Abstract
Fanconi anemia (FA) is a heritable human cancer-susceptibility disorder, delineating a genetically heterogenous pathway for the repair of replication-blocking lesions such as interstrand DNA cross-links. Here we demonstrate that one component of this pathway, FANCJ, is a structure-specific DNA helicase that dissociates guanine quadruplex DNA (G4 DNA) in vitro. Moreover, in contrast with previously identified G4 DNA helicases, such as the Bloom's helicase (BLM), FANCJ unwinds G4 substrates with 5′–3′ polarity. In the FA-J human patient cell line EUFA0030 the loss of FANCJ G4 unwinding function correlates with the accumulation of large genomic deletions in the vicinity of sequences, which match the G4 DNA signature. Together these findings support a role for FANCJ in the maintenance of potentially unstable genomic G/C tracts during replication.
FANCJ (also called BACH1 and BRIP1) plays an important role in the maintenance of genome integrity in humans. Although individuals with monoallelic mutations in FANCJ have increased susceptibility to breast cancer (1), inheritance of biallelic FANCJ mutations results in the chromosome instability disease Fanconi anemia (2–4).
Fanconi anemia (FA)3 defines a genetic pathway for dealing with DNA interstrand cross-links (reviewed in Ref. 5). Consequently, cells from FA patients are characteristically sensitive to cross-linking agents such as mitomycin C and cisplatin. Exposure to these agents causes increased chromosomal aberrations, including DNA breaks and chromatid interchanges, which may lead to mis-segregation of chromosomes during cell division and ultimately to cell death. However, because the level of DNA cross-links in the cell is usually low, it is unlikely that these lesions are the normal substrate for this pathway.
Most of the 13 known FA proteins provide few clues to their biochemical function. However, FANCJ is an exception as it resembles a member of the DEAH family of DNA helicases and has been shown to unwind forked DNA and D-loops substrates in vitro (6, 7).
Genetic studies in Caenorhabditis elegans suggest that FANCJ may be required to deal with DNA secondary structures that form in guanine-rich regions of the genome. Worms defective for the FANCJ homolog, DOG-1, were found to accumulate deletions in regions of the genome characterized by long stretches of polyguanine (8). Moreover these deletions were not dependent on the non-homologous end-joining or homologous recombination pathways for DNA repair (9, 10). Characteristically these deletions contain one breakpoint close to the 3′ end of the polyguanine tract and extend various distances 5′ of the tract.
Recent studies report similar deletions triggered by sequences that match the consensus signature, G3–5N1–3-G3–5N1–3G3–5N1–3G1–3, for the formation of G4 DNA (11). G4 DNA is an unusual DNA secondary structure formed by the pairing of four guanines in a planar array and stabilized through Hoogsteen bonds. These structures may be widespread in the human genome with more than 360,000 DNA sequences conforming to the G4 DNA signature (19). Consequently, it is possible that G4 DNA has a biological function, for example, in transcriptional regulation. Nevertheless, the exceptional stability of G4 DNA suggests it is likely to present a significant obstacle during DNA replication and may require the action of specific enzymes to resolve it.
Here we have investigated the role of FANCJ in dealing with G4 DNA structures. We demonstrate that FANCJ is a structure-specific DNA helicase able to resolve G4 DNA in vitro. We also report that cells from human FA-J patients, like dog-1 mutants in C. elegans, accumulate large genomic deletions in regions of the genome matching the G4 DNA signature. These data suggest that FANCJ contributes to an evolutionarily conserved pathway for the maintenance of genomic G/C tracts.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—Baculovirus expressing FANCJ-FLAG-HIS12 protein was produced from plasmid pDEST8-BRIP1 using the Bac to Bac Baculovirus Expression System protocols (Invitrogen). Expression and nickel affinity purification was essentially as described (12). Peak fractions were pooled and diluted with 20 mm Tris-HCl, pH 7.6, 10% glycerol to reduce the KCl concentration to 150 mm and incubated with FLAG antibody resin (Sigma) for 2 h at 4 °C. FLAG purification was carried out as described previously (13). BLM protein was purified as described previously (14).
