Abstract
Collagens are the most abundant proteins in marine animals and their degradation is important for the recycling of marine nitrogen. However, it is rather unclear how marine collagens are degraded because few marine collagenolytic proteases are studied in detail. Deseasins are a new type of multidomain subtilases. Here, the collagenolytic activity of deseasin MCP-01, the type example of deseasins, was studied. MCP-01 had broad substrate specificity to various type collagens from terrestrial and marine animals. It completely decomposed insoluble collagen into soluble peptides and amino acids, and was more prone to degrade marine collagen than terrestrial collagen. Thirty-seven cleavage sites of MCP-01 on bovine collagen chains were elucidated, showing the cleavage is various but specific. As the main extracellular cold-adapted protease from deep-sea bacterium Pseudoalteromonas sp. SM9913, MCP-01 displayed high activity at low temperature and alkaline range. Our data also showed that the C-terminal polycystic kidney disease (PKD) domain of MCP-01 was able to bind insoluble collagen and facilitate the insoluble collagen digestion by MCP-01. Site-directed mutagenesis demonstrated that Trp-36 of the PKD domain played a key role in its binding to insoluble collagen. It is the first time that the structure and function of a marine collagenolytic protease, deseasin MCP-01, has been studied in detail. Moreover, the PKD domain was experimentally proven to bind to insoluble protein for the first time. These results imply that MCP-01 would play an important role in the degradation of deep-sea sedimentary particulate organic nitrogen.
Collagens, which distribute in skin, scale, bone, tendon, teeth, and blood vessels are the major protein constituents of the extracellular matrix and the most abundant proteins in all higher organisms including marine animals. It is an important fraction of marine organic nitrogen, and its degradation is important for marine nitrogen recycling.
Because the tightly coiled triple helical collagen molecule assembles into water-insoluble fibers, collagens are resistant to most proteases except for a limited number of collagenolytic proteases. The mechanism of marine collagen degradation in marine nitrogen recycling is rather unclear because there are only several reports about marine collagenolytic enzyme-producing bacteria and collagenases (1–5). Collagenolytic proteases include metalloproteinases, serine proteases, and other proteases. Of all the collagenolytic proteases, mammalian collagenases, which are mammalian matrix metalloproteinases have been investigated in more detail than collagenolytic proteases from bacteria (6). Studies on the collagenolytic proteases from marine bacteria are rather minor and preliminary. Some marine collagenolytic enzyme-producing bacteria have been reported (1, 5). The collagenase from marine Vibro B-30 was purified and primarily characterized, which was reported at 1978 and 1980 (2–4). Since then, there has been no report on the characterization of marine bacterial collagenolytic proteases. The lack of knowledge of marine collagenolytic proteases is a huge holdback on the elucidation of the mechanism of marine collagen degradation.
Collagens are water-insoluble fibers and thus a collagen-binding domain occurs in collagenolytic proteases, which has been proven in mammalian matrix metalloproteinases and bacterial collagenases from land (6). Because there is no report on the gene and structure of a marine collagenolytic protease, it is unknown whether marine collagenolytic proteases have collagen-binding domains. Deseasins are a new type of subtilases mainly secreted by bacteria in deep-sea or lake sediment (7). Compared with other subtilases, deseasins are all multidomain enzymes with a polycystic kidney disease (PKD)3 domain at their C terminus (7). PKD domain has been found in some chitinases (8–10), cellulases (11), proteases (12), and collagenases (13). Except that the PKD domain of the chitinase ChiA from Alteromonas sp. O-7 was reported to have binding ability to chitin (10), the function of the PKD domain of other enzymes has not yet been experimentally proven. Genomic sequence analysis of marine Gramella forsetii revealed that many exported proteins contain PKD domains (14), indicating that PKD domains might widely lie in the hydrolases of marine heterotrophic bacteria. Therefore, elucidation of the function of the PKD domain is of universal importance.
Deseasin MCP-01, the main extracellular protease of the psychrotolerant bacterium Pseudoalteromonas sp. SM9913 from 1855 meter deep-sea sediment, is the type example of deseasins studied because it is the first deseasin that has been purified and characterized. The catalytic characters of MCP-01 with casein as substrate have been studied (7, 15–17). Moreover, it was noticed that the multidomain structure of MCP-01 is similar to that of some hydrolases capable of digesting insoluble biopolymer, such as celluloses (11), chitinases (8–10), and collagenolytic proteases (13), and that the C-terminal PKD domain of MCP-01 looks like a substrate-binding domain just as those in the hydrolases capable of digesting insoluble biopolymer. Thus, it was speculated that deseasin MCP-01 is probably able to hydrolyze insoluble proteins in deep-sea. In this article, deseasin MCP-01 was proven to be able to digest insoluble collagens into soluble peptides and its collagenolytic characters were studied. Moreover, the C-terminal PKD domain of MCP-01 was proven to function as a binding domain during insoluble collagen digestion, and its binding properties, as well as its function in the digestion of insoluble collagen by MCP-01 was studied.
EXPERIMENTAL PROCEDURES
Materials—Deseasin MCP-01 was purified from Pseudoalteromonas sp. SM9913 as previously described (7). Fish collagen was extracted from Pseudosciaena polyactis with the method described by Song et al. (18). Collagenase from Clostridium histolyticum and insoluble type I collagen fiber (bovine achilles tendon) was purchased from Worthington Biochemical Co., type II and IV collagen from BD Biosciences, acid-dissolved type I collagen (calf skin), elastin (bovine neck ligament), Pz-peptide (4-phenylazobenzyloixycarbonyl-Pro-Leu-Gly-Pro-o-Arg), and Su-AAA (N-succinyl-Ala-Ala-Ala-p-nitroanilide) from Sigma, and gelatin from Boston Biomedical Inc. Escherichia coli DH5α and pET-22b(+) (Novagen) were used as host and plasmid for the construction of expression vectors, and E. coli BL21(DE3) (Transgen) was used as expression host.
Protein Expression and Purification of the Catalytic Domain of MCP-01—The genome DNA of Pseudoalteromonas sp. SM9913 was prepared with the previous method (7). A DNA fragment coding for the catalytic domain (CD) of MCP-01 (from M1 to G419) was amplified by PCR with the genome DNA of Pseudoalteromonas sp. SM9913, Pfu polymerase (Fermentas), and two primers (P1, 5′-GCTCATATGAAAACAAAATTATCTATT-3′, and P2, 5′-CGCTCGAGTACCTCACCAATAGCGC-3′). The obtained fragment was then ligated into the NdeI-XhoI sites of vector pET-22b(+) to construct a recombined vector for the expression of CD, which was then transformed into E. coli BL21(DE3). The transferred strain was grown on LB agar containing 100 μg/ml ampicillin overnight. The culture was diluted 100-fold and grown on LB-ampicillin at 37 °C to A600 = 1.2. Then expression was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside, at 170 rpm at 15 °C for 72 h. After a centrifugation of the fermented broth at 10,000 × g at 4 °C for 5 min, the proteins in the supernatant were precipitated by 70% saturation of ammonium sulfate. The precipitate was dissolved in Buffer A (50 mm Tris-HCl, pH 7.5) and dialyzed against the same buffer. The sample was subjected onto a DEAE-Sepharose Fast Flow column equilibriated with buffer A. The column was eluted with a linear gradient of 0–0.8 m NaCl in buffer A and the fraction with protease activity was collected and dialyzed against Buffer B (50 mm Tris-HCl, pH 9.0). Then, the sample was further purified by DEAE-Sepharose Fast Flow equilibrated with buffer B with a linear gradient elution of 0–0.8 mm NaCl in buffer B and the fraction with protease activity was collected. Its purity was detected by SDS-PAGE with the method of Laemmli (19).
Expression and Purification of the Recombinant PKD and Enhanced Green Fluorescent Protein Fusion PKD—A DNA fragment coding for the PKD domain of MCP-01 was amplified by PCR with the genome DNA of Pseudoalteromonas sp. SM9913, Pfu polymerase, and two primers (P3, 5′-GGGACGCCATATGAGCCCACAACCACCACAA-3′, and P4, 5′-CCGCTCGAGAACAACCACTGTTTGAGTGAATGTA-3′). A recombined vector for the expression of recombinant PKD was constructed by a ligation of the fragment into the NdeI-XhoI sites of vector pET-22b(+)(DE3). To analyze the binding ability of the PKD domain to insoluble collagen, the EGFP was used to fuse with the PKD domain. The EGFP gene egfp was amplified with an overlapping sequence from vector pEGFP-N1 (Clontech) by PCR. The DNA fragment encoding PKD with the same overlapping sequence was also amplified. The two fragments were overlapped by mutagenesis by overlapping extension PCR (20). The chimeric gene was subcloned into pET-22b(+) for the expression of fusion protein PKD-EGFP. Both recombinant vectors were transformed into E. coli BL21(DE3). The recombinant PKD and PKD-EGFP were all expressed as C-terminal His6-tagged proteins by an inducement of 0.6 mm isopropyl β-d-thiogalactopyranoside at 20 °C for 16 h, and purified with His·Bind metal chelating column.
Site-directed Mutagenesis of the PKD Domain—Alignment of PKD domains were performed by Clustal X 1.83 (21). Site-directed mutagenesis was carried out by mutagenesis by overlapping extension-PCR with the vector PKD-EGFP as template. Mutated sites were introduced by the primers with single or double mutation. The mutated genes were subcloned into pET-22b(+) and transformed into E. coli BL21(DE3). All mutations were confirmed by enzyme digestion and nucleotide sequencing. Fusion proteins were all expressed and purified with the same condition as PKD-EGFP.
Protein Determination and Enzyme Assays—Proteins were assessed by the method of Lowry et al. (22) with bovine serum albumin as standard. The collagenolytic activities of deseasin MCP-01 and its CD against collagen were determined with the method provided by Worthington Biochemical Co. (23). The reaction time was 5 h for fish-insoluble collagen fiber and bovine-insoluble type I collagen fiber, and 0.5 h for bovine-soluble types I, II, and IV collagen and gelatin. For insoluble collagen, 1 unit equals 1 nmol of l-leucine equivalents released from collagen in 1 h. For soluble collagens and gelatin, 1 unit equals 1 nmol of l-leucine equivalents from collagen in 1 min. The activity of MCP-01 to Pz-peptide was carried out with the method described by Miyake et al. (24). The enzyme activity to casein and Su-AAA (N-succinyl-Ala-Ala-Ala-p-nitroanilide) was measured with the methods previously described (7). The activity of the collagenase from C. histolyticum to all of the above substrates was measured with the same method as for MCP-01.
To determine the effect of temperature on the activity of MCP-01 and the collagenase from C. histolyticum to bovine-insoluble type I collagen, the enzyme activities were measured from 0 to 80 °C. 50 mm Tris-HCl from pH 6.0 to 9.0 was used to determine the preferable pH profile of MCP-01 in the digestion of bovine-insoluble type I collagen fiber. Various metal ions and inhibitors were added into buffer A to determine their influence on the activity of MCP-01 to bovine-insoluble type I collagen fiber.
Digestion Pattern Analysis of Insoluble Collagen Fiber by MCP-01—Fish-insoluble collagen fiber and bovine-insoluble type I collagen fiber were digested by MCP-01 at 60 °C for 5 h, respectively. After centrifugation, the supernatant was assayed by SDS-PAGE with 12.5% acrylamide, and transferred to Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). N-terminal amino acid sequences of the peptides released from bovine-insoluble type I collagen fiber by MCP-01 were analyzed with Edman degradation by PROCISE491 (Applied Biosystems) at Beijing University, China.
Collagen Binding Assay—Collagen binding assay was performed by adding various concentrations of PKD-EGFP or its mutants into 5 mg of bovine-insoluble type I collagen fiber (or insoluble elastin) in 0.5 ml of double distilled water in a 1.5-ml microcentrifuge tube. Samples were incubated for 1 h at 40 °C with mixing and then centrifuged at 10,000 × g for 5 min. The free fluorescence intensity in the solution before and after incubation was determined with FP-6500 spectrofluorometer (Jasco, Japan). EGFP was used as control in the experiment. The concentration of the initial and unabsorbed proteins was also determined.
Structural Analysis of PKD Domain and Its Mutant W36A with CD—The PKD domain and its mutant W36A was expressed in E. coli BL21(DE3) and purified with His·Bind metal chelating column. CD spectra of the purified PKD domain and W36A in the same concentration (0.5 mg/ml) in 20 mm boric buffer (pH 8.0) was measured on a Jasco J810 spectropolarimeter (Japan) with a bandwidth of 4 nm, a response time of 1 s, and a scan speed of 200 nm/min. Each spectrum was an average of three scans monitored between 190 and 250 nm. The path length of the cuvette was 0.1 cm. The raw CD data were converted into mean residue ellipticity (deg cm2 dmol–1) at the entire wavelength using the relation: [θ]λ = θλM0/10/c.
RESULTS
Characterization of the Collagenolytic Activity of Deseasin MCP-01
Substrate Specificity—Deseasin MCP-01 purified from the culture of Pseudoalteromonas sp. SM9913 was used to hydrolyze some proteins and synthetic peptides to determine its substrate specificity. All the collagens used fish collagen and bovine type I (both insoluble and acid-dissolved), II, and IV collagens, as well as Pz-peptide were all suitable substrates for deseasin MCP-01. For the insoluble collagens from different sources, MCP-01 had higher activity to fish collagen and lower activity to bovine collagen than the collagenase from C. histolyticum. For soluble collagens, the order of preferred substrate for MCP-01 was gelatin > acid-dissolved type I collagen > type IV collagen > type II collagen, and the order for the collagenase from C. histolyticum was gelatin > type II collagen > acid-dissolved type I collagen > type IV collagen. MCP-01 had nearly the same specific activity to Pz-peptide as the collagenase from C. histolyticum. For other proteins, MCP-01 had obvious activity to casein, as well as a slight activity to elastin according to its slight activity to Su-AAA, whereas the collagenase from C. histolyticum had no activity to casein and Su-AAA (Table 1). These results showed that deseasin MCP-01 has broad specificity to various collagens. Moreover, as a marine protease, MCP-01 preferably degrades the collagen from marine animals than that from terrestrial animals. Collagens are the most abundant proteins of marine animals and are an important fraction of marine organic nitrogen. Therefore, it could be suggested that MCP-01 would have high ability to degrade various collagens in deep-sea sediments and therefore play an important role in the recycling of marine nitrogen.
TABLE 1.
The substrate specificity of deseasin MCP-01 compared with that of the collagenase from C. histolyticum
Note, the activities of MCP-01 and the collagenase from C. histolyticum to various substrates at 40 °C were measured with the methods described under “Experimental Procedures.” The data represent the mean of three experimental repeats with S.D. ≤ 5%.
| Substrate | MCP-01 | Collagenasea | |
|---|---|---|---|
| units/mg | |||
| Fish-insoluble collagen fiber | 10434 | 4456 | |
| Bovine-insoluble type I collagen fiber | 1608 | 5274 | |
| Bovine acid-dissolved type I collagen | 136.7 | 113.1 | |
| Bovine type II collagen | 67.6 | 157.8 | |
| Bovine type IV collagen | 89.3 | 97.0 | |
| Gelatin | 151.0 | 181.5 | |
| Casein | 0.68b | 0 | |
| Pz-PLGPRc | 24.8 | 24.6 | |
| Su-AAAd | 2.94b | 0 | |
The collagenase from C. histolyticum purchased from Worthington Biochemical Corp.
Data from our previous work [7].
4-Phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-Arg.
N-Succinyl-Ala-Ala-Ala-p-nitroanilide.
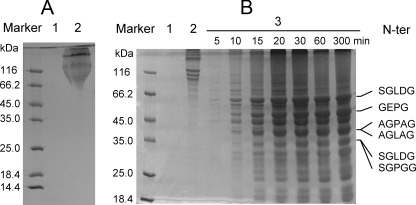
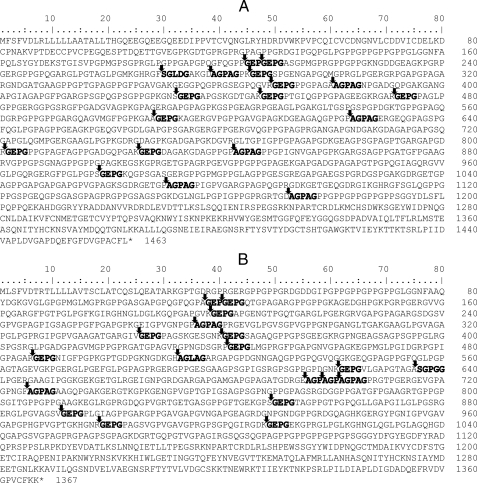
Digestion Pattern of Insoluble Collagen Fiber by MCP-01—The digestion pattern of fish and bovine-insoluble collagen fiber by MCP-01 was analyzed by SDS-PAGE. After a 5-h digestion by MCP-01 at 60 °C, all fish-insoluble collagen fiber in the tube disappeared, whereas those in the control tubes without MCP-01 had only a little decrease. No peptide could be detected from the digestion solution of fish collagen by 12.5% SDS-PAGE (Fig. 1A), showing that MCP-01 decomposed fish-insoluble collagen fiber completely into amino acids and small peptides lower than 10 kDa. As for bovine type I collagen fiber, after a 5-h digestion, almost all the collagen fiber in the tube with MCP-01 disappeared, whereas the fiber in the control tube without MCP-01 had no visible change. SDS-PAGE analysis showed that a large amount of soluble peptides with molecular mass lower than 100 kDa were released from bovine collagen digestion by MCP-01 (Fig. 1B). These results further showed that MCP-01, as a marine collagenolytic protease, preferably decomposed fish collagen more than bovine collagen. Because the sequences of chain α1 and chain α2 of bovine type I collagen were clear, we tried to determine the cleavage sites of MCP-01 on them. The peptides released from bovine collagen were separated in SDS-PAGE and submitted to N-terminal sequence analysis. Finally, the N-terminal sequences of 6 released peptides in the peptide bands in SDS-PAGE were determined (Fig. 1B). A time course analysis of the digestion pattern of bovine type I collagen fiber by MCP-01 showed that the peptide with the N-terminal sequence of SGLDG and molecular mass of about 58 kDa was preferentially derived during the digestion (Fig. 1B). According to the determined N-terminal sequences, 37 possible cleavage sites of MCP-01 on type I collagen chain α1 and chain α2 were deduced (Fig. 2), suggesting that MCP-01 had various but specific cleavage sites on type I collagen fiber. This digestion pattern was similar to that of other bacterial collagenolytic enzymes (28), but different from that of the mammalian collagenases. Mammalian collagenases only had one specific cleavage site on collagens to produce three-quarter and one-quarter fragments (29).
FIGURE 1.
SDS-PAGE analysis of the digestion patterns of deseasin MCP-01 on fish- and bovine-insoluble collagens and the N-terminal sequences of some peptides released from bovine-insoluble type I collagens. A, digestion result of fish insoluble collagen fiber from Pseudosciaena polyactis by MCP-01. Lane 1, the result of 5 mg of fish-insoluble collagen fiber incubated with 18 μg of MCP-01 at 60 °C for 5 h; lane 2, control, the result of fish-insoluble collagen fiber incubated in 50 mm Tris-HCl without MCP-01 at 60 °C for 5 h. B, digestion result of bovine-insoluble type I collagen fiber by MCP-01. Lane 1, MCP-01 (18 μg) incubated in 50 mm Tris-HCl without collagen at 60 °C for 5 h; lane 2, 5 mg of bovine-insoluble type I collagen fiber incubated in 50 mm Tris-HCl without MCP-01 at 60 °C for 5 h; lanes 3–9, the digestion results of 5 mg of bovine insoluble type I collagen fiber incubated with 18 μg of MCP-01 at 60 °C for 5, 10, 15, 20, 30, 60, and 300 min, respectively. The N-terminal sequences of 6 released peptides are indicated on the right side. They were determined by Edman degradation using PROCISE491.
FIGURE 2.
The possible cleavage sites of deseasin MCP-01 on bovine collagen chain α1 (A) and chain α2 (B) deduced according to the N-terminal sequences of 6 released peptides from bovine-insoluble type I collagen by MCP-01. The cleavage sites are indicated by arrows. The N-terminal sequences determined are shown in bold letters.
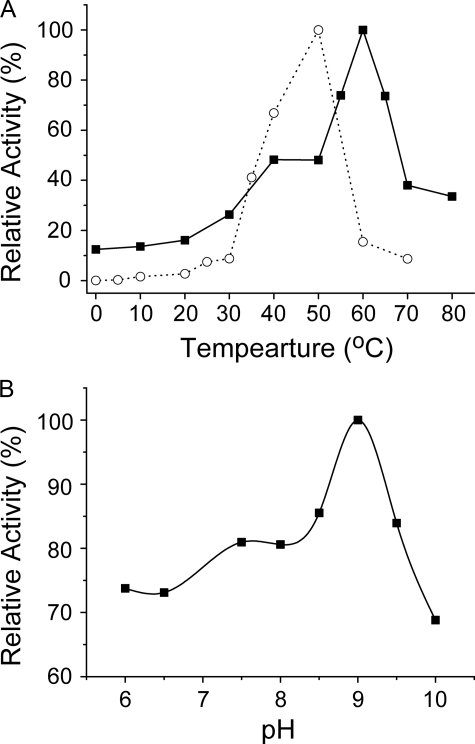
Effects of Temperature and pH on the Collagenolytic Activity of MCP-01—With bovine-insoluble type I collagen fiber as substrate, MCP-01 was most active at 60 °C, and kept 12.4% of the highest activity at 0 °C. In contrast, the collagenase from C. histolyticum was most active at 50 °C, and almost no activity at 0 °C could be detected (Fig. 3A). This showed that, as a cold-adapted enzyme from a deep-sea psychrotolerant bacterium, MCP-01 has high activity at low temperature to be adapted to the cold environment. Cold-adapted enzymes usually have an optimum temperature lower than 40 °C because of their thermolability. Cold-adapted MCP-01 had an unusual high optimum temperature of 60 °C in the digestion of insoluble type I collagen fiber. Because type I collagen can be converted to gelatin at 60 °C and MCP-01 had high gelatinolytic activity (Table 1), most of the activity of MCP-01 at 60 °C might result from its gelatinolytic activity.
FIGURE 3.
Effect of temperature (A) and pH (B) on the collagenolytic activity of deseasin MCP-01. A, deseasin MCP-01 (▪) and the collagenase from C. histolyticum (○) was incubated with 5 mg of bovine-insoluble type I collagen fiber in 50 mm Tris-HCl (0.36 mm CaCl2 added, pH 7.5) at various temperatures for 5 h, respectively. Collagenase activity was determined with the method described under “Experimental Procedures.” The enzyme activity of deseasin MCP-01 at 60 °C and that of the collagenase from C. histolyticum at 50 °C were taken as 100%. B, the collagenolytic activities of deseasin MCP-01 in 50 mm Tris-HCl from pH 6.0 to 10.0 were determined as described under “Experimental Procedures.” The highest activity was taken as 100%. The data are the mean of three experimental repeats with S.D. ≤ 5%.
With bovine-insoluble type I collagen fiber as substrate in Tris-HCl buffer, deseasin MCP-01 had an optimum at pH 9.0, and over 60% activity remained between pH 6.0 and 10.0 (Fig. 3B). Our experimental results showed MCP-01 had no collagenolytic activity in acetate, phosphate, carbonate, and a broad pH buffer containing sodium citrate. Therefore, the activity of MCP-01 to type I collagen fiber below pH 6.0 and over pH 10.0 could not be carried out.
Effects of Metal Ions and Protease Inhibitors on the Collagenolytic Activity of MCP-01—Effects of 13 metal ions on the activity of deseasin MCP-01 to insoluble type I collagen fiber were measured. Among these metal ions, only Ca2+ (4 mm) markedly increased the enzyme activity by 174.1%. Zn2+, Ni2+, Cu2+, and Fe2+ severely inhibited the enzyme activity by more than 50%. Co2+, Mg2+, Ba2+, Sn2+, and Mn2+ moderately inhibited the enzyme activity by 38.7, 20.1, 17.4, 11.1, and 10.7% at 4 mm, respectively. Sr2+ and Li+ had a slight inhibitory effect and K+ had no effect on the enzyme activity. As a serine protease, the activity of MCP-01 to type I collagen fiber was conspicuously inhibited by the serine protease inhibitor phenylmethylsulfonyl fluoride (96.1% inhibition, 5 mm). In addition, It was also inhibited by the divalent cation chelators EDTA (92.1%, 1 mm) and EGTA (73.6%, 1 mm), o-phenanthroline (36.6%, 5 mm), and sodium citrate (71.2%, 5 mm) probably because the Ca2+ in the catalytic domain of MCP-01 was deprived by these chelators (Table 2). These results showed that some metal ions and inhibitors had different and significant effects on the collagenolytic activity of MCP-01, which should be considered in the study and application of MCP-01.
TABLE 2.
Effects of metal ions and protease inhibitors on the collagenolytic activity of deseasin MCP-01 to bovine-insoluble type I collagen fiber
Note, the enzyme activity of MCP-01 in buffer A without any metal ion or inhibitor was taken as control (100%). The data represent the mean of three experimental repeats with S.D. ≤ 5%.
|
Metal ion
|
Relative activity (%)
|
Metal ion
|
Relative activity (%)
|
Inhibitors
|
Residual activity (%)
|
|||
|---|---|---|---|---|---|---|---|---|
| 2 mm | 4 mm | 2 mm | 4 mm | 1 mm | 5 mm | |||
| Control | 100 | 100 | Mn2+ | 85.7 | 89.3 | Control | 100 | 100 |
| Li+ | 86.2 | 94.5 | Fe2+ | 51.4 | 44.6 | PMSFa | 9.0 | 3.9 |
| K+ | 100.4 | 100.2 | Co2+ | 71.8 | 61.3 | EDTA | 7.9 | 11.6 |
| Mg2+ | 79.4 | 79.9 | Ni2+ | 40.8 | 31.2 | EGTA | 26.4 | 58.9 |
| Ca2+ | 249.4 | 274.1 | Zn2+ | 31.2 | 24.1 | SC | 43.6 | 28.8 |
| Sr2+ | 92.2 | 98.7 | Cu2+ | 45.2 | 37.4 | o-P | 69.3 | 63.4 |
| Ba2+ | 90.3 | 82.6 | Sn2+ | 99.1 | 88.9 | |||
PMSF, phenylmethylsulfonyl fluoride; SC, sodium citrate; o-P, o-phenanthroline.
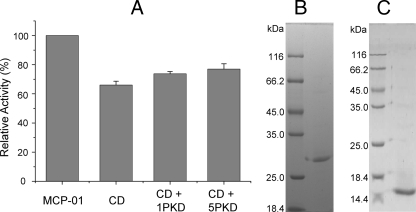
Effects of the PKD Domain on the Collagenolytic Efficiency of the Catalytic Domain of MCP-01—Mature MCP-01 is composed of a CD, a linker, a P-proprotein domain, and a PKD domain (7). The CD and PKD of MCP-01 were recombined, expressed, and purified, respectively (Fig. 4). The recombined CD was able to digest insoluble collagen, but its collagenolytic efficiency was 36.0% lower than that of wild MCP-01 (Fig. 4). To evaluate the function of the PKD domain in the insoluble collagen fiber digestion by MCP-01, the effect of the PKD domain on the collagenolytic efficiency of CD was measured. After the insoluble collagen fiber was incubated with 1 or 5 molar eq of the recombined PKD domain for 5 h, 1 molar eq of the recombined CD was added and incubated for 5 h to digest the collagen fiber. The collagenolytic efficiency of CD could be enhanced by 20.9 and 33.2% by 1 and 5 molar eq of the recombined PKD domain (Fig. 4). Therefore, the PKD domain of MCP-01 may function on the collagen fiber to facilitate the insoluble collagen fiber digestion by MCP-01.
FIGURE 4.
A, effect of exogenous PKD domain on the digestion of bovine-insoluble type I collagen fiber by the CD of MCP-01. B, the purified recombinant CD. C, the purified recombinant PKD. Insoluble type I collagen fiber (5 mg) was first incubated in the absence and presence of PKD (1 or 5 molar eq) in 50 mm Tris-HCl (with 0.36 mm CaCl2, pH 7.5) for 5 h at 40 °C, and then equivalents (200 pmol) of MCP-01 (first column) and CD (second column) were added, respectively, and incubated for another 5 h for activity assay. Enzyme activity was determined as mentioned under “Experimental Procedures.” First column, the activity of MCP-01 without PKD treatment; second column, the activity of CD without PKD treatment; third column, the activity of CD with 1 molar eq of PKD treatment; fourth column, the activity of CD with 5 molar eq of PKD treatment.
Collagen-binding Properties of the PKD Domain of MCP-01
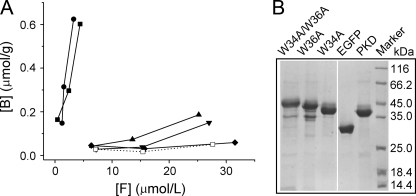
Collagen Binding Analysis of the PKD Domain of MCP-01—The PKD domain were expressed in E. coli as an EGFP fusion protein (PKD-EGFP) and purified with His·Bind metal chelating column (Fig. 5). Then its binding ability to the insoluble collagen fiber was assayed by a spectrofluorometer with EGFP as control. As shown in Fig. 5, PKD-EGFP displayed a significant binding ability to the insoluble collagen fiber, whereas EGFP had no binding ability, showing that the PKD domain of MCP-01 has binding ability to the insoluble collagen fiber. Therefore, the PKD domain of MCP-01 functions as a binding domain in the insoluble collagen fiber digestion by MCP-01. Besides, the PKD domain of MCP-01 had no binding ability to insoluble elastin, which may be another important component of deep-sea sedimentary particulate organic nitrogen (PON) (Fig. 5).
FIGURE 5.
A, binding ability of PKD domain and its mutants to bovine-insoluble type I collagen fiber and elastin. B, purified EGFP and fusion proteins of PKD and its three mutants. Various concentrations of EGFP, and fusion proteins of PKD and its mutants were incubated with a fixed amount of collagen (or elastin) in double distilled water at 40 °C for 1 h with mixing. The free fluorescence intensity in the solution before and after incubation was determined with spectrofluorometer. EGFP was used as the control. The figure shows the equilibrium adsorption isotherms (B versus F) by measuring the bounded protein amount (B) and the unbounded protein amount (F). The data represent the mean of three experimental repeats with S.D. ≤ 5%. ▪, PKD incubated with collagen; •, W34A incubated with collagen; ▴, W36A incubated with collagen; ▾, W34A/W36A incubated with collagen; ♦, EGFP incubated with collagen; □, PKD incubated with elastin.
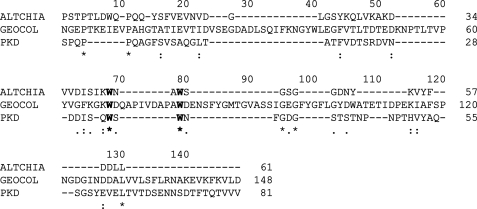
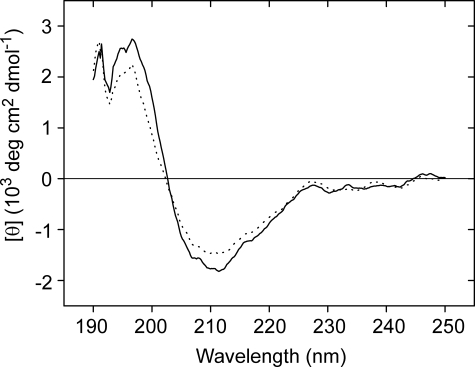
Determination of the Key Residue in PKD Domain for Collagen Binding—To clarify the important amino acid residues in PKD domain for collagen binding, side-directed mutagenesis was introduced. The PKD domain sequences of MCP-01, a chitinase from Alteromonas sp. O-7 (10), and a collagen-binding region of a collagenolytic protease from Geobacillus collagenovorans MO-1 (25), were aligned to determine the residues to be mutated. The alignment revealed 7 completely conserved sites (Fig. 6). Because aromatic residues usually play key roles in the binding of the binding domain to insoluble substrate (26, 27), the two aromatic residues, Trp-34 and Trp-36, among the conserved residues were chosen to be mutated. Two single mutants, W34A and W36A, and one double mutant, W34A/W36A, were constructed and expressed as EGFP fusion proteins (Fig. 5). Then, the binding ability of the three mutants was measured and compared with that of the PKD domain (Fig. 5). Mutant W34A almost had the same binding ability to the insoluble collagen fiber as the PKD domain, showing that Trp at position 34 plays little role in the binding. The binding abilities of W36A and W34A/W36A to the insoluble collagen fiber reduced 83.6 and 97.6%, respectively, compared with that of the PKD domain, indicating that mutation of Trp-36 to Ala-36 caused a significant reduction of the collagen binding ability of the PKD domain. A comparison of the CD spectra of the PKD domain and W36A indicated that mutation of Trp-36 to Ala-36 caused little structural changes in the PKD domain (Fig. 7). So the low collagen binding ability of W36A mainly resulted from the mutation of Trp to Ala rather than the structural change. These results showed that the Trp residue at position 36 played a key role in the binding of the PKD domain to the insoluble collagen fiber.
FIGURE 6.
Alignment of the amino acid sequence of the PKD domain of deseasin MCP-01 with those from the chinase ChiA and the protease from G.collagenovorans MO-1. ALTCHIA, the PKD domain of chitinase ChiA from Alteromonas sp. strain O-7 (AB063629); GEOCOL, the intervening region of the collagenolytic protease from G. collagenovorans MO-1 (AB260948); PKD, the PKD domain of deseasin MCP-01 from Pseudoalteromonas sp. SM9913. Identical residues are indicated by asterisks. Conserved aromatic residues are indicated by bold letters. Sequences were aligned using Clustal X 1.83 (21).
FIGURE 7.
CD spectra of PKD domain and its W36A mutant. The spectra were measured on a Jasco J-810 spectropolarimeter (Japan) with the method described under “Experimental Procedures.” The straight line represents PKD and the dotted line represents its W36A mutant.
DISCUSSION
Deseasin MCP-01 is the main extracellular protease of the deep-sea bacterium Pseudoalteromonas sp. SM9913 (15). It has been proven to be a cold-adapted multidomain subtilase with high autolytic activity, whose enzymatic characters have been studied with casein as substrate (7, 15–17). In this article, it was proven that deseasin MCP-01 is a collagenolytic serine protease, capable of digesting various collagens from both terrestrial and marine animals. Moreover, as a marine collagenolytic protease, it is more prone to degrade marine collagen than terrestrial collagen. Collagens are the major protein constituents of the extracellular matrix and the most abundant proteins in all higher organisms including marine animals. Therefore, it is an important fraction of marine organic nitrogen, and its degradation is important for marine nitrogen recycling. The broad specificity of MCP-01 to collagens suggests that MCP-01 would have a high ability to degrade various collagens in deep-sea sediments and therefore play an important role in the recycling of marine nitrogen.
Of all the collagenolytic proteases, mammalian collagenase that belong to matrix metalloproteinases have been investigated in detail. Mammalian collagenases have the same modular structure with an N-terminal domain, a linker peptide, and a C-terminal domain. The N-terminal domain is a catalytic domain belonging to M10, and the C-terminal domain is a collagen-binding domain with a unique four-bladed β-propeller structure (30). Although it is a serine protease, deseasin MCP-01 has the same modular structure as mammalian collagenases, which contains an N-terminal catalytic domain belonging to Ser-8, a linker peptide and a C-terminal PKD domain (7). The PKD domain has similar structure and function as the C-terminal domain of mammalian collagenases. It has a β-sandwich fold with 5 parallel β-sheets (7, 31) and has collagen-binding ability proven in this article. Mammalian collagenases hydrolyze various collagens with significant quantitative differences in activity and specificity. They cleave collagens at a single site to produce characteristic three-quarter and one-quarter fragments (29, 30). Deseasin MCP-01 had broad specificity to various collagens. However, unlike mammalian collagenases, deseasin MCP-01 had various but specific cleavage sites on type I collagen to produce various fragments as shown in Fig. 1B.
Among collagenolytic proteases from bacteria, metalloproteases are the most frequently occurring, whereas the number of serine proteases is rather minor (6). Among collagenolytic serine proteases, the thermostable protease of G. collagenovorans MO-1 isolated from soil is studied in most detail. It is a 210-kDa protease consisting of two identical subunits (25, 28). Deseasin MCP-01 is a cold-adapted protease of 65.84 kDa from the deep-sea sedimentary bacterium Pseudoalteromonas sp. SM9913 (7). The identity of their sequences is only 22.7%. Despite these differences, these two proteases have some similar characteristics on their collagenolysis. Similar to the protease from G. collagenovorans MO-1, MCP-01 exhibited broad substrate specificity to various type collagens. Moreover, MCP-01 had various but specific cleavage sites on insoluble collagen, and thus insoluble collagen fiber was digested into dissolved peptides smaller than 100 kDa. According to the N-terminal sequences of some released peptides, 37 possible cleavage sites of MCP-01 on bovine collagen chains were deduced. Unlike the thermostable protease from G. collagenovorans MO-1, MCP-01 is a cold-adapted enzyme and remained as 12.4% collagenolytic activity at 0 °C. In addition, its collagenolytic activity exhibited an alkaline profile. These reflect the adaptation of the collagenolytic activity of MCP-01 to the cold and alkaline deep-sea environment. MCP-01 had a high optimum temperature of 60 °C, which may result from its high gelatinolytic activity.
It is well known that many enzymes responsible for digestion of insoluble polymers, such as xylanases, cellulases, chitinases, and collagenases, contain substrate-binding domains (6, 10, 32, 33). Collagenolytic metalloproteases from bacteria have been reported to have collagen-binding domains (6). Among collagenolytic serine proteases, only the protease from G. collagenovorans MO-1 has been experimentally proven to have a collagen-binding region (25). MCP-01 and other deseasins all have a C-terminal PKD domain (7). PKD domain widely lies in chitinases (8–10), cellulases (11), collagenases (13), and proteases (12). The PKD domain of chitinase ChiA from Alteromonas sp. O-7 has been proven to have chitin-binding ability (10). However, the function of the PKD domain in other enzymes has not been reported. In this article, the PKD domain of deseasin MCP-01 was proven to function as a binding domain to bind insoluble collagen fiber during its digestion. Not only did it bind insoluble collagen fiber, but it also functioned on collagen fiber to facilitate fiber digestion by MCP-01, a mechanism that needs to be investigated further. Aromatic residues usually play key roles in the binding of binding domains to insoluble substrates (26, 27). Site-directed mutagenesis proved that Trp-36 plays a key role in the binding of the PKD domain of MCP-01 to collagen fiber. To our knowledge, PKD domain was experimentally proven to bind a protein for the first time. Genomic sequence analysis of marine G. forsetii revealed that 14 susCD-like operons frequently encode exported proteins with PKD domains (14), which suggests that PKD domains probably widely lie in the hydrolases of marine heterotrophic bacteria. The result in this article and that in the report by Orikoshi (10) demonstrate that the PKD domains in marine hydrolases function as a binding domain binding not only insoluble organic carbon such as chitin but also insoluble organic nitrogen such as collagen. Therefore, they are important for the digestion of marine insoluble biopolymers.
There is yet no gene or structure of a marine collagenolytic protease elucidated, and consequentially the digestion mechanism of marine animal collagens by collagenases is unclear. This article demonstrated that the marine collagenolytic protease MCP-01 has similar structural architecture to those from the land, having a collagen-binding domain, and a similar mechanism for collagen digestion, binding insoluble collagens by its binding domain, and then degrading them into soluble peptides and amino acids.
The total input of PON from seawater to the deep-sea sediment is 36 ± 21 μmol m–2 d–1 (34). Considering the vast area of deep-sea floor and such a huge input of PON, the recycling of PON in deep-sea sediment would be a non-negligible part of global nitrogen cycling. However, it is still unclear how these PON were degraded because the identification of the microorganisms and the enzymes participating in this process are scarce (34, 35). Because MCP-01 has specific activity to insoluble protein, MCP-01 would play an important role in the degradation of deep-sea sedimentary PON. Besides MCP-01, most deseasins are from bacteria in deep-sea or lake sediment. Therefore, deseasins and their original bacteria may play an important role in the decomposition of sedimentary PON, which still needs further confirmation by experiments on other deseasins.
The work was supported by Hi-Tech Research and Development Program of China Grants 2006AA09Z414 and 2007AA091903, National Natural Science Foundation of China Grant 30770040, Program for New Century Excellent Talents in University Grant NCET-06-0578, Foundation for Young Excellent Scientists in Shandong Province Grant 2006BS02002, and COMRA Program Grant DYXM-115-02-2-6. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKD domain, polycystic kidney disease domain; PON, particulate organic nitrogen; Pz-peptide, 4-phenylazobenzyloixycarbonyl-Pro-Leu-Gly-Pro-o-Arg; Su-AAA, N-succinyl-Ala-Ala-Ala-p-nitroanilide; CD, catalytic domain; EGFP, enhanced green fluorescent protein.
References
- 1.Merkel, J. R., Dreisbach, J. H., and Ziegler, H. B. (1975) Appl. Microbiol. 29 145–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreisbacht, J. H., and Merkel, J. R. (1978) J. Bacteriol. 135 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merkel, J. R., and Dreisbach, J. H. (1978) Biochemistry 17 2857–2863 [DOI] [PubMed] [Google Scholar]
- 4.Waite, J. H., Tamer, M. L., and Merkel, J. R. (1980) J. Biol. Chem. 255 3596–3599 [PubMed] [Google Scholar]
- 5.Kurata, A., Miyazaki, M., Kobayashi, T., Nogi, Y., and Horikoshi, K. (2007) Int. J. Syst. Evol. Microbiol. 57 1549–1553 [DOI] [PubMed] [Google Scholar]
- 6.Watanabe, K. (2004) Appl. Microbiol. Biotechnol. 63 520–526 [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. L., Xie, B. B., Lu, J. T., He, H. L., and Zhang, Y. Z. (2007) Microbiol-SGM 153 2116–2125 [DOI] [PubMed] [Google Scholar]
- 8.Perrakis, A., Tews, I., Dauter, Z., Oppenheim, A. B., Chet, I., Wilson, K. S., and Vorgias, C. E. (1994) Structure 2 1169–1180 [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto, K., Nukui, E., Itoh, H., Sato, T., Kobayashi, T., Imada, C., Watanabe, E., Inamori, Y., and Tsujibo, H. (2002) J. Bacteriol. 184 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orikoshi, H., Nakayama, S., Hanato, C., Miyamoto, K., and Tsujibo, H. (2005) J. Appl. Microbiol. 99 551–557 [DOI] [PubMed] [Google Scholar]
- 11.Ahsan, M. M., Kimura, T., Karita, S., Sakka, K., and Ohmiya, K. (1996) J. Bacteriol. 178 5732–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda, K., Ito, M., Uchida, K., Shibano, Y., Fukuhara, K., and Takahashi, S. (1996) J. Biochem. (Tokyo) 120 564–572 [DOI] [PubMed] [Google Scholar]
- 13.Matsushita, O., Jung, C. M., Katayama, S., Minami, J., Takahashi, Y., and Okabe, A. (1999) J. Bacteriol. 181 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer, M., Kube, M., Teeling, H., Richter, M., Lombardot, T., Allers, E., Würdemann, C. A., Quast, C., Kuhl, H., Knaust, F., Woebken, D., Bischof, K., Mussmann, M., Choudhuri, J. V., Meyer, F., Reinhardt, R., Amann, R. I., and Glöckner, F. O. (2006) Environ. Microbiol. 8 2201–2213 [DOI] [PubMed] [Google Scholar]
- 15.Chen, X. L., Zhang, Y. Z., Gao, P. J., and Luan, X. W. (2003) Mar. Biol. 143 989–993 [Google Scholar]
- 16.Chen, X. L., Sun, C. Y., Zhang, Y. Z., and Gao, P. J. (2003) Biotech. Lett. 25 1763–1767 [DOI] [PubMed] [Google Scholar]
- 17.Chen, X. L., Zhang, Y. Z., Lu, J. T., Xie, B. B., and Sun, C. Y. (2007) Biochem. Biophys. Res. Commun. 358 704–709 [DOI] [PubMed] [Google Scholar]
- 18.Song, E., Kim, S. Y., Chun, T., Byun, H.-J., and Lee, Y. M. (2006) Biomaterials 27 2951–2961 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 20.An, Y. F., and Ji, J. F. (2005) Appl. Microbiol. Biotechnol. 68 774–778 [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265–275 [PubMed] [Google Scholar]
- 23.Worthington Biochemical Corp. (1972) Worthington Enzyme Manual, pp. 43–45, Freehold, NJ
- 24.Miyake, R., Shigeri, Y., Tatsu, Y., Yumoto, N., Umekawa, M., Tsujimoto, Y., Matsui, H., and Watanabe, K. (2005) J. Bacteriol. 187 4140–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoi, Y., Horinaka, M., Tsujimoto, Y., Matsui, H., and Watanabe, K. (2006) J. Bacteriol. 188 6572–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, T., Simpson, P., Williamson, M. P., Hazlewood, G. P., Gilbert, H. J., and Orosz, L. (1998) FEBS Lett. 429 312–316 [DOI] [PubMed] [Google Scholar]
- 27.Ferrandon, S., Sterzenbach, T., Mersha, F. B., and Xu, M. Q. (2003) Biochim. Biophys. Acta 1621 31–40 [DOI] [PubMed] [Google Scholar]
- 28.Okamoto, M., Yonejima, Y., Tsujimoto, Y., Suzuki, Y., and Watanabe, K. (2001) Appl. Microbiol. Biotechnol. 57 103–108 [DOI] [PubMed] [Google Scholar]
- 29.Cawston, T. E. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawling, N. D., and Woessner, J. F., eds) 2nd Ed., pp. 472–480, Elsevier, London
- 30.Henriet, P., and Eeckhout, Y. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawling, N. D., and Woessner, J. F., eds) 2 Ed., pp. 486–494, Elsevier, London
- 31.Bycroft, M., Bateman, A., Clarke, J., Hamill, S. J., Sandford, R., Thomas, R. L., and Chothia, C. (1999) EMBO J. 18 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomme, P., Warren, R. A. J., Miller, R. C., Jr., Kilburn, D. G., and Gilkes, N. R. (1995) in Enzymatic Degradation of Insoluble Polysaccharides (Saddler, J. N., and Penner, M. H., eds) Vol. 618, pp. 142–161, American Chemical Society, Washington, DC [Google Scholar]
- 33.Tam, E. M., Wu, Y. I., Butler, G. S., Stack, M. S., and Overall, C. M. (2002) J. Biol. Chem. 277 39005–39014 [DOI] [PubMed] [Google Scholar]
- 34.Brunnrgard, J., Grandel, S., Stahl, H., Tengberg, A., and Hall, P. O. J. (2004) Prog. Oceanogr. 63 159–181 [Google Scholar]
- 35.Jorgensen, B. B., and Boetius, A. (2007) Nature 5 770–781 [DOI] [PubMed] [Google Scholar]