Abstract
ATP-DnaA is the initiator of chromosomal replication in Escherichia coli, and the activity of DnaA is regulated by the regulatory inactivation of the DnaA (RIDA) system. In this system, the Hda protein promotes DnaA-ATP hydrolysis to produce inactive ADP-DnaA in a mechanism that is mediated by the DNA-loaded form of the replicase sliding clamp. In this study, we first revealed that hda translation uses an unusual initiation codon, CUG, located downstream of the annotated initiation codon. The CUG initiation codon could be used for restricting the Hda level, as this initiation codon has a low translation efficiency, and the cellular Hda level is only ∼100 molecules per cell. Hda translated using the correct reading frame was purified and found to have a high RIDA activity in vitro. Moreover, we found that Hda has a high affinity for ADP but not for other nucleotides, including ATP. ADP-Hda was active in the RIDA system in vitro and stable in a monomeric state, whereas apo-Hda formed inactive homomultimers. Both ADP-Hda and apo-Hda could form complexes with the DNA-loaded clamp; however, only ADP-Hda-DNA-clamp complexes were highly functional in the following interaction with DnaA. Formation of ADP-Hda was also observed in vivo, and mutant analysis suggested that ADP binding is crucial for cellular Hda activity. Thus, we propose that ADP is a crucial Hda ligand that promotes the activated conformation of the protein. ADP-dependent monomerization might enable the arginine finger of the Hda AAA+ domain to be accessible to ATP bound to the DnaA AAA+ domain.
The initiation of chromosomal replication is strictly regulated during the cell cycle. In Escherichia coli, a crucial target for this regulation is the formation of an active initiation complex, including the ATP-bound DnaA protein (ATP-DnaA) and the chromosomal replication origin, oriC (1, 2). The DiaA protein directly stimulates formation of this complex, which leads to the unwinding of duplex DNA within the oriC (3, 4). DnaB helicase then expands the unwound region to allow the loading of DnaG primase and DNA polymerase (pol)5 III holoenzyme (5). The pol III holoenzyme consists of the pol III core, clamp (β subunit), and γ complex (clamp loader). The clamp forms a ring-shaped structure as a homodimer and is loaded onto the primed DNA by the γ complex to tether the pol III core onto DNA during DNA synthesis. After synthesis of the Okazaki fragments, the pol III core dissociates, and the sliding clamp remains on the DNA (5).
There are at least three systems that repress extra initiations (2, 6, 7). First, binding of SeqA to hemimethylated oriC temporarily inhibits initiation complex formation (8–10). The minimal oriC region contains 11 GATC sequence repeats that are available for modification by the DNA-adenine methyltransferase. Nascent strand synthesis transiently generates a hemimethylated oriC that is the target of SeqA. In the second system, there is a reduction of DnaA molecules that are accessible to the oriC. The datA locus is located near the oriC and can bind to a considerable number of DnaA molecules (11). Titration of DnaA molecules onto datA locus is enhanced by duplication of the locus following initiation.
The third system involves the regulatory inactivation of DnaA (RIDA) and promotes the hydrolysis of DnaA-bound ATP, which yields inactive ADP-DnaA (12, 13). RIDA depends on the Hda protein and the DNA-loaded form of the clamp. The requirement of the DNA-loaded clamp can ensure timely inactivation of DnaA during the replication cycle of the chromosome. The cellular ATP-DnaA level increases prior to the initiation of replication and decreases depending on chromosomal replication (12, 14). Hda-deficient cells or cells bearing a RIDA-resistant mutant of DnaA exhibit accumulation of ATP-DnaA levels and cause extra initiation (12, 15–17).
The clamp-mediated coupling of replication and initiation regulation appears to be widely distributed among prokaryotes and eukaryotes. Bacillus subtilis YabA acts as a negative regulator of initiation by forming a complex with DnaA and the clamp; however, this protein shares no obvious homology with the primary sequence of Hda (18–20). The eukaryotic sliding clamp, proliferating cell nuclear antigen, interacts with the replication licensing factor Cdt1 (21–23). This interaction promotes ubiquitin-mediated proteolysis of Cdt1, which is associated with the blockage of re-replication.
In a reconstituted RIDA assay, Hda catalytically promotes DnaA-ATP hydrolysis in the presence of the DNA-loaded clamp (24, 25). This reaction requires the clamp binding to Hda via its short N-terminal region that carries the QL(S/D)LF motif conserved among clamp-binding proteins (24–27). Besides the N-terminal region, Hda carries an AAA+ domain that is homologous with the DnaA AAA+ domain (12). This domain bears Walker-type nucleotide-binding motifs and several specific motifs that are conserved in the AAA+ family (28). A common feature of typical AAA+ proteins is the formation of multimers in which ATP binding and hydrolysis events occur at the inter-subunit interface and mediate a conformational change. In this process, a conserved arginine residue called an arginine finger plays a crucial role in the recognition of ATP bound to the adjacent protomer (29, 30). Based on our biochemical and mutational analyses of DnaA, Hda, and the DNA-loaded clamp, the following model is provided (24, 25). The Hda arginine finger directly promotes DnaA-bound ATP hydrolysis upon interaction between the AAA+ domains of DnaA and Hda. The DNA-loaded clamp mediates this interaction as a platform, on which the DnaA N-terminal domain supportively interacts with the Hda-clamp complex, and the DnaA DNA-binding domain recognizes the duplex DNA flanking the clamp.
Here we found that Hda specifically binds to ADP but not ATP in vitro and in vivo. Apo-Hda formed a homomultimer that is likely to consist of several protomers in the absence of ADP. ADP binding promoted dissociation of the Hda homomultimers to monomers and activated Hda function in the RIDA system in vitro. Further in vitro and in vivo analyses using the Walker motif mutants of Hda suggested that ADP binding is crucial for Hda function in vivo. ADP-Hda but not apo-Hda formed a functional complex with ATP-DnaA and the DNA-loaded clamp. The Hda arginine finger could be buried into the inter-subunit interface of apo-Hda homomultimers, and ADP-dependent monomerization might expose the arginine finger on the surface, promoting interaction with ATP-DnaA.
EXPERIMENTAL PROCEDURES
Construction and Expression of Hda Derivatives for the N-terminal Analyses—Plasmids pBAD/Hda and pHda4 (an intermediate pBAD/Hda construction) were constructed previously (25). pBAD/Hda-cHis was constructed by HindIII digestion and self-ligation of a PCR fragment amplified using pHda4 as a template and the primers 5′-GTCGGATGCGGAAGCTTGGC-3′ and 5′-CCCAAGCTTCAGTGATGGTGATGGTGATGCAACTTCAGAATTTCTTTCACAAACGG-3′. For overproduction of Hda, KA450 [ΔoriC1071::Tn10 dnaA107 (Am) rnhA199 (Am)] cells (15) harboring pHda4, pBAD/Hda, or pBAD/Hda-cHis were grown at 37 °C in LB medium containing 100 μg/ml ampicillin and 50 μg/ml thymine. When the absorbance (A600) of the culture reached 0.5, arabinose (0.5%) was added, and cells were incubated for an additional 2 h.
Determination of the N-terminal Sequence of Hda-cHis—Purification of Hda-cHis expressed using pBAD/Hda-cHis was performed using the same method as that previously described for a His-tagged Hda (25). Hda-cHis was separated by 15% SDS-PAGE, blotted onto a polyvinylidene difluoride membrane (Millipore), and sequenced by an Edman degradation method using a Procise 494HT protein sequencer (Applied Biosystems).
Construction and Purification of Hda-cHis—A fragment bearing the Hda-cHis coding region was amplified by PCR using pBAD/Hda-cHis as a template and the primers 5′-CGTCTAGAAGGAGATATACATATGAACACACCGGCACAGCTCTC-3′ and 5′-CTCTCATCCGCCAAAACAGCCAAG-3′, and cloned into the NdeI and HindIII sites of pET21a (Novagen), resulting in pET/Hda-cHis. BL21(DE3) cells harboring pET/Hda-cHis were grown at 37 °C in LB medium containing 100 μg/ml ampicillin until the A600 of the culture reached 0.8; isopropyl 1-thio-β-d-galactopyranoside (1 mm) was added, and cells were further incubated for 2 h. All the following procedures were performed at 0–6 °C. Isolation of a soluble fraction from cell lysates and nickel column chromatography were performed by the same method as that previously described for a His-tagged Hda (25), yielding Hda-cHis Fr II.
To isolate the multimers and monomers, Hda-cHis Fr II (55 μg) was loaded using a flow rate of 5 μl/min onto a Superdex-200 PC 3.2/30 column equilibrated with buffer A containing 20 mm Tris-HCl (pH 7.5), 10% glycerol, 8 mm dithiothreitol, 0.01% Brij-58, 0.5 mm EDTA, 5 mm magnesium acetate, and 120 mm potassium glutamate.
For dissociation of ADP, Hda-cHis Fr II (3.5 mg/ml, 220 μl) was incubated on ice for 30 min in the presence of 5 mm EDTA. Precipitates were collected by centrifugation, dissolved in buffer A containing 6 m urea, and loaded onto a Superdex-200 HR 10/30 column equilibrated with buffer A at a flow rate of 0.5 ml/min, yielding Hda-cHis Fr III.
Purification of Hda-cHis and Its Mutant Derivatives from an Insoluble Fraction—Hda-cHis was purified from an insoluble fraction of BL21(DE3) cell lysate harboring pET/Hda-cHis. The insoluble cell lysate fraction was washed, and protein pellets were obtained by the same method as that previously described for nontagged Hda (25). The pellets were dissolved in buffer A containing 6 m urea and loaded onto a Superdex-200 HR 10/30 column using the same method as that described for Hda-cHis Fr III. Plasmids for overexpressing mutant Hda-cHis were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) and pET/Hda-cHis. Purification of these derivatives was performed using an insoluble cell lysate fraction and the same method as described above.
Construction and Purification of nHis-Hda—The plasmid for overexpressing nHis-Hda was constructed by XhoI digestion and self-ligation of a PCR fragment amplified using pBAD/His-Hda as a template and the primers 5′-CCACTCTCGAGCTCGGATCC-3′ and 5′-GGCTCGAGAAACACACCGGCACAGC-3′. Expression and purification were performed by the same method as that previously described for His-tagged Hda (25), yielding nHis-Hda Fr II. nHis-Hda Fr II (28 μg) was loaded onto a Superose-12 PC 3.2/30 column equilibrated with buffer A at a flow rate of 40 μl/min, and fractions including the multimers or monomers were pooled.
Western Blot Analysis—Western blot analyses were performed as described previously (25), except that SDS-PAGE was performed until the 6.5-kDa prestained marker protein had migrated out of the gel.
ADP Binding Assay—Hda proteins were incubated for 20 min at 30 °C or on ice in buffer A (12.5 or 25 μl) containing 0.1 mg/ml bovine serum albumin and the indicated concentrations of [3H]ADP. Samples were passed through nitrocellulose membranes (Millipore HA, 0.45 μm), followed by washing and quantification of the radioactivity that was retained on the membranes (31).
Purification of a Purchased ATP Sample—ATP (Sigma) was applied onto a Mini-Q column equilibrated with buffer containing 20 mm Tris-HCl (pH 8.0), 7 m urea, and 50 mm NaCl and eluted with a linear gradient of NaCl from 50 to 400 mm. Elution of ATP and ADP was monitored at A280. The peak fraction of ATP was pooled, diluted in water, and applied onto a Mini-Q column equilibrated with 50 mm NH4HCO3. ATP was eluted with 1 m NH4HCO3 and dried under a vacuum.
Reconstituted RIDA Assay—The staged RIDA reconstituted system was used essentially as described previously (25). In the first stage, the clamp (50 pmol) and φX174 nicked circular DNA (1.75 μg, 0.5 pmol, circular) were incubated for 20 min at 30 °C in 25 μl of buffer B (20 mm Tris-HCl (pH 7.5), 10% glycerol, 8 mm dithiothreitol, 0.01% Brij-58, 8 mm magnesium acetate, and 20 mm potassium glutamate) containing 1 mm ATP and γ complex (225 ng). The resulting DNA-loaded clamp was isolated in a void volume by a spin-column method using Sephacryl S-400 HR (0.63 ml; GE Healthcare) equilibrated with buffer B. In the second stage, the isolated DNA-loaded clamp (100 molecules of the clamp per DNA circle) was incubated for 20 min at 30 °C in buffer B (25 μl) containing 120 mm potassium glutamate, 0.1 mg/ml bovine serum albumin, [α-32P]ATP-bound DnaA (0.5 pmol), and Hda in the presence or absence of ATP (2 mm) or ADP (30 or 200 μm). Nucleotides bound to DnaA were recovered on a nitrocellulose filter and analyzed using polyethyleneimine TLC and a BAS2500 Bio-imaging analyzer (Fuji Film).
In Vivo Analysis of Hda Activity in DnaA-ATP Hydrolysis—A fragment containing an hda-coding region and its upstream sequence (1.5 kb) was amplified by PCR using genome DNA and the primers 5′-CGGGATCCCGCTCTCTTTCGCAATGGG-3′ and 5′-CCGCTCGAGCCGCATCCGACAATAAACACC-3′, before being cloned into the BamHI and XhoI sites of pACYC177, resulting in pHCS4-1. The kan gene derived from pUC4K (GE Healthcare) was inserted into the BamHI site of pHCS4-1, resulting in pACYC/ntHda. pACYC/ntHdaA1B1, pACYC/ntHdaB2, or pACYC/ntHdaA1B2 was constructed using the QuikChange site-directed mutagenesis kit (Stratagene). These hda mutants bear substitutions of R56A/D102A (for A1B1), D102A/N103A (for B2), and R56A/D102A/N103A (for A1B2). MK86 [rnhA17::Tn3 ΔoriC::TcR Δhda::CmR] cells (12) harboring pACYC177, pACYC/ntHda, pACYC/ntHdaA1B1, pACYC/ntHdaB2, or pACYC/ntHdaA1B2 were grown at 37 °C in modified TG medium containing [32P]orthophosphate (0.4 mCi/ml) and kanamycin (100 μg/ml) but lacking NaCl and Na2SO4 to an A600 of 0.15 as described previously (13). DnaA was isolated by immunoprecipitation from cleared lysates, and the nucleotides bound to the recovered DnaA were separated using polyethyleneimine TLC, followed by BAS2500 analysis.
In Vivo Detection of ADP-Hda—Labeled cells and cell lysates were prepared using the method described above for the in vivo analysis of DnaA nucleotide forms. MC1061 cells harboring pBAD/nHis-Hda were grown at 37 °C in modified TG medium containing [32P]orthophosphate (0.1 mCi/ml), ampicillin (100 μg/ml), and 0.2% glycerol instead of glucose. When A600 of the culture reached 0.1, arabinose (0.5%) was added and incubation continued for 10 min. Cells in the culture (0.5 ml) were collected by a brief centrifugation, washed in 0.9% NaCl (0.5 ml), incubated on ice for 15 min in the presence of lysozyme (5 mg/ml) in buffer C (0.2 ml) containing 20 mm Tris-HCl (pH 7.5), 500 mm NaCl, 0.1% Triton X-100, 10% glycerol, 10 mm
2-mercaptoethanol, 5 mm magnesium acetate, 10 mm imidazole, 0.1 mm ATP, and 0.1 mm ADP, and frozen in liquid nitrogen. These lysates were thawed on ice, clarified by centrifugation, and incubated at 4 °C for 15 min with rotation in the presence of MagneHis nickel particles (7.5 μl, Promega). The beads were collected and washed four times in buffer C (50 μl), and the bead-bound materials were recovered in buffer C (5 μl) containing 1 m imidazole. Radiolabeled nucleotides in the recovered fraction were separated using polyethyleneimine TLC and a solvent consisting of 1 m HCOOH and 0.7 m LiCl or one consisting of 0.5 m KH2PO4, followed by BAS2500 analysis.
Spin-column Assay for Association of DnaA and Hda with the DNA-Clamp Complex—For a large scale preparation of the DNA-loaded clamp, the clamp (2 nmol) and M13mp18 nicked circular DNA (95 μg, 20 pmol, circular) were incubated for 20 min at 30 °C in buffer B (1 ml) containing 1 mm ATP and 9 μg of γ complex. The reaction was loaded onto a Superose-6 10/30 column equilibrated with buffer B at a flow rate of 0.2 ml/min, yielding the DNA-loaded clamp in the void fraction (60 molecules of the clamp per DNA circle). The isolated DNA-loaded clamp was incubated on ice for 15 min in buffer A (12.5 or 25 μl) containing Hda, followed by a brief incubation in the presence of ATP-DnaA (2 pmol or the indicated amount) and immediate spin-column isolation as follows. Buffer A (12.5 μl) was added only when the reaction was 12.5 μl, and the sample was applied onto a Sephacryl S-400 HR spin column (0.63 ml; GE Healthcare) equilibrated with buffer B, and the DNA and bound proteins were isolated into the void by a brief centrifugation (3000 × g, 3 min) at 4 °C.
Pulldown Assay for Association of DnaA and Hda with the DNA-Clamp Complex—The DNA-loaded clamp was prepared using the same method as described for the spin-column analysis. nHis-Hda (5 pmol or the indicated amount) was incubated on ice for 15 min in the presence of Co2+-conjugated magnet beads TALON (120 μg, Invitrogen) in buffer D (12.5 μl) containing 5 mm Tris-HCl (pH 7.5), 10% glycerol, 8 mm 2-mercaptoethanol, 0.01% Brij-58, 8 mm magnesium acetate, and 100 mm potassium glutamate. The Hda-bound beads were further incubated on ice for 5 or 15 min in the presence of the DNA-loaded clamp and 20 mm imidazole, followed by incubation for 10 min in the presence of ATP-DnaA, collection by magnetic force, and washing in buffer D (25 μl) containing 20 mm imidazole. The bead-bound materials were eluted in buffer (10 μl) containing 10 mm Tris (pH 8.0), 1 mm EDTA, and 1% SDS.
RESULTS
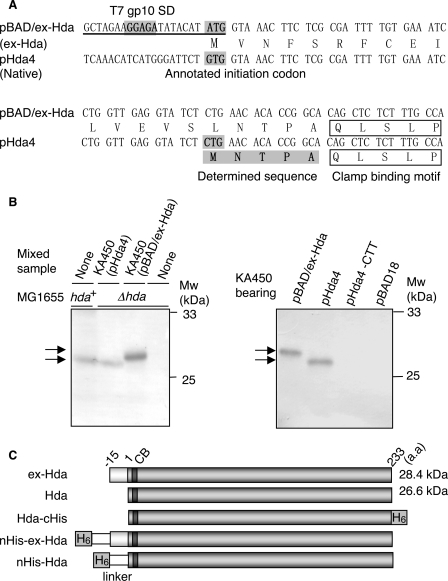
The hda Initiation Codon CUG Is Located Downstream of the Annotated GUG Initiation Codon—Previously, we constructed an Hda-overproducing plasmid (pBAD/Hda) based on the annotated coding region of the hda gene (Fig. 1A) (25). In the course of analyses, we noticed that the size of Hda expressed by pBAD/Hda (the protein and the plasmid are designated ex-Hda and pBAD/ex-Hda) was slightly higher than that of native Hda expressed from the chromosomal gene (Fig. 1B). We could not detect this slight size difference under the previous Western blot conditions; however, in this study, we extended the electrophoresis time and were able to observe small variations. pBAD/ex-Hda carried modifications in the hda upstream region to enhance expression as follows: the annotated initiation codon GUG (32) and the original Shine-Dalgarno (SD) sequence were replaced with AUG and the T7 phage gp10 SD sequence, respectively (Fig. 1A) (25). To determine whether the size difference was attributed to these modifications, we analyzed the sizes of Hda expressed by the chromosomal gene, pBAD/ex-Hda and pHda4, which bears the original chromosomal upstream region of the hda gene (Fig. 1A). The size of pHda4-derived Hda was similar to that of the chromosomal gene-derived Hda and lower than that of ex-Hda (Fig. 1B). In these experiments, Hda was overexpressed in KA450 cells [ΔoriC rnhA dnaA]. Overexpression of Hda inhibits normal cell growth, which is suppressed by deletions of oriC and rnhA (12). The rnhA mutation activates alternative origins to replicate the chromosome in a manner independent of DnaA and oriC (33). To determine the N-terminal residues of the native Hda, we constructed pBAD/Hda-cHis, a pHda4 derivative expressing a C-terminally hexahistidine-tagged form of Hda (Hda-cHis; Fig. 1C). Edman degradation using purified Hda-cHis revealed that the N-terminal amino acid sequence was MNTPA, which suggests that the initiation codon used in vivo is the CUG sequence that is located 45 nucleotides downstream from the annotated GUG initiation codon (Fig. 1A). A substitution of the CUG sequence to CUU inhibited Hda expression (Fig. 1B). Like CUU, CUG usually codes for leucine. The size of the Hda translated from the CUG sequence was calculated to be 26.6 kDa; this means that the size of the native Hda was ∼2 kDa smaller than the size of ex-Hda (Fig. 1C), which is consistent with the results of the Western blot analysis (Fig. 1B). These results support the idea that the CUG sequence uniquely functions as the initiation codon of Hda. The extra N terminus in ex-Hda is rich in hydrophobic residues (Fig. 1A), which might expedite the migration of this protein in SDS-PAGE and might result in a reduced difference in the apparent molecular sizes between ex-Hda and the native Hda.