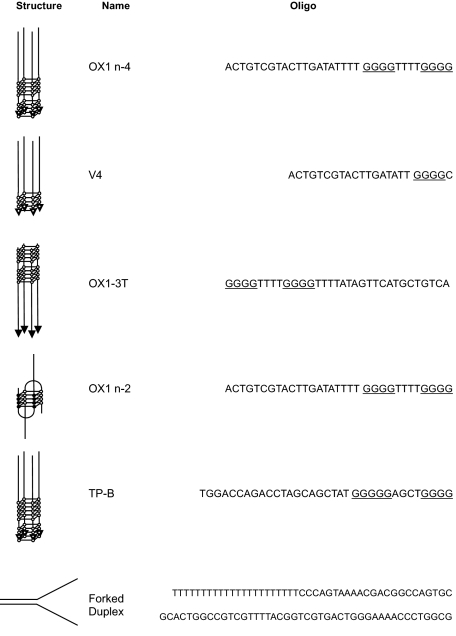
DNA Substrates and DNA Labeling—Synthetic oligonucleotides used this study are listed in Table 1. The G-quadruplex forming guanines are underlined.
TABLE 1.
Oligonucleotide substrates
Except for OX1-n2, quadruplex DNA substrates were formed by heating 300 mm of the appropriate oligo in 20 mm Tris-HCl, pH 7.6, 1 mm EDTA, and 1 m NaCl to 95 °C for 5 min followed by incubation overnight at 60 °C essentially as described previously (15–18). The bimolecular G4 substrate, OX1-n2, was formed as above but using 1 m KCl instead of NaCl. Branched and duplex substrates were prepared by mixing equimolar ratios of FK1 and FK2 (branched DNA) and OX1 and OX1C (Duplex), respectively, in buffer containing 10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm EDTA, heating to 95 °C for 4 min and cooling to room temperature over 1 h.
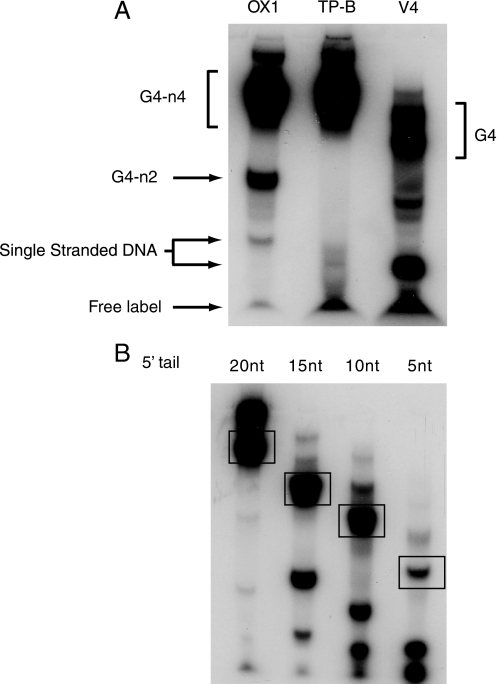
Single-stranded oligos (50 pmol) and G4 DNA (300 pmol) were 5′ end-labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (GE Healthcare) in reactions containing 70 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 5 mm dithiothreitol, 30 μCi of [γ-32P]ATP, and 10 units of kinase at 37 °C for 1 h. The labeled reaction DNAs were resolved on 10% TBE polyacrylamide gels and the bands excised following exposure to Super RX film (Fuji) (Fig. 1). The radiolabeled DNA was eluted from the excised bands overnight in TE buffer containing 10 mm KCl. The polyacrylamide was removed by centrifugation and the DNA in the supernatant precipitated with isopropyl alcohol. The purified substrates were resuspended in TE with 10 mm KCl and stored at –20 °C. G4 DNA made from OX1-3T was 3′-end labeled using terminal deoxynucleotide transferase (Roche) and [α-32P]ddATP, according to the manufacturer's instructions.
FIGURE 1.
Purification of DNA substrates. DNA substrates were annealed and end-labeled as described under “Experimental Procedures.” Substrates were next purified after separation by preparative polyacrylamide gel electrophoresis. A, OX-1 (bimolecular (n-2) and tetramolecular (n-4) forms as indicated), TP-B, and V4 G4 DNA substrates are shown. Unannealed single-stranded DNA is also marked. B, G4 DNA substrates with shortened 5′ single-stranded tails are indicated. Excised are denoted by the boxes.
DNA Unwinding Assays—Helicase reactions (20 μl) contained 40 mm Tris-HCl, pH 7.6, 50 mm NaCl, 2 mm dithiothreitol, 5 mm ATP, 5 mm MgCl2, 2% glycerol, 100 μg/ml bovine serum albumin and the indicated amounts of protein and nucleic acid substrate. Reactions were incubated at 37 °C for 30 min unless otherwise indicated, then terminated with 25 mm EDTA, 2 mg/ml proteinase K, and 0.5% SDS. Samples were analyzed on 10% nondenaturing TBE PAGE.
For competition assays, 2 nm FANCJ protein was incubated with the indicated concentrations of competitor DNA in reaction buffer without ATP and incubated for 10 min at 37 °C. We then added 0.5 nm 32P-labeled OX1 DNA and 5 mm ATP and incubated for a further 30 min at 37 °C before resolving reaction products by PAGE.
ATP Hydrolysis Assays—Proteins (10 pmol) were incubated with the indicated concentrations of OX1 G4 DNA, forked DNA, or DNA duplex for 1 h at 37 °C in helicase buffer containing 1 μmol of unlabeled ATP and 2 μmol of [γ-32P]ATP. Reactions were quenched with 17.5 mm EDTA. 1-μl samples were spotted onto polyethyleneimine-cellulose F thin layer chromatography plates (Merck) and resolved in 0.75 m KH2PO4. Free phosphate and ADP signals were visualized using a Typhoon PhosphorImager and quantified using ImageQuant software (Molecular Dynamics). Error bars represent the standard deviation of three independent experiments.
Comparative Genome Hybridization—Comparative genome hybridization (CGH) was performed on genomic DNA extracted from EUFA0030 fibroblasts (FA-J), PD20 fibroblasts (FA-D2), and human fetal fibroblast (control) cells. We used single array CGH design for whole human genome (hg18; NCBI Build 36), with isothermal probes selected from the Nimblegen hg18_tiling data base using a 6000-bp interval (mean 6253 bp, median 6270 bp) and a 250-bp window. Probe length varied from 50 to 75 bp. CGH was performed pairwise (FA-J versus human fetal fibroblast, FA-J versus FA-D2, and FA-D2 versus human fetal fibroblast) and in duplicate. Virtual arrays were created to summarize the raw data, and deleted regions were defined as a negative fold change between the datasets where the log-ratio was less than –0.5. To minimize the risk of false positives, these regions of interest were further refined to only include those in which more than 2 consecutive probes were deleted. This analysis identified 213 regions that are uniquely deleted in FA-J cells. To determine whether the breakpoints of regions that are uniquely deleted in FA-J cells are associated with G4 DNA signatures, we utilized the quadparser algorithm to detect sequences of the form d(G3+N1–7G3+N1–7-G3+N1–7G3+) (19). The exact breakpoints must lie in the 6 kb flanking the first and last deleted probe of each region, based on the average interval between probes. Hence quadparser analysis was performed on the sequence 6 kb up- and downstream of each of the known deleted regions. G4 DNA signatures were classified according to the strand orientation, as deletions are expected to initiate 3′ to a G4 signature and extend for variable lengths 5′ from this sequence. Statistical analysis was performed using a Hidden Markov model to show significance (p < 0.05) for the correlation between G4 DNA signatures oriented 3′ of the FA-J-specific deleted regions, compared with G4 signatures situated with the opposite orientation.
RESULTS
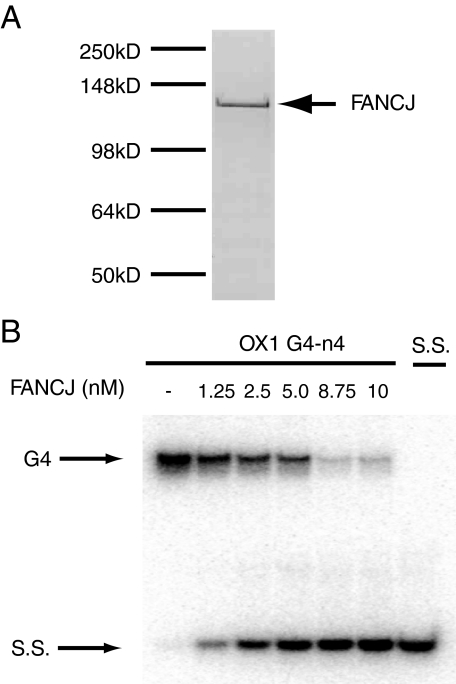
FANCJ Helicase Unwinds G4 DNA—To investigate whether human FANCJ is able to unwind G-quadruplex DNA structures we first expressed recombinant human FANCJ from baculovirus-infected Sf9 cells and purified the protein to homogeneity using nickel and FLAG affinity chromatography (Fig. 2A).
FIGURE 2.
FANCJ is an ATP-dependent G4 unwinding enzyme. A, recombinant FANCJ protein was expressed and purified from Sf9 cells. Protein was resolved by SDS-PAGE and visualized using simply blue stain (Invitrogen). B, FANCJ unwinds G4 DNA substrates. 32P-End-labeled OX1 G4 DNA was incubated with increasing concentrations of FANCJ (as indicated) in the presence of ATP. Unwinding of G4 DNA is detected by the production of a single-stranded oligonucleotide DNA product (S.S.). Reaction products were analyzed by polyacrylamide gel electrophoresis and visualized using a PhosphorImager (GE Healthcare).
G4 DNAs can form from a variety of DNA sequences matching the consensus signature (G3–5,N1–3G3–5,N1–3,G3–5N1–3-G3–5). In this study we used oligonucleotide G4 DNA substrates based on the G-rich sequences of the Oxytrichia nova telomere (OX1), the murine immunoglobulin switch region (TP-B), and a four-stranded G4 structure (V4) (16, 18, 20). After annealing to form different G4 DNA structures (Table 1), all substrates were end-labeled with 32P and purified to homogeneity using polyacrylamide gel electrophoresis (Fig. 1).
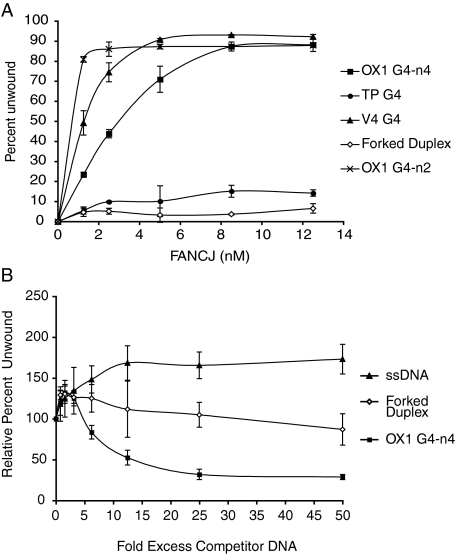
We incubated different G4 DNA substrates with various concentrations of FANCJ for 30 min at 37 °C, in the presence of ATP and resolved the reaction products using PAGE. We found that FANCJ unwound the tetramolecular G4 DNA substrates, OX1-n4 and V4, very efficiently, dissociating more than 80% of the substrate into single-stranded oligonucleotide product (Figs. 2B and 3A).
FIGURE 3.
FANCJ preferentially unwinds G4 structure. A, FANCJ unwinds G4 DNA but not forked duplex DNA. The graph shows the efficiency at which FANCJ unwinds different DNA substrates (as indicated). Reactions were performed as described above. B, unwinding of OX1 DNA is not competed by preincubation with forked duplex DNA or single-stranded DNA competitors. Experiments were performed using 2 nm FANCJ as described under “Experimental Procedures.” G4 unwinding was normalized to reactions containing no competitor (100%).
When annealed in the presence of potassium rather than sodium the OX1 oligonucleotide can also form a bimolecular G4 structure, which we refer to as OX1-n2, which is resolved from its tetramolecular form using PAGE (Table 1 and Fig. 1) (20). OX1-n2 was unwound by FANCJ with even greater efficiency than OX1-n4. Dissociation of all these substrates was complete with no intermediate products observed.
On the other hand only 10% of the TP-B DNA substrate was resolved to single-stranded DNA by FANCJ in the same period (Fig. 3A). It is known that G4 DNAs occur in multiple forms, depending on DNA sequence, length of G-tract, and available cation. Although the structure of TP DNA is unknown, the reduced efficiency at which FANCJ unwinds TP relative to OX1 most likely reflects differences in structure and stability of these G4 DNA substrates.
Substrate Specificity—We next compared the efficiency at which FANCJ unwinds G4 DNA compared with duplex and forked DNA substrates. We were surprised to find that whereas FANCJ unwound ∼90% of the G4 OX1-n4 substrate in 30 min, we were unable to detect unwinding of either duplex DNA or forked DNA substrates using a wide range of protein concentrations (Fig. 3A).
This result is at odds with Gupta et al. (7) who reported efficient unwinding of forked DNAs by FANCJ. Therefore, we repeated our experiment using substrates and conditions identical to those employed in this previous study. However, even under these conditions we failed to observe any unwinding of forked DNA by FANCJ. Therefore, in our hands FANCJ unwinds G4 DNA but not duplex or forked DNAs.
To investigate this further we performed competition assays in which 2 nm FANCJ was preincubated with either unlabeled forked, single-stranded or G4 competitor DNA. After 10 min 32P-labeled OX1-n4 DNA and ATP were added and reactions were incubated for a further 30 min at 37 °C to enable unwinding of OX1-n4 to occur. As expected, preincubation of FANCJ with increasing concentrations of unlabeled OX1-n4 competed efficiently for unwinding of 32P-labeled OX1-n4 substrate (Fig. 3B). On the other hand very little competition was observed with either ssDNA or forked DNA competitors, present at up to 50-fold molar excess (Fig. 3B). From this we conclude that FANCJ is indeed a highly specific DNA helicase, with a marked preference for unwinding G4 DNA.
We also note that unwinding of 32P-labeled G4 substrate by FANCJ was stimulated in the presence of low concentrations of all competitor DNAs. This might occur if the “active” form of FANCJ is a multimeric complex whose assembly is enhanced in the presence of DNA. This is currently under investigation.
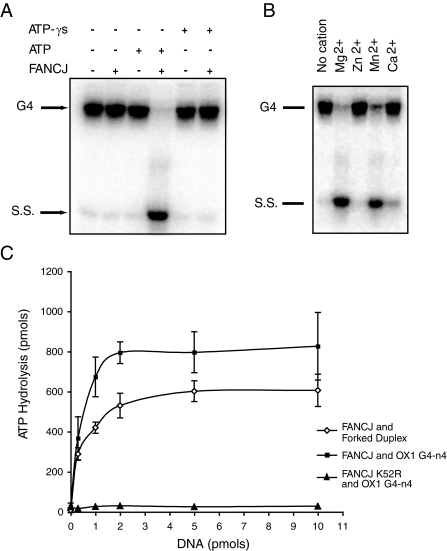
Uncoupled DNA Unwinding and ATP Hydrolysis—Unwinding of G4 DNA by FANCJ requires hydrolysis of ATP and a divalent cation, preferably Mg2+ (Fig. 4, A and B). Accordingly, we observed no dissociation of OX1-n4 substrate in the absence of ATP, in the presence of the slowly hydrolyzable ATP analog ATPγS, or with a mutant form of FANCJ (FANCJK52R) that is unable to hydrolyze ATP by virtue of a lysine to arginine substitution at residue 52 in its Walker A motif (Fig. 4C).
FIGURE 4.
G4 DNA unwinding requires ATP hydrolysis and magnesium ions. A, ATP requirement. Unwinding assays were performed as described in the legend Fig. 1, with 2 nm FANCJ but with no nucleotide, with 5 mm ATP or 5 mm ATPγS as indicated. B, cation requirement. G4 unwinding assay were performed as before with various cations (5 mm) as indicated. C, the ATPase function of FANCJ is stimulated by G4 and forked duplex DNA. ATP hydrolysis by FANCJ (10 pmol) was measured using thin layer chromatography and [γ-32P]ATP in the presence of different concentrations of forked duplex or OX1 G4 DNA as indicated. ATP hydrolysis of FANCJK52R protein in which the ATP binding site has been mutated was also measured.
Previous studies showed that ATP hydrolