pronounced. In this case, deletion of the distal DAD had little effect on F-actin bundling (Fig. 10E), although further deletion of the more proximal motif reduced, but did not eliminate, F-actin bundling (Fig. 10F). The chimeric construct FRL2.FH2.2+3 was also able to efficiently bundle F-actin in this assay. Thus, the proximal WH2/DAD-like motif is playing a role in F-actin bundling rather than autoregulation, and therefore we designate the proximal motif as WH2.
FIGURE 10.
The FH2 domains of FRL2 and FRL3 are able to bind and bundle actin filaments. A, alignment of WH2-like domains of FRL2 and FRL3 with the WH2 domains of INF2, N-WASP1, N-WASP2, WASP, and WAVE. B, FRL3.FH2; C, FRL3.FH2.1045; D, FRL3.FH2.1023; E, FRL2.FH2; F, FRL2.FH2.986; G, FRL2.FH2.965; H, FRL2.FH2.2+3 (0, 0.15, 0.3, and 0.6 μm) were incubated with F-actin (2 μm) and then centrifuged at 16,000 × g for 10 min to pellet F-actin bundles. Equivalent samples of the supernatant and pellet were subjected to SDS-PAGE, and the proteins were visualized with Coomassie Blue.
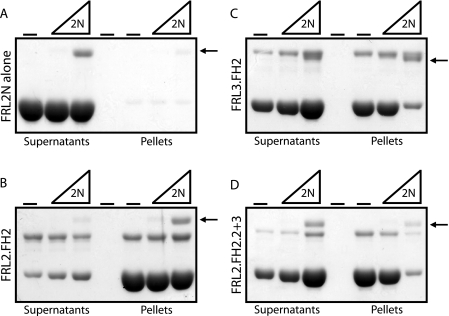
We used the F-actin bundling assay to investigate directly the inhibitory interaction between the N- and C-terminal regulatory domains of FRL2 and FRL3. FRL2N protein on its own did not induce F-actin bundling in vitro and did not pellet (Fig. 11A). Incubation of FRL2N with FRL3.FH2 or FRL2.FH2.2+3 greatly inhibited the ability of either FH2 domain to bundle F-actin (Fig. 11, C and D). In contrast, incubation with FRL2N had essentially no effect on F-actin bundling by FRL2.FH2. However, although there was no inhibition of FRL2.FH2-induced actin bundling, FRL2N was still recruited to the actin pellet by FRL2.FH2 (Fig. 11B).
FIGURE 11.
FRL2N inhibits actin bundling by FRL3.FH2 and FRL2.FH2.2+3. FRL2N (0, 0.1, and 0.5 μm) was incubated with F-actin (2 μm) (A) and FRL2.FH2 (0.5 μm) (B), FRL3.FH2 (0.5 μm) (C), or FRL2.2+3 (0.5 μm) (D) and then centrifuged at 16,000 × g for 10 min to pellet F-actin bundles. Equivalent samples of the supernatant and pellet were subjected to SDS-PAGE, and the proteins were visualized with Coomassie Blue. The arrow indicates the FRL2N protein band.
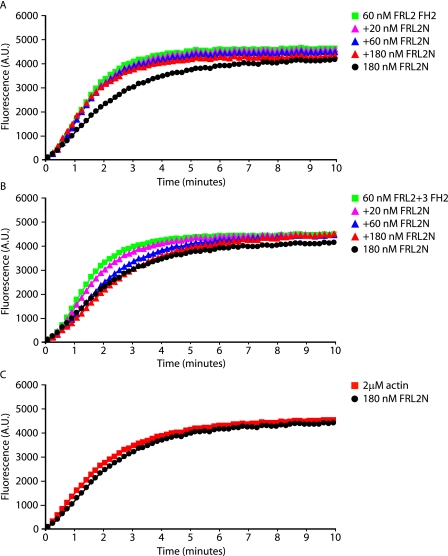
These results were confirmed using the pyrene-actin polymerization assay (Fig. 12). In this assay, both FRL2.FH2 and FRL2. FH2.2+3 are able to induce actin polymerization. A 3-fold molar excess of FRL2N has no effect on FRL2 FH2-induced actin polymerization (Fig. 12A) but is able to completely inhibit FRL2.FH2.2+3 activity (Fig. 12B). Thus, as was the case in vivo, FRL2.FH2 binds directly to FR2N but in a noninhibitory complex.
FIGURE 12.
FRL2N inhibits actin polymerization induced by FRL2.FH2.2+3 but not by FRL2.FH2. Purified FRL2.FH2 and FRL2.FH2.2+3 derivatives were tested in pyrene-actin polymerization assays at the indicated concentrations in the presence or absence of purified FRL2N. A, FRL2 FH2 (60 nm) induces actin polymerization. This polymerization is not inhibited by preincubation with FRL2N (20, 60, and 180 nm). FRL2N alone (180 nm) has no effect on polymerization. B, FRL2.FH2.2+3 (60 nm) induces actin polymerization and is inhibited by preincubation with FRL2N (20, 60, and 180 nm). C, FRL2N alone (180 nm) does not affect actin polymerization. G-actin concentration was 2 μm. A.U., absorbance unts.
DISCUSSION
We report here an initial structure-function analysis of the Diaphanous-related formins FRL2 and FRL3. We find that the C-terminal regions of both FRL2 and FRL3 contain two WH2/DAD-like motifs. The more proximal motif we have designated as “WH2” based on homology and because of the role it plays in F-actin bundling. The more distal motif we have designated “DAD,” since it is required for autoregulation of FRL3. We also show that these proteins are able to form hetero-oligomers through the dimerization of either their FH2 or N-terminal domains.
F-actin Bundling—We found that both FRL2 and FRL3 are able to bind and to bundle F-actin in vitro. This activity is apparently dependent upon a WH2-like motif found immediately C-terminal to FH2 and proximal to the regulatory DAD motif. It has been noted previously that WH2 and DAD domains are quite similar at the sequence level. Indeed, the INF2 DAD is also a G-actin-binding WH2 domain that antagonizes the activity of the INF2 FH2 domain (19). The proximal WH2/DAD sequence in both FRL2 and FRL3 shares extensive homology with the WH2 motif found in INF2, N-WASP, and other proteins (Fig. 10A). Functionally, this sequence also exhibits more WH2-like behavior in that it is absolutely required for F-actin bundling by FRL3 and is required for efficient F-actin bundling by FRL2. This result is reminiscent of the ability of espin to bind and bundle F-actin in a WH2-dependent fashion (31). In this case, F-actin bundling is dependent upon G-actin binding by WH2 and the presence of two additional F-actin binding sites in espin (31). By analogy, our results would suggest a model where F-actin bundling by FRL3 is dependent on barbed end binding by the FH2 domain and an additional actin binding surface on WH2. Despite the overall homology between FRL2 and FRL3, the WH2 independence of F-actin bundling by FRL2 suggests that the two proteins are bundling actin filaments by distinct mechanisms. We favor a model where F-actin bundling by FRL2 is likely to be similar to FRL1 and dependent on dissociation of the FH2 dimer (20, 28), whereas the WH2-dependent F-actin bundling by FRL3 is more similar to F-actin bundling by espins (31). Further experimentation is required to determine if the WH2 motif found in FRL2 and FRL3 is itself sufficient to confer F-actin binding when incorporated into a heterologous protein. A more complete analysis of how the C-terminal WH2 and DAD motifs of these proteins affect FH2-induced actin polymerization will be reported elsewhere.3
Autoregulation—Our results show that full-length FRL2 is constitutively active when expressed by transient transfection in NIH 3T3 cells. This could result either from an inability of the DID and DAD domains to interact or a failure of this interaction to inhibit FH2 activity. Both in vivo and in vitro we were able to demonstrate that the DID of FRL2 is able to bind to DAD, but this association does not inhibit FH2 activity (Figs. 4, 6, 11B, and 12A). The inability of this interaction to inhibit FH2 activity involves a deficit within the C-terminal regulatory domain despite the presence of two DAD-like motifs in this region. Numerous lines of evidence suggest that this result is not an artifact of our system; nor is it an inherent property of the FH2 domain of FRL2. First, the N terminus of FRL2 is able to inhibit the activity of the FRL3 C terminus, demonstrating that FRL2 contains a functional DID (Fig. 3). Second, the C-terminal derivatives of FRL2 that contain the DAD of FRL3 are inhibited by either FRL2N or FRL3N in trans (Figs. 4, 6, 11B, and 12A). Third, a chimeric full-length FRL2 protein containing the DAD of FRL3 is autoinhibited (supplemental Fig. 1, E and G). Therefore, FRL2 FH2 activity can be inhibited through a DID/DAD interaction. Fourth, replacement of the DAD of FRL3 with that from FRL2 partially relieves FRL3 autoinhibition, although not to the same extent as deletion of the FRL3 DAD (supplemental Fig. 1, B, C, and F). The observation that, in the context of full-length FRL3, the DAD of FRL2 is able to support some level of autoinhibition suggests that the constitutive activity of FRL2 in vivo is not due to the absence of some unknown regulatory factor in NIH 3T3 cells. Instead, we favor the idea that the constitutive activity of FRL2 results from a combination of a suboptimal DAD sequence and an FH2 domain that is intrinsically more refractive to DID/DAD inhibition (see below).
FHOD1, like FRL2 and FRL3, also possesses two C-terminal DAD-like motifs, with the more distal DAD mediating FHOD1 autoregulation (32). For FHOD1 and for mDia2, efficient DID/DAD dimerization and autoinhibition is dependent upon a cluster of basic residues C-terminal to DAD (32, 33). Examination of the FRL2 and FRL3 primary sequences reveals that the number of basic residues varies significantly between the various alternative splice forms of each of these proteins (34). The FRL2b isoform used in this study has the least (3 arginines), whereas the FRL3b isoform used here has the most (9 arginines) (Fig. 4A). However, we found that the C terminus of FRL2 bound strongly to FRL2N in a DAD-dependent manner both in vivo and in vitro (Figs. 6 and 11). Therefore, it seems that in this case, the basic residues are not required so much for the DID/DAD interaction as for allowing this interaction to inhibit FH2 activity.
Current models suggest that DID and DAD inhibit FH2 activity in the autoinhibited conformation by simply masking the FH2 domain. However, this cannot be the case for FRL2, since we show here that the direct interaction of DID with the DAD of FRL2 does not inhibit FH2 activity. Regardless of whether or not an additional factor is required to mediate FRL2 inhibition in vivo, our results are most consistent with a model where the functional DID/DAD interaction induces a conformational change in the FH2 domain to inhibit FH2 activity. Additional support for this model comes from the recently solved structure of the DAAM1 FH2 domain (35). This study found the isolated DAAM1 FH2 to be in an apparently autoinhibited conformation. Thus, our results lay the foundation for future studies into how DID/DAD mediates inhibition of formin activity.
Heterodimerization—Recently, we showed that the DID/DAD interaction is able to mediate heterodimerization of mDia1 and mDia2 (27). However, we were unable to detect heterodimerization of FH2 with FH2 or of the N-terminal dimerization domains despite the extensive homology shared by these proteins (FH2 60% identical, 78% similar; dimerization domains 68% identical, 81% similar). Overall, the FRL2 and FRL3 proteins are highly homologous (74% identical, 87% similar). As with mDia1 and mDia2, we showed that the DID of FRL2 is able to bind to the DAD of FRL3 and vice versa. In addition, and unlike mDia1 and mDia2, both the FRL2 and FRL3 N-terminal dimerization domains (Fig. 7A) and FH2 domains (Fig. 7B) are also able to form hetero-oligomers in vivo, as did the full-length proteins (Fig. 7C). It is not clear if mDia1 and mDia2 or if FRL2 and FRL3 provide the more general model for formin protein dimerization. The ability of FRL2 and FRL3 to form heterodimers represents a possible regulatory mechanism for both proteins. Perhaps a constitutively active FRL2 becomes inhibited when bound to an autoinhibited FRL3, although the opposite may be equally likely (i.e. FRL3 is activated when bound to FRL2). Therefore, it will be of interest to determine how FRL2 and FRL3 expression overlaps in different tissues and cell types.
In summary, our results describe a novel WH2-dependent mechanism for F-actin bundling by formin proteins and suggest that the current model of Diaphanous-related formin autoregulation must be revised.
Supplementary Material
Acknowledgments
We thank Henry Higgs for insightful discussion of the results and members of the laboratory for critical reading of the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviation used is: SRF, serum response factor.
D. C. Vaillant and J. W. Copeland, manuscript in preparation.
References
- 1.Higgs, H. N., and Peterson, K. J. (2005) Mol. Biol. Cell 16 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, F., Drubin, D., and Nurse, P. (1997) J. Cell Biol. 137 169-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evangelista, M., Pruyne, D., Amberg, D. C., Boone, C., and Bretscher, A. (2002) Nat. Cell Biol. 4 260-269 [DOI] [PubMed] [Google Scholar]
- 4.Feierbach, B., and Chang, F. (2001) Curr. Biol. 11 1656-1665 [DOI] [PubMed] [Google Scholar]
- 5.Kobielak, A., Pasolli, H. A., and Fuchs, E. (2004) Nat. Cell Biol. 6 21-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellegrin, S., and Mellor, H. (2005) Curr. Biol. 15 129-133 [DOI] [PubMed] [Google Scholar]
- 7.Sagot, I., Rodal, A. A., Moseley, J., Goode, B. L., and Pellman, D. (2002) Nat. Cell Biol. 4 626-631 [DOI] [PubMed] [Google Scholar]
- 8.Schirenbeck, A., Bretschneider, T., Arasada, R., Schleicher, M., and Faix, J. (2005) Nat. Cell Biol. 7 619-625 [DOI] [PubMed] [Google Scholar]
- 9.Severson, A. F., Baillie, D. L., and Bowerman, B. (2002) Curr. Biol. 12 2066-2075 [DOI] [PubMed] [Google Scholar]
- 10.Ishizaki, T., Morishima, Y., Okamoto, M., Furuyashiki, T., Kato, T., and Narumiya, S. (2001) Nat. Cell Biol. 3 8-14 [DOI] [PubMed] [Google Scholar]
- 11.Sotiropoulos, A., Gineitis, D., Copeland, J., and Treisman, R. (1999) Cell 98 159-169 [DOI] [PubMed] [Google Scholar]
- 12.Miralles, F., Posern, G., Zaromytidou, A. I., and Treisman, R. (2003) Cell 113 329-342 [DOI] [PubMed] [Google Scholar]
- 13.Copeland, J. W., and Treisman, R. (2002) Mol. Biol. Cell 13 4088-4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallar, B. J., and Alberts, A. S. (2003) Trends Cell Biol. 13 435-446 [DOI] [PubMed] [Google Scholar]
- 15.Kovar, D. R. (2006) Curr. Opin. Cell Biol. 18 11-17 [DOI] [PubMed] [Google Scholar]
- 16.Romero, S., Le Clainche, C., Didry, D., Egile, C., Pantaloni, D., and Carlier, M. F. (2004) Cell 119 419-429 [DOI] [PubMed] [Google Scholar]
- 17.Kovar, D. R., Harris, E. S., Mahaffy, R., Higgs, H. N., and Pollard, T. D. (2006) Cell 124 423-435 [DOI] [PubMed] [Google Scholar]
- 18.Gasteier, J. E., Madrid, R., Krautkramer, E., Schroder, S., Muranyi, W., Benichou, S., and Fackler, O. T. (2003) J. Biol. Chem. 278 38902-38912 [DOI] [PubMed] [Google Scholar]
- 19.Chhabra, E. S., and Higgs, H. N. (2006) J. Biol. Chem. 281 26754-26767 [DOI] [PubMed] [Google Scholar]
- 20.Harris, E. S., Rouiller, I., Hanein, D., and Higgs, H. N. (2006) J. Biol. Chem. 281 14383-14392 [DOI] [PubMed] [Google Scholar]
- 21.Michelot, A., Derivery, E., Paterski-Boujemaa, R., Guerin, C., Huang, S., Parcy, F., Staiger, C. J., and Blanchoin, L. (2006) Curr. Biol. 16 1924-1930 [DOI] [PubMed] [Google Scholar]
- 22.Rose, R., Weyand, M., Lammers, M., Ishizaki, T., Ahmadian, M. R., and Wittinghofer, A. (2005) Nature 435 513-518 [DOI] [PubMed] [Google Scholar]
- 23.Nezami, A. G., Poy, F., and Eck, M. J. (2006) Structure 14 257-263 [DOI] [PubMed] [Google Scholar]
- 24.Goode, B. L., and Eck, M. J. (2007) Annu. Rev. Biochem. [DOI] [PubMed]
- 25.Geneste, O., Copeland, J. W., and Treisman, R. (2002) J. Cell Biol. 157 831-838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Copeland, J. W., Copeland, S. J., and Treisman, R. (2004) J. Biol. Chem. 279 50250-50256 [DOI] [PubMed] [Google Scholar]
- 27.Copeland, S. J., Green, B. J., Burchat, S., Papalia, G. A., Banner, D., and Copeland, J. W. (2007) J. Biol. Chem. 282 30120-30130 [DOI] [PubMed] [Google Scholar]
- 28.Harris, E. S., Li, F., and Higgs, H. N. (2004) J. Biol. Chem. 279 20076-20087 [DOI] [PubMed] [Google Scholar]
- 29.Grosse, R., Copeland, J. W., Newsome, T. P., Way, M., and Treisman, R. (2003) EMBO J. 22 3050-3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posern, G., Sotiropoulos, A., and Treisman, R. (2002) Mol. Biol. Cell 13 4167-4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loomis, P. A., Kelly, A. E., Zheng, L., Changyaleket, B., Sekerkova, G., Mugnaini, E., Ferreira, A., Mullins, R. D., and Bartles, J. R. (2006) J. Cell Sci. 119 1655-1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonichen, A., Alexander, M., Gasteier, J. E., Cuesta, F. E., Fackler, O. T., and Geyer, M. (2006) J. Biol. Chem. 281 5084-5093 [DOI] [PubMed] [Google Scholar]
- 33.Wallar, B. J., Stropich, B. N., Schoenherr, J. A., Holman, H. A., Kitchen, S. M., and Alberts, A. S. (2006) J. Biol. Chem. 281 4300-4307 [DOI] [PubMed] [Google Scholar]
- 34.Katoh, M. (2003) Int. J. Oncol. 22 1161-1168 [PubMed] [Google Scholar]
- 35.Lu, J., Meng, W., Poy, F., Maiti, S., Goode, B. L., and Eck, M. J. (2007) J. Mol. Biol. 369 1258-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.