FIGURE 1.
Identification of the Hda initiation codon. A, hda upstream DNA sequences within pBAD/ex-Hda and pHda4. pHda4 carries the chromosome-derived original upstream region. pBAD/ex-Hda carries the modified upstream region (underlined). SD sequence derived from the T7 phage gp10, the positions corresponding to the annotated GUG, and identified CUG initiation codons are shaded. The N-terminal amino acid sequence determined by Edman degradation is also shaded. The clamp-binding motif is indicated. B, Western blot analysis of Hda. KA450 [ΔoriC rnhA dnaA] cells carrying the indicated plasmids were grown, followed by induction of Hda as described under the “Experimental Procedures,” and Western blot analysis was performed (0.013 μl for pBAD/ex-Hda or 2.5μl for pHda4, pHda4-CTT, and pBAD18) (left and right panels). Left panel, MG1655 (hda+) cells and MG1655 derivative MK86 (Δhda) cells were grown at 37 °C in LB medium until the A600 was equal to 0.5; aliquots (80 μl) of the cultures were also used for the indicated lanes with or without mixing with the indicated KA450 sample. As the amount of the KA450 sample was minimal, Hda expressed from the KA450 chromosome was below the detectable level. MK86 was used as a negative control. Also the MK86 sample was mixed with the KA450 samples to normalize the total protein amount. The migration positions of the Hda proteins (arrows) and molecular weight standards (Mw) are indicated. In pHda4-CTT, the CUG initiation codon of Hda was substituted to CUU. pBAD18 was a vector used. C, structures of Hda derivatives used in this study. CB and H6 indicate the clamp-binding motif and hexahistidine tag, respectively. Hda is the same as ntHda.
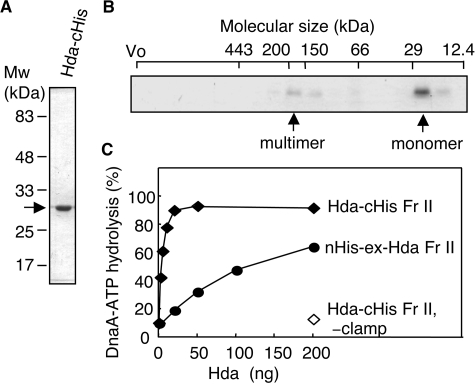
RIDA Activity of Hda-cHis—To analyze the activity of Hda-cHis in the RIDA system, we overproduced Hda-cHis using pET/Hda-cHis. Approximately 50% of the expressed Hda-cHis was recovered in a soluble cell lysate fraction and was used for purification by nickel-affinity column chromatography. The purity of the resultant fraction (Hda-cHis Fr II) was >90% (Fig. 2A). Gel filtration analysis of Hda-cHis Fr II revealed major and minor peaks with a ratio of ∼9:1 (Fig. 2B). Taking into consideration the calculated mass (27.5 kDa) of Hda-cHis, the major peak (∼25 kDa) corresponded to the size of monomers, and the minor peak (150–200 kDa) corresponded to the size of homomultimers consisting of 6–7 protomers.
FIGURE 2.
Purification and RIDA activity of Hda-cHis. A, Hda-cHis Fr II (1 μg) was analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. B, Hda-cHis Fr II (55 μg) was analyzed using Superdex-200 PC 3.2/30 gel filtration. Eluted proteins were collected in 30-μl fractions, and portions (0.5 μl) were analyzed by SDS-PAGE and silver staining. The positions of the void volume (Vo) and molecular size markers were determined under the same conditions and are indicated at the top of the gel. Arrows indicate the peak fractions of Hda-cHis. C, activity for DnaA-ATP hydrolysis was assessed using a staged RIDA reconstituted system as described under “Experimental Procedures.” [α-32P]ATP-DnaA (0.5 pmol) was incubated for 20 min at 30 °C in the presence or absence (–clamp) of the DNA-loaded clamp (20 fmol as clamp) in buffer containing 2 mm ATP and the indicated amounts of Hda-cHis Fr II or nHis-ex-Hda Fr II.
We next assessed the activity of Hda-cHis Fr II for DnaA-ATP hydrolysis in a staged RIDA-reconstituted system (25). To eliminate the possible action of Hda in a clamp-loading process, we first prepared the DNA-loaded form of the clamp. Gel filtration column chromatography was performed after a clamp-loading reaction using the clamp, the clamp loader, and nicked circular form of φX174 DNA as described previously (25). The DNA-loaded clamp was then added to a reaction containing ATP-DnaA, Hda, and 2 mm ATP. Hda-cHis Fr II efficiently promoted DnaA-ATP hydrolysis depending on the DNA-loaded clamp (Fig. 2C). The specific activity of Hda-cHis Fr II was ∼30-fold higher than that of nHis-ex-Hda Fr II (Fig. 2C). nHis-ex-Hda is an ex-Hda derivative bearing an N-terminal hexahistidine tag; this protein was used previously and was designated His-Hda (25). The extra N terminus could be detrimental to Hda activity in nHis-ex-Hda (see below). nHis-ex-Hda Fr II was purified by a similar method to Hda-cHis Fr II.
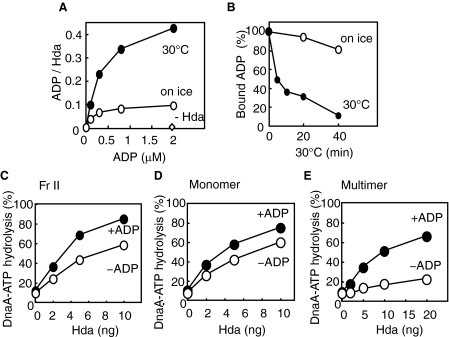
ADP Specifically Binds to Hda—Most proteins carrying AAA+ domains possess an activity for nucleotide binding. We therefore examined the nucleotide binding activity of Hda-cHis Fr II using a filter-retention assay. When a binding reaction was performed at 30 °C for 20 min, considerable binding for ADP was detected (Fig. 3A). The dissociation constant and the maximum binding stoichiometry were deduced from a Scatchard plot, resulting in ∼0.4 μm and ∼0.5 per Hda monomer, respectively. Efficient binding of ADP requires incubation at 30 °C and is considerably inhibited on ice (Fig. 3A). When [3H]ADP-bound Hda was formed and further incubated in the presence of nonradioactive ADP, dissociation of [3H]ADP was observed in a temperature (30 °C)-dependent manner (Fig. 3B). These results indicate that the association and/or dissociation of Hda from ADP requires heat, which might be related to a conformational change in Hda. To analyze the binding specificity of Hda for ADP, competition of various nucleotides was assessed by a filter-retention assay using Hda-cHis Fr II. The concentration of competitor nucleotides was 200-fold higher than that of [3H]ADP. There were no obvious competitor nucleotides with the exception of ADP itself (Table 1). Approximately 55% inhibition of ADP binding was observed in the presence of 100 μm ATP, which suggests that the affinity of Hda for ATP is ∼200-fold lower than that of ADP. We used a purified ATP sample with a contaminated ADP level of <0.5%, but we were not able to rule out the possibility that inhibition was attributed to an undetectable level of contaminated ADP. Given that the intracellular concentration of ATP is only ∼12-fold higher than that of ADP (34), the high specificity of Hda in binding is consistent with the idea that even in vivo Hda molecules would predominantly complex with ADP (see below).
FIGURE 3.
Role for ADP in Hda-cHis activity. A, ADP binding activity of Hda. [3H]ADP was incubated at 30 °C or on ice for 20 min in the presence or absence (–Hda) of Hda-cHis Fr II (1 pmol as a monomer), followed by retention on a nitrocellulose filter. The bound ADP molecules per Hda monomer are presented. B, dissociation of Hda-cHis-bound ADP. Hda-cHis Fr II (18 pmol) was incubated at 30 °C for 20 min in buffer (5 μl) containing 5 μm [3H]ADP. A portion (0.28 μl) of the reaction was diluted in buffer (25 μl) containing 1 μm nonradiolabeled ADP and was further incubated at 30 °C or on ice for the indicated time, followed by the filter-retention analysis. C–E, DnaA-ATP hydrolysis activity was analyzed in a staged RIDA reconstituted system. The DNA-loaded clamps (20 fmol as clamp) were incubated in buffer containing [α-32P]ATP-DnaA (0.5 pmol) in the presence (+ADP) or absence (–ADP) of 30 μm ADP. Hda-cHis monomers and multimers were isolated by gel filtration of Hda-cHis Fr II (see Fig. 2B). C, Hda-cHis Fr II. D, Hda-cHis monomers. E, Hda-cHis multimers.
TABLE 1.
Nucleotide specificity of Hda-cHis Fr II
Hda-cHis Fr II was incubated at 30 °C for 20 min in buffer containing [3H]ADP (0.5 μm) and competitor nucleotide (100 μm), before a filter retention assay was performed. The error range with each experiment was 0.5–6.5%.
| Competitor | Bound ADP | Competitor | Bound ADP |
|---|---|---|---|
| % control | % control | ||
| None | (100) | AMP | 84 |
| ADP | 2.0 | dATP | 88 |
| ATP | 44 | dGTP | 105 |
| GTP | 107 | dCTP | 100 |
| CTP | 106 | dTTP | 88 |
| UTP | 97 | ppGpp | 93 |
| GDP | 99 | cAMP | 103 |
| CDP | 92 | ||
| UDP | 104 |
ADP Promotes the Activity of Hda Multimers—To analyze the effect of ADP on Hda activity in DnaA-ATP hydrolysis, we performed the reconstituted RIDA assay. The isolated DNA-loaded clamps were transferred and incubated in buffer containing Hda and ATP-DnaA in the presence or absence of ADP. When Hda-cHis Fr II was used, Hda activity was stimulated ∼2-fold depending on ADP (Fig. 3C). We further examined the effect of ADP using Hda multimers and monomers that were isolated from Hda-cHis Fr II (Fig. 2B and Fig. 3, D and E). The specific activity of the monomers was similar to that of ADP-stimulated Hda-cHis Fr II (Fig. 3, C and D) and was not significantly stimulated in the presence of ADP (Fig. 3D). In contrast, the specific activity of the multimers was ∼20-fold lower than that of the monomers in the absence of ADP (Fig. 3E). Moreover, the presence of ADP stimulated the activity of the multimers by ∼10-fold. These results suggest an idea that the ADP form of Hda is monomeric and active in the DnaA-ATP hydrolysis, whereas the multimers are the ADP-free inactive form. This idea is consistent with the observation that the monomer form was predominant in Hda-cHis Fr II (Fig. 2B) and was basically active in DnaA-ATP hydrolysis. It is conceivable that the multimers contained in Fr II were activated by ADP, resulting in a 2-fold stimulation (Fig. 3C). A slight reduction in the activity of the monomers was seen in the absence of ADP, which might be due to the dissociation of ADP during the reaction (Fig. 3D). The specific activity of the multimers in the presence of ADP was slightly lower than that of the monomers (Fig. 3, D and E), which might be due to the fact that the ADP association to Hda multimers takes a certain time even at 30 °C (see below).
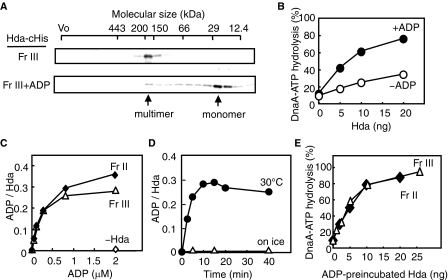
ADP Dissociates the Hda Multimers—The above results implied that the Hda multimers consisted of apo-Hda, whereas the monomers were the ADP form and active in RIDA. This presumption is consistent with the fact that ADP-Hda complexes are stable at 0 °C (Fig. 3), and also with the idea that ADP-Hda complex formed in vivo was preserved during the purification that was performed at 0–6 °C. To further clarify this presumption, we prepared apo-Hda using Hda-cHis Fr II. As the monomers were predominantly included, Hda-cHis Fr II was incubated on ice in the presence of 5 mm EDTA to dissociate any ADP that was potentially associated with the Hda monomers. During this incubation, most of the Hda molecules formed insoluble aggregates, which were dissolved in buffer including 6 m urea. To refold Hda, the dissolved protein was loaded on a gel filtration column equilibrated with buffer excluding ADP and urea. Hda was predominantly eluted in a peak fraction corresponding to ∼70 kDa (2–3-mer as Hda-cHis) and was stored at –80 °C (Hda-cHis Fr III). When we analyzed the stored Hda-cHis Fr III using gel filtration, Hda was eluted as a peak at a position corresponding to ∼200 kDa (Fig. 4A, upper panel). In the first gel filtration, the time for protein association would not be sufficient, resulting in the elution of an intermediate, unstable form of Hda oligomers. The size of the multimers (∼200 kDa) corresponds to 6–7 Hda-cHis protomers, but the multimers might consist of fewer number of Hda molecules if Hda in the multimers is partially unfolded.
FIGURE 4.
ADP dissociates Hda-cHis multimers to monomers. A, gel filtration analysis. Hda-cHis Fr III (Fr III, 0.5 μg) was analyzed using a Superdex-200 PC 3.2/30 column. Hda-cHis Fr III (0.5 μg) was incubated at 30 °C for 20 min in the presence of 100 μm ADP and analyzed in a similar manner (Fr III+ADP). Eluted proteins were collected in 30-μl fractions, followed by analysis by SDS-PAGE and silver staining. Arrows indicate the peak fractions of Hda-cHis. B, DnaA-ATP hydrolysis activity of Hda-cHis Fr III was analyzed in a staged RIDA reconstituted system in the presence (+ADP) or absence (–ADP) of 30 μm ADP, as described for Fig. 3C. C, ADP binding activities of Hda-cHis Fr II and Hda-cHis Fr III. These fractions were incubated at 30 °C for 20 min in the presence of the indicated concentration of [3H]ADP, before the filter-retention assay was performed. Bound ADP molecules per Hda monomer are presented. D, Hda-cHis Fr III (55 ng or 2 pmol as monomers) was incubated at 30 °C or on ice for the indicated time in the presence of 0.8 μm [3H]ADP, followed by the filter-retention assay. E, Hda-cHis Fr II and Hda-cHis Fr III were incubated at 30 °C for 20 min in the presence of 30 μm ADP, followed by a DnaA-ATP hydrolysis activity assay using a staged RIDA reconstituted system in the presence of the DNA-loaded clamp (20 fmol as clamp), [α-32P]ATP-DnaA (0.5 pmol), and 30 μm ADP.
Like the multimers obtained from Fr II (Fig. 3E), Hda-cHis Fr III was basically inactive and profoundly stimulated in an ADP-dependent manner (Fig. 4B). When Hda-cHis Fr III was incubated in the presence of ADP and analyzed by gel filtration, the majority of Hda was eluted as a peak in the position of the monomers (Fig. 4A, lower panel). These observations are consistent with the notion that apo-Hda was formed by treatment with EDTA, urea and refolding, resulting in Hda-cHis Fr III. Similar results in ADP-dependent DnaA-ATP hydrolysis were obtained when the multimers isolated in the second gel filtration were used (data not shown).
ADP binding and the ADP-dependent RIDA activities of Hda-cHis Fr III were further assessed in comparison with those of Fr II (Fig. 4, C–E). Hda-cHis Fr III retained ADP binding activity at a level similar to that of Fr II (Fig. 4C). Efficient binding of ADP was observed at 30 °C but took ∼10 min (Fig. 4D). These results are consistent with the data obtained using Fr II (Fig. 3, D and E). In contrast, ADP binding was severely inhibited on ice (Fig. 4D). As ADP-Hda complex was stable on ice (Fig. 3B), at the low temperature Hda would take an inactive conformation that inhibits association of ADP in addition to dissociation of ADP once bound at 30 °C. Unlike Hda-cHis Fr III, Hda-cHis Fr II showed a slight ADP binding activity even on ice (Fig. 3A), which might attributed to a predominance of Hda monomers in Fr II. The specificity for ADP was similar to that of Fr II (supplemental Table S1). When Hda-cHis Fr III was preincubated at 30 °C in the presence of ADP and was next incubated with ATP-DnaA and the DNA-loaded clamps, the activity was retained at a level similar to that of Fr II (Fig. 4E). These results demonstrate that the EDTA and urea treatments did not irreversibly affect Hda and that Hda-cHis Fr III is comparable in RIDA activity to Fr II. Taken together, these results support the notion that apo-Hda forms homomultimers that are inactive in RIDA and that ADP binding activates Hda by dissociating the homomultimers into monomers.
Extra N Terminus Is Inhibitory to ADP Binding—To investigate the detrimental effect of the extra N terminus of nHis-ex-Hda, we constructed nHis-Hda that was an nHis-ex-Hda derivative lacking the extra region (Fig. 1C). We purified nHis-Hda Fr II using a similar method to that of nHis-ex-Hda Fr II and Hda-cHis Fr II. The ADP binding activity of nHis-Hda Fr II was significantly higher than that of nHis-ex-Hda Fr II (supplemental Fig. S1A), indicating that ADP binding is inhibited by the extra region. This result was supported by the observation that nHis-Hda Fr II but not nHis-ex-Hda Fr II contained monomers and multimers (supplemental Fig. S1, B and C, upper panel). The isolated nHis-Hda multimers retained the activity for DnaA-ATP hydrolysis at a level comparable with that of Hda-cHis Fr III in the presence of ADP (supplemental Fig. S1D).
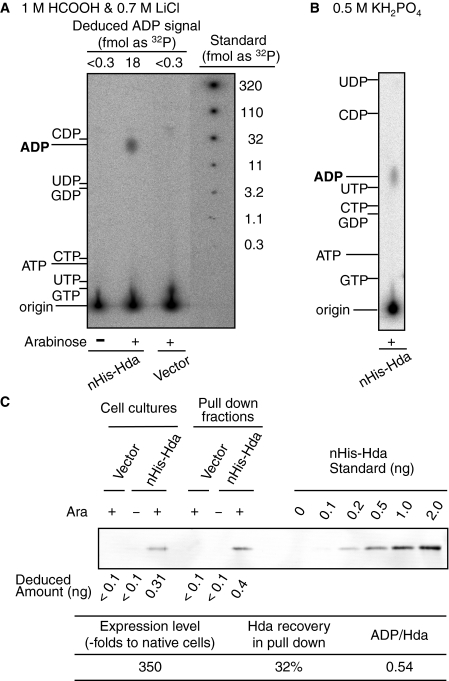
Nucleotide-bound Form of Cellular Hda Protein—We next attempted to detect nucleotides bound to Hda proteins in vivo. Cells bearing the nHis-Hda expression plasmid (pBAD/nHis-Hda) were grown in 32P-labeled culture and further incubated for 10 min in the presence of 0.5% arabinose. nHis-Hda was collected using a nickel bead pulldown method, and the Hda-bound nucleotides were analyzed by TLC. The presence of a single spot corresponding to ADP correlated with nHis-Hda expression (Fig. 5, A and B). This result is consistent with the in vitro data showing that Hda specifically binds to ADP rather than ATP and the other nucleotides (Table 1). The Hda content in nHis-Hda expressing cells was ∼350-fold higher than that in the native cells as determined by Western blot analysis (Fig. 5C) (25). Recovery of nHis-Hda from the expressing cells was deduced to be 30–35%, and the ratio of the recovered ADP per Hda monomer was deduced to be ∼0.54 (Fig. 5C). In the pulldown experiments used for Fig. 5C, Hda was specifically isolated (supplemental Fig. S2). Taken together, these results suggest that Hda complexed with ADP is present in vivo.
FIGURE 5.
Nucleotide-bound state of Hda in vivo. MC1061 cells harboring pBAD/HisB (Vector) or pBAD/nHis-Hda (nHis-Hda) were grown at 37 °C in a minimum medium containing [32P]orthophosphate. Aliquots (0.5 ml) were collected before (–) or after (+) incubation for 10 min in the presence of 0.5% arabinose, and used for a pulldown analysis (A and B) and Western bolt analysis (C). A and B, nHis-Hda was recovered from the cell lysates using a nickel pulldown method, resulting in the pulldown fractions (5 μl). Portions (0.25 μl) of the fractions were analyzed for Hda-bound nucleotides by TLC using a solvent consisting of 1 m HCOOH and 0.7 m LiCl (A) or 0.5 m KH2PO4 (B). The migration positions of various nucleotides were analyzed on the same TLC plate and detected by UV exposure and are indicated. Signals corresponding to ADP were quantified, and the amounts as a [32P]orthophosphate were deduced using a quantitative standard of [32P]orthophosphate spotted on the same TLC plate (Deduced ADP signal)(A). C, expression level and recovered amounts of nHis-Hda were determined by Western blot analysis. Labeled cells in the culture (5 μl) were used to determine the expression level, and pulldown fractions (0.2 μl) were isolated and used to determine the amount of recovered nHis-Hda. Intensities corresponding to nHis-Hda were quantified, and the amounts were deduced using a quantitative standard of purified nHis-Hda (Deduced amount). The deduced amount of nHis-Hda in the cells was used to calculate a relative value (Expression level) compared with the cellular amounts of Hda in MG1655 (25). The deduced amount of nHis-Hda in the pulldown fraction was used to calculate the recovery (Hda recovery in pull down), which is also shown. Based on the amount of [32P]ADP determined in A, the ratio of ADP molecules per nHis-Hda monomer in the pulldown fraction was determined (ADP/Hda).
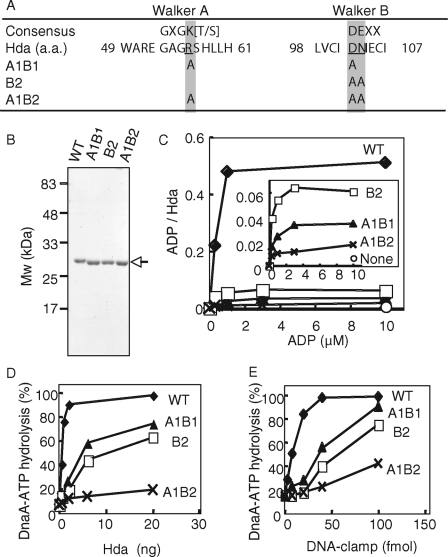
Walker Motif Mutants of Hda Are Defective in ADP Binding in Vitro—To investigate the in vivo significance of ADP binding to Hda, we constructed three Hda mutants bearing amino acid substitutions in the Walker A and B motifs. Hda A1B1, Hda B2, and Hda A1B2 bear mutations of R56A/D102A, D102A/N103A, and R56A/D102A/N103A, respectively (Fig. 6A). These Hda mutants were expressed as forms equivalent to Hda-cHis using the pET/Hda-cHis-derivative plasmids. The expressed mutant Hda proteins predominantly formed insoluble aggregates, whereas ∼50% of wild-type Hda-cHis were recovered in a soluble fraction. The wild-type and mutant Hda-cHis proteins in the insoluble fractions were dissolved with urea, and urea in the samples was excluded by gel filtration. In the resulting fractions, the purity of these proteins was >90% (Fig. 6B). Most likely the expedited migration rate of the mutant proteins in SDS-PAGE was caused by substitutions of the charged amino acid residues to those with smaller and non-charged side chains rather than degradation of protein termini, as these proteins retained in the N-terminal clamp binding activity (see below) and the C-terminal nickel beads binding activity (data not shown). Further gel filtration analyses showed that the wild-type and mutant proteins formed multimers (∼200 kDa) (data not shown). The purified wild-type Hda-cHis was active in DnaA-ATP hydrolysis and ADP binding at levels comparable with those of Hda-cHis purified from a soluble fraction (supplemental Fig. S3).
FIGURE 6.
In vitro analysis of Walker motif mutants of Hda. A, amino acid (a.a.) sequence of the Hda Walker A and B motifs and their consensus sequences are shown. The amino acid substitutions in the mutant Hda used in this study are also indicated. B, wild-type (WT) and mutant (A1B1, B2, and A1B2) Hda-cHis proteins (0.5 μg) were purified from insoluble cell lysate fractions (see “Experimental Procedures”) and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. C, indicated concentrations of [3H]ADP were incubated at 30 °C for 20 min in the presence or absence (None) of wild-type (WT) or mutant (A1B1, B2, or A1B2) Hda-cHis proteins (20 pmol), followed by retention on a nitrocellulose filter. The bound ADP molecules per Hda monomer are presented. The inset shows an expanded view of the data from the mutant Hda-cHis proteins. D and E, DnaA-ATP hydrolysis activity of the wild-type Hda-cHis protein (WT) and the mutant Hda-cHis proteins (A1B1, B2, and A1B2) was assessed in a staged RIDA reconstituted system (30 °C, 20 min) in the presence of 0.5 pmol of [α-32P]ATP-DnaA and 200 μm ADP. D, DNA-loaded clamp (100 fmol as clamp) was included. E, indicated amounts (as clamp) of the DNA-loaded clamp were incubated in the presence of 20 ng of wild-type Hda-cHis protein (WT) or mutant Hda-cHis proteins (A1B1, B2, or A1B2).
The filter-retention assay revealed that Hda A1B2 was severely impaired in ADP binding (Fig. 6C). In accordance with this, DnaA-ATP hydrolysis activity of Hda A1B2 in the presence of 0.2 mm ADP was severely inhibited (Fig. 6, D and E). The clamp binding activity was retained in Hda A1B2 (see below), indicating the possibility that complete denaturation of this mutant protein is unlikely.
In contrast to Hda A1B2, Hda A1B1 and Hda B2 had residual activities in ADP binding and DnaA-ATP hydrolysis (Fig. 6, C–E). The ADP binding activity of Hda A1B1 was ∼10-fold lower than that of the wild-type protein (Fig. 6C). Concurrent with this, the specific activity of Hda A1B1 in DnaA-ATP hydrolysis was ∼10-fold lower than that of the wild-type protein (Fig. 6, D and E). In addition, the ADP binding activity of Hda B2 was inhibited, but its residual activity was only ∼2-fold higher than that of Hda A1B1 (Fig. 6, C–E). The DnaA-ATP hydrolysis activity of Hda B2 was inhibited, but a residual activity was seen at a level similar to that of Hda A1B1. These results suggest a correlation between the ADP binding and RIDA activity of Hda.
Role for ADP Binding to Hda in Vivo—The overall ATP-DnaA level is repressed to ∼20% of the total ATP/ADP-DnaA in an Hda-dependent manner in a culture containing asynchronously dividing cells (12, 14). hda disruption leads to accumulation of the ATP-DnaA level up to ∼70%. To investigate whether ADP is required for the function of Hda in vivo, we assessed the ATP-DnaA levels in cells bearing the Walker motif mutations within hda. pACYC177 derivatives carrying the nontagged form (ntHda) of wild-type or mutant hda allele were introduced into MK86 [Δhda ΔoriC rnhA] cells. pACYC177, a low copy plasmid, was used to minimize expression of Hda. As expected, the derivative plasmids did not cause growth inhibition of the host cells.
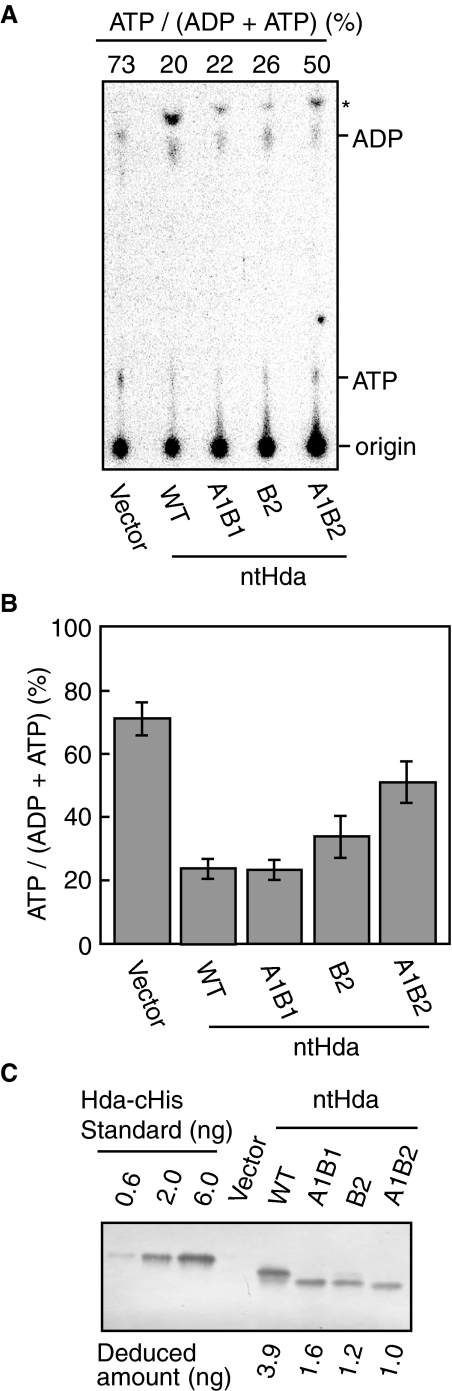
In comparison with cells lacking Hda, the cellular ATP-DnaA level was repressed to ∼20% in a manner that was dependent on the cellular wild-type ntHda (Fig. 7, A and B, 1st and 2nd lanes). In cells expressing ntHda A1B2, the ATP-DnaA level was elevated to ∼50% (Fig. 7, A and B, 5th lane), indicating that this Hda mutant was defective in DnaA-ATP hydrolysis in vivo. This is consistent with the in vitro results indicating severe defects in ADP binding and DnaA-ATP hydrolysis in this mutant Hda (Fig. 6). Cells expressing either ntHda A1B1 or ntHda B2 showed an ATP-DnaA level that was similar to that seen in cells expressing wild-type Hda (Fig. 7, A and B, 3rd and 4th lanes). It is likely that the residual activities of Hda A1B1 and Hda B2 that were observed in vitro (as shown in Fig. 6) support their function in cells. The Hda levels in the strains bearing the hda plasmids were elevated compared with that in a wild-type strain (Fig. 7C). Furthermore, cells contain chaperone proteins and 200–300 μm levels of ADP (34).
FIGURE 7.
In vivo analysis of the Walker motif mutants of Hda. A, MK86 [Δhda ΔoriC rnhA] cells harboring pACYC177 (Vector), pACYC/ntHda (WT), pACYC/ntHdaA1B1 (A1B1), pACYC/ntHdaB2 (B2), or pACYC/ntHdaA1B2 (A1B2) were grown at 37 °C in the presence of [32P]orthophosphate. Nucleotide-bound DnaA protein was immunochemically isolated from cell lysates, and the recovered nucleotides were analyzed as described under “Experimental Procedures.” Proportions (%) of ATP-DnaA included in the total amount of ATP/ADP-DnaA molecules are shown (ATP/(ADP+ATP)). The origin and the migration positions of ATP and ADP are indicated. The asterisk indicates a nonspecific signal, which was seen in the samples prepared using pre-immune serum. B, experiments shown in A were independently repeated three times, and the results with standard deviations are shown. C, expression levels of cells used in this experiment. Western blotting was performed using the indicated cells (A600 of 0.15, 67μl). The amount of ntHda was quantified and deduced using a quantitative standard of purified Hda-cHis (Deduced amount). ntHda is the nontagged form of Hda. The relative level of wild-type ntHda was 180-fold increased compared with that of MG1655 (25).
The cellular content of ntHda A1B2 was comparable with that of ntHda B2 and ntHda A1B1 (Fig. 7C). Thus, the higher ATP-DnaA level in ntHda A1B2-expressing cells could not be attributed to a reduction in cellular Hda molecules. Taken together, these results support the idea that Hda requires ADP for its cellular activity.
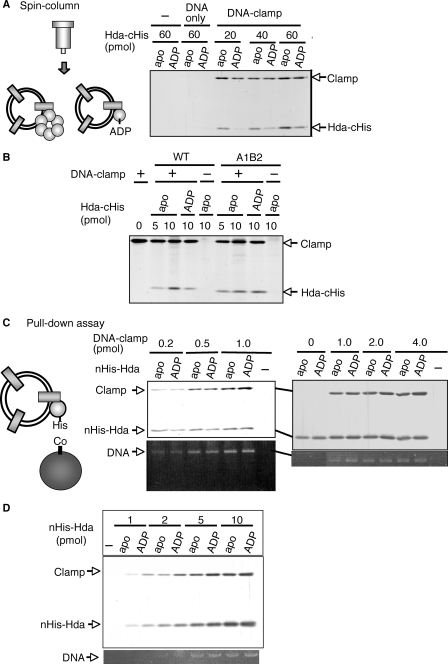
Effect of ADP on Complex Formation of Hda with the DNA-loaded Clamp—We next addressed the mechanism by which ADP promotes Hda activity in the RIDA system. First, we used a spin column to analyze the activities of ADP-Hda and apo-Hda in a complex formation with the DNA-loaded clamp. In this assay, we used wild-type apo-Hda-cHis and Hda A1B2 that were obtained in the experiments shown in Fig. 6. ADP-Hda-cHis was formed by incubating apo-Hda-cHis in the presence of ADP. In a spin-column analysis, both apo-Hda-cHis and ADP-Hda-cHis were recovered in a clamp-dependent manner (Fig. 8A). The recovered number of apo-Hda-cHis per clamp molecule was 2–4-fold higher than that of ADP-Hda-cHis. Most likely this difference is attributed to the fact that apo-Hda-cHis forms homomultimers, whereas ADP-Hda-cHis is monomeric (Fig. 4A). The spin column-isolated complexes of ADP-Hda-cHis and the DNA-loaded clamp were active in the DnaA-ATP hydrolysis, whereas those of apo-Hda-cHis and the DNA-loaded clamp were inactive (data not shown).
FIGURE 8.
Interaction of apo-Hda and ADP-Hda with the DNA-loaded clamp. A, spin-column analysis. Hda-cHis multimers (apo-Hda-cHis, apo) were preincubated at 30 °C for 20 min in the presence of 100 μm ADP, resulting in ADP-Hda-cHis (ADP). The indicated amounts of apo-Hda-cHis or ADP-Hda-cHis were incubated on ice for 15 min in the presence (DNA-clamp) or absence (–) of the DNA-loaded clamp (4 pmol as clamp, 320 ng as M13mp18 nicked circular DNA). Proteins associated with the DNA were isolated by gel filtration spin column into the void fraction. 1/10th of the void fraction was analyzed by SDS-PAGE and silver staining. The same experiment was performed using M13mp18 nicked circular DNA (320 ng) instead of the DNA-loaded clamp (DNA only). B, clamp binding activity of the Hda A1B2 mutant. Wild-type (WT) and A1B2 mutant (A1B2) Hda-cHis multimers were used as the apo-form (apo) or were preincubated in the presence of ADP as above (ADP). The indicated amounts of apo-Hda-cHis or ADP-preincubated Hda-cHis were incubated on ice for 15 min in the presence (+) or absence (–) of the DNA-loaded clamp (1 pmol as clamp). The void fraction was isolated by a gel filtration spin column and analyzed as described for A. C, pulldown assay. nHis-Hda multimers were used as the apo-form (apo) or were preincubated in the presence of ADP as above (ADP). TALON beads were incubated in the presence or absence (–) of 5 pmol of apo-nHis-Hda or ADP-nHis-Hda, followed by incubation on ice for 15 min in buffer containing the indicated amounts of the DNA-loaded clamp. The bead-bound materials were recovered, washed, and eluted in 1% SDS. An aliquot of the elution was analyzed by SDS-PAGE and silver staining (upper panel). The remaining elution was used for analysis of DNA recovery by 1% agarose gel electrophoresis and ethidium bromide staining (lower panel). D, various amounts of apo-nHis-Hda or ADP-nHis-Hda were analyzed by a similar pulldown assay in the presence of the DNA-loaded clamp (1 pmol).
To further assess the role of ADP, we performed a similar spin-column analysis using Hda A1B2. Hda A1B2 exhibited clamp binding activity at a level similar to that of wild-type Hda (Hda-cHis multimers) in the absence of ADP (Fig. 8B). The recovery of wild-type Hda, but not Hda A1B2, was decreased when Hda was preincubated with ADP. This is consistent with the fact that Hda A1B2 is inactive in ADP binding and thus stably forms multimers.
We further performed a pulldown assay using the DNA-loaded clamp and Hda-conjugated beads. In this assay, we used nHis-Hda because of inefficient binding of Hda-cHis multimers to Co2+-conjugated TALON beads. nHis-Hda multimers (apo-nHis-Hda) (supplemental Fig. S1B) were incubated at 30 °C for 20 min in the presence of ADP, resulting in the ADP form. Recovery of the clamp and DNA were comparable between the apo-nHis-Hda and ADP-nHis-Hda (Fig. 8, C and D). These results suggest that ADP does not significantly affect the affinity of Hda for the DNA-loaded clamp.
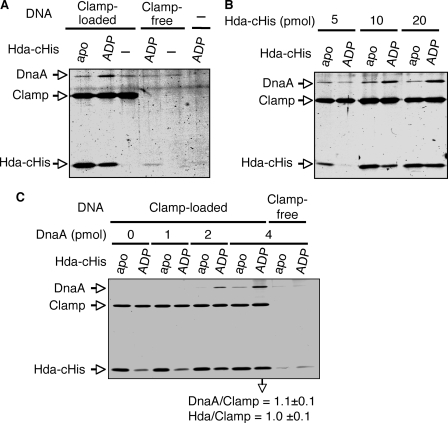
ADP-dependent Complex Formation of Hda with DnaA Mediated by the DNA-loaded Clamp—Based on analyses of the RIDA reaction, we have implicated that DnaA-ATP hydrolysis takes place upon the formation of a dynamic complex in which ATP-DnaA interacts with Hda in a manner dependent on the DNA-loaded clamp. To assess the role of ADP binding to Hda during complex formation, we first performed spin-column analysis. To reduce the rate of the catalytic ATP-hydrolysis reaction, the DNA-loaded clamp and Hda-cHis were incubated on ice, and ATP-DnaA was added, followed by immediate spin-column analysis. DnaA was recovered in a manner that was dependent on the DNA-loaded clamp and ADP-Hda-cHis (Fig. 9). Apo-Hda-cHis was impaired in the DnaA recovery. The recovered amounts of apo-Hda-cHis were higher than those of ADP-Hda-cHis. These differences were similarly observed for the data of Fig. 8, A and B, and are most likely attributed to the multimer formation of apo-Hda. Only a slight recovery of DnaA and Hda was detected even when the clamp-free DNA was used (Fig. 9, A and C), which is most likely attributed to a very weak, nonspecific DNA binding activity of these proteins. These results suggest that ADP stimulates Hda activity in clamp-dependent interactions with DnaA.
FIGURE 9.
Spin-column analysis for complex formation of ADP-Hda, ATP-DnaA, and the DNA-loaded clamp. Apo-Hda-cHis (apo) and ADP-Hda-cHis (ADP) were prepared as described for Fig. 8A. A, apo-Hda-cHis or ADP-Hda-cHis (10 pmol) was incubated on ice for 5 min in the presence of the DNA-loaded clamp (Clamp-loaded) (1 pmol as clamp, 80 ng as M13mp18 nicked circular DNA) or M13mp18 nicked circular DNA (Clamp-free) (80 ng). ATP-DnaA (2 pmol) was then added to the reaction, followed by immediate spin-column gel filtration, isolation of the void fraction, and analysis by SDS-PAGE described for Fig. 8A. –, no DNA nor protein. B, indicate amounts of apo-Hda-cHis or ADP-Hda-cHis were similarly analyzed using the DNA-loaded clamp (1 pmol as clamp) and ATP-DnaA (2 pmol). C, indicated amounts of ATP-DnaA were similarly analyzed using the DNA-loaded clamp (1 pmol as clamp) and apo-Hda-cHis or ADP-Hda-cHis (20 pmol). Two independent experiments were preformed, and similar data were obtained. Representative data are shown. In the experiments using 4 pmol of ATP-DnaA, the mean molar ratios of recovered DnaA/Clamp (β dimer) and Hda/Clamp were deduced using quantitative standards and are indicated below the gel image.
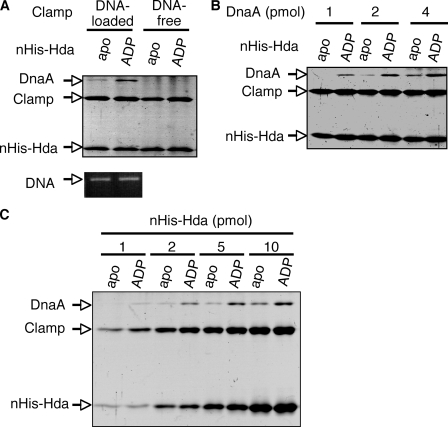
To provide independent evidence, we further performed a pulldown assay using nHis-Hda. ATP-DnaA was incubated on ice in buffer containing apo-nHis-Hda or ADP-nHis-Hda in the presence or absence of the DNA-loaded clamp, followed by the recovery of nHis-Hda using TALON beads. ADP-nHis-Hda but not apo-nHis-Hda assisted the recovery of DnaA (Fig. 10), consistent with the above observation. DnaA was recovered in a manner depending on the DNA-loaded clamp but not the DNA-free clamp, consistent with the fact that RIDA requires the DNA-loaded clamp. The recovered amounts of the clamp, DNA, and Hda were not affected by ADP binding to Hda. Altogether, these results demonstrate that ADP binding of Hda stimulates stable interaction with ATP-DnaA when Hda is complexed with the DNA-loaded clamp.
FIGURE 10.
Pulldown assay for complex formation of ADP-Hda, ATP-DnaA, and the DNA-loaded clamp. Apo-nHis-Hda (apo) and ADP-nHis-Hda (ADP) were conjugated to TALON beads as described for Fig. 8C. A, bead-conjugated apo-nHis-Hda or ADP-nHis-Hda (5 pmol) was incubated on ice for 5 min in buffer containing the DNA-loaded or DNA-free clamp (2 pmol as clamp), and ATP-DnaA (4 pmol) was included and incubated for 10 min. The bead-bound materials were analyzed as described for Fig. 8C. B, indicated amounts of DnaA were similarly analyzed using the pulldown assay in the presence of the DNA-loaded clamp (2 pmol as clamp) and nHis-Hda (5 pmol). C, indicated amounts of nHis-Hda were similarly analyzed using the pulldown assay in the presence of the DNA-loaded clamp (1 pmol as clamp) and ATP-DnaA (2 pmol).
DISCUSSION
In this study, we demonstrate that Hda is associated with the unusual property of specific binding to ADP but not ATP, and that ADP is the crucial activator of Hda. Unlike apo-Hda, ADP-Hda functions to hydrolyze DnaA-ATP. This activation is mediated by dissociation of apo-Hda multimers into ADP-Hda monomers. In addition, we have demonstrated that ADP-Hda can be formed in vivo and that an Hda mutant severely deficient in ADP binding was inactive in repression of the cellular ATP-DnaA level. Moreover, for the first time we have demonstrated in vitro that ATP-DnaA, ADP-Hda, and the DNA-loaded clamp can physically form a complex. Like ADP-Hda, apo-Hda can form a complex with the DNA-loaded clamp. In contrast, ADP-Hda, but not apo-Hda, is the active form for the subsequent ATP-DnaA interaction with the Hda-clamp complex. Thus, we have revealed ADP-dependent activation of Hda and its underlying mechanism. Hda might be able to act as a regulatory switch for the RIDA system in vivo via the ADP-mediated monomerization of inactive multimers.
One of the findings in this study is that the hda initiation codon is CUG (Fig. 1). In E. coli, the usage of AUG, GUG, UUG, and AUU as initiation codons is approximately ∼90, ∼8, ∼1, and <1%, respectively (35). In contrast, no gene has been previously reported to use CUG as an initiation codon in E. coli. The artificially introduced CUG initiation codon starts translation inefficiently; experiments using the lacZ fusion indicate that the activity of CUG in translational initiation is 1–3% that of AUG (36, 37). In eukaryotic cells and viruses, several genes are reported to use CUG as an initiation codon. For example, the 34-kDa isoform of human fibroblast growth factor 2 and the p27 coat protein of Hibiscus chlorotic ringspot virus are naturally translated from the CUG initiation codon (38, 39). In these cases, the CUG initiation codon is known to be used for restrained expression of the proteins. We have previously determined that the cellular content of the Hda monomer is only ∼100 per cell (25). The CUG initiation codon might therefore be used to maintain the low level of cellular Hda molecules as overexpression of Hda is detrimental to cell viability (12, 40).
We have previously used nHis-ex-Hda bearing the N-terminal extra 15 amino acid residues that were derived from a previously annotated coding region (Fig. 1). nHis-ex-Hda exhibited a lower affinity for ADP than nHis-Hda, indicating that the N-terminal extra region was inhibitory in ADP binding (supplemental Fig. S1A). The crystal structure of several AAA+ proteins has revealed that the N terminus of the AAA+ domain is located at a position close to the nucleotide-binding pocket (41). The N-terminal extra region is rich in hydrophobic amino acid residues (42), which might cause improper interactions with the nucleotide-binding pocket, thereby resulting in the inhibition of ADP binding. Like Hda-cHis, the DnaA-ATP hydrolysis activity of nHis-ex-Hda depends on ADP (data not shown). Thus, unlike the tight affinity for ADP, ADP-dependent activation of Hda was preserved in nHis-ex-Hda. In previous in vitro RIDA assays, 2 mm ATP was constantly contained (12, 13, 15, 16, 24, 25). The ATP sample contained a slight ADP contamination (4%) that was probably generated by degradation of ATP. Thus, the previous RIDA assays included 80 μm
ADP, which was high enough to support ADP binding of Hda (Fig. 3A; supplemental Fig. S1A). nHis-ex-Hda predominantly formed insoluble aggregates when overproduced in cells, (25), unlike Hda-cHis and nHis-Hda. The hydrophobic moiety of the extra N terminus might be related to the insoluble feature. In the presence of 0.1% Triton X-100 and the absence of ADP, nHis-ex-Hda formed complexes predominantly containing homodimers (supplemental Fig. S1C), as also shown in our previous study (25). This detergent was required for the stable maintenance of nHis-ex-Hda activity in storage solution (25). In the presence of 0.1% Triton X-100 and the absence of ADP, Hda predominantly formed homodimers regardless of the extra N terminus (data not shown).
Unlike most other AAA+ proteins, Hda can only bind ADP but not ATP (Table 1 and Figs. 3 and 5). This unique feature could be consistent with the structure within the Hda Walker A motif, in that Hda bears the sequence GRS instead of the typical sequence GKT. The conserved lysine residue within GKT interacts with the β and γ phosphate oxygens (43). The corresponding residue of Hda is arginine, and its bulky side chain might interfere with ATP entry into the binding pocket.
Among AAA+ proteins, the δ′ subunit of pol III γ complex lacks the conserved Walker A motif as well as affinity for ATP and ADP (44). The δ′ subunit carries an arginine residue corresponding to the AAA+ arginine finger motif and supplies this residue for ATP that is bound on the neighboring γ subunit included in the γ complex, leading to ATP hydrolysis (45). The arginine finger of Hda is crucial for catalyzing DnaA-ATP hydrolysis (25). In this sense, the δ′ subunit and Hda might share a common feature that they do not bind ATP and supply the arginine finger with ATP bound to another protein (γ subunit or DnaA). Energy from ATP is not required for the function of these proteins.
ADP binding of Hda was required for the stable monomeric state. In typical cases of AAA+ proteins, the inter-protomer interface includes a surface of the nucleotide-binding pocket. ADP binding might cause a physical obstacle or a conformational change in this interface, resulting in inhibition of the interaction between Hda protomers. In several AAA+ proteins, ADP inhibits the formation of multimers. For example, the ADP form of Katanin, a microtubule-severing factor, and Vps4p, an endosomal sorting factor, are composed of homodimers, whereas their ATP forms are homomultimers (46, 47). Origin recognition complex-Cdc6 association is supported by ATPγS but not ADP (48).
In vitro, apo-Hda is inactive for DnaA-ATP hydrolysis, whereas ADP-Hda is active (Figs. 3 and 4). Apo-Hda assembles into homomultimers consisting of several protomers; this is similar to many AAA+ proteins that are able to form multimers, including 5–7 protomers. As seen for typical AAA+ protein multimers, the arginine finger of apo-Hda multimers might be buried in the interface of the protomers, resulting in inhibition of the interaction with DnaA. Upon ADP binding, the Hda homomultimers dissociate into monomers, exposing the arginine finger on the protein surface, thereby allowing interaction with DnaA.
Mutant analysis suggested that ADP binding is crucial for Hda activity in vitro and in vivo (Figs. 6 and 7). There is thus a possibility that Hda activity might be controlled by association and dissociation of ADP in vivo. As the cellular ADP concentration has been determined to be 200–300 μm (34), there is no reason to assume the presence of a factor stimulating ADP binding to Hda. In contrast, it would be worthwhile to consider whether a factor inhibiting ADP binding of Hda is present. In rapidly growing E. coli cells, a new round of replication initiation takes place before completion of the previous round (49). Under these situations, RIDA might be restrained even in the presence of the DNA-loaded clamps for assisting an increase in the ATP-DnaA level. It is possible that a factor dissociating the ADP-Hda complex plays a regulatory role by inhibiting RIDA in a timely manner. It would be interesting to search for such a factor and to investigate whether the cellular ratio of apo-Hda/ADP-Hda varies depending on the cell cycle in rapidly growing cells.
Supplementary Material
Acknowledgments
We are grateful to Dr. Norie Fujikawa, Dr. Wataru Kagawa, Dr. Hitoshi Kurumizaka, and Dr. Shigeyuki Yokoyama for structural analysis and discussion on Hda.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
Footnotes
The abbreviations used are: pol, DNA polymerase; RIDA, regulatory inactivation of DnaA; SD, Shine-Dalgarno; ATPγS, adenosine 5′-O-(thiotriphosphate); Fr, fraction; ntHda, nontagged form of Hda.
References
- 1.Messer, W. (2002) FEMS Microbiol. Rev. 26 355–374 [DOI] [PubMed] [Google Scholar]
- 2.Kaguni, J. M. (2006) Annu. Rev. Microbiol. 60 351–375 [DOI] [PubMed] [Google Scholar]
- 3.Ishida, T., Akimitsu, N., Kashioka, T., Hatano, M., Kubota, T., Ogata, Y., Sekimizu, K., and Katayama, T. (2004) J. Biol. Chem. 279 45546–45555 [DOI] [PubMed] [Google Scholar]
- 4.Keyamura, K., Fujikawa, N., Ishida, T., Ozaki, S., Su'etsugu, M., Fujimitsu, K., Kagawa, W., Yokoyama, S., Kurumizaka, H., and Katayama, T. (2007) Genes Dev. 21 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donnell, M. (2006) J. Biol. Chem. 281 10653–10656 [DOI] [PubMed] [Google Scholar]
- 6.Katayama, T. (2001) Mol. Microbiol. 41 9–17 [DOI] [PubMed] [Google Scholar]
- 7.Zakrzewska-Czerwinska, J., Jakimowicz, D., Zawilak-Pawlik, A., and Messer, W. (2007) FEMS Microbiol. Rev. 31 378–387 [DOI] [PubMed] [Google Scholar]
- 8.Lu, M., Campbell, J. L., Boye, E., and Kleckner, N. (1994) Cell 77 413–426 [DOI] [PubMed] [Google Scholar]
- 9.Slater, S., Wold, S., Lu, M., Boye, E., Skarstad, K., and Kleckner, N. (1995) Cell 82 927–936 [DOI] [PubMed] [Google Scholar]
- 10.Nievera, C., Torgue, J. J., Grimwade, J. E., and Leonard, A. C. (2006) Mol. Cell 24 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitagawa, R., Ozaki, T., Moriya, S., and Ogawa, T. (1998) Genes Dev. 12 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, J., and Katayama, T. (2001) EMBO J. 20 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, T., Kubota, T., Kurokawa, K., Crooke, E., and Sekimizu, K. (1998) Cell 94 61–71 [DOI] [PubMed] [Google Scholar]
- 14.Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K., and Katayama, T. (1999) EMBO J. 18 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida, S., Fujimitsu, K., Sekimizu, K., Ohmura, T., Ueda, T., and Katayama, T. (2002) J. Biol. Chem. 277 14986–14995 [DOI] [PubMed] [Google Scholar]
- 16.Su'etsugu, M., Kawakami, H., Kurokawa, K., Kubota, T., Takata, M., and Katayama, T. (2001) Mol. Microbiol. 40 376–386 [DOI] [PubMed] [Google Scholar]
- 17.Camara, J. E., Breier, A. M., Brendler, T., Austin, S., Cozzarelli, N. R., and Crooke, E. (2005) EMBO Rep. 6 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noirot-Gros, M. F., Dervyn, E., Wu, L. J., Mervelet, P., Errington, J., Ehrlich, S. D., and Noirot, P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, M., Ogura, Y., Harry, E. J., Ogasawara, N., and Moriya, S. (2005) FEMS Microbiol. Lett. 247 73–79 [DOI] [PubMed] [Google Scholar]
- 20.Noirot-Gros, M. F., Velten, M., Yoshimura, M., McGovern, S., Morimoto, T., Ehrlich, S. D., Ogasawara, N., Polard, P., and Noirot, P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias, E. E., and Walter, J. C. (2006) Nat. Cell Biol. 8 84–90 [DOI] [PubMed] [Google Scholar]
- 22.Nishitani, H., Sugimoto, N., Roukos, V., Nakanishi, Y., Saijo, M., Obuse, C., Tsurimoto, T., Nakayama, K. I., Nakayama, K., Fujita, M., Lygerou, Z., and Nishimoto, T. (2006) EMBO J. 25 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senga, T., Sivaprasad, U., Zhu, W., Park, J. H., Arias, E. E., Walter, J. C., and Dutta, A. (2006) J. Biol. Chem. 281 6246–6252 [DOI] [PubMed] [Google Scholar]
- 24.Su'etsugu, M., Takata, M., Kubota, T., Matsuda, Y., and Katayama, T. (2004) Genes Cells 9 509–522 [DOI] [PubMed] [Google Scholar]
- 25.Su'etsugu, M., Shimuta, T., Ishida, T., Kawakami, H., and Katayama, T. (2005) J. Biol. Chem. 280 6528–6536 [DOI] [PubMed] [Google Scholar]
- 26.Dalrymple, B. P., Kongsuwan, K., Wijffels, G., Dixon, N. E., and Jennings, P. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurz, M., Dalrymple, B., Wijffels, G., and Kongsuwan, K. (2004) J. Bacteriol. 186 3508–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. V. (1999) Genome Res. 9 27–43 [PubMed] [Google Scholar]
- 29.Davey, M. J., Jeruzalmi, D., Kuriyan, J., and O'Donnell, M. (2002) Nat. Rev. Mol. Cell Biol. 3 826–835 [DOI] [PubMed] [Google Scholar]
- 30.Ogura, T., Whiteheart, S. W., and Wilkinson, A. J. (2004) J. Struct. Biol. 146 106–112 [DOI] [PubMed] [Google Scholar]
- 31.Sekimizu, K., Bramhill, D., and Kornberg, A. (1987) Cell 50 259–265 [DOI] [PubMed] [Google Scholar]
- 32.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., Gregor, J., Davis, N. W., Kirkpatrick, H. A., Goeden, M. A., Rose, D. J., Mau, B., and Shao, Y. (1997) Science 277 1453–1474 [DOI] [PubMed] [Google Scholar]
- 33.Kogoma, T. (1997) Microbiol. Mol. Biol. Rev. 61 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhard, J., and Nygaard, P. (1987) in Escherichia coli and Salmonella: Cellular and Molecular Biology, (Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Schaechter, M., and Unbarger, H. E., eds), pp. 1579–1601, American Society for Microbiology, Washington, D. C.
- 35.Schneider, T. D., Stormo, G. D., Gold, L., and Ehrenfeucht, A. (1986) J. Mol. Biol. 188 415–431 [DOI] [PubMed] [Google Scholar]
- 36.Sussman, J. K., Simons, E. L., and Simons, R. W. (1996) Mol. Microbiol. 21 347–360 [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell, S. M., and Janssen, G. R. (2001) J. Bacteriol. 183 1277–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnaud, E., Touriol, C., Boutonnet, C., Gensac, M. C., Vagner, S., Prats, H., and Prats, A. C. (1999) Mol. Cell. Biol. 19 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh, D. C., Wang, X., Wong, S. M., and Liu, D. X. (2006) Virus Res. 122 35–44 [DOI] [PubMed] [Google Scholar]
- 40.Banack, T., Clauson, N., Ogbaa, N., Villar, J., Oliver, D., and Firshein, W. (2005) J. Bacteriol. 187 8507–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanson, P. I., and Whiteheart, S. W. (2005) Nat. Rev. Mol. Cell Biol. 6 519–529 [DOI] [PubMed] [Google Scholar]
- 42.Kawakami, H., Su'etsugu, M., and Katayama, T. (2006) J. Struct. Biol. 156 220–229 [DOI] [PubMed] [Google Scholar]
- 43.Ogura, T., and Wilkinson, A. J. (2001) Genes Cells 6 575–597 [DOI] [PubMed] [Google Scholar]
- 44.Jeruzalmi, D., O'Donnell, M., and Kuriyan, J. (2001) Cell 106, 429–441 [DOI] [PubMed] [Google Scholar]
- 45.Johnson, A., and O'Donnell, M. (2003) J. Biol. Chem. 278 14406–14413 [DOI] [PubMed] [Google Scholar]
- 46.Babst, M., Wendland, B., Estepa, E. J., and Emr, S. D. (1998) EMBO J. 17 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartman, J. J., and Vale, R. D. (1999) Science 286 782–785 [DOI] [PubMed] [Google Scholar]
- 48.Speck, C., Chen, Z., Li, H., and Stillman, B. (2005) Nat. Struct. Mol. Biol. 12 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper, S., and Helmstetter, C. E. (1968) J. Mol. Biol. 31 519–540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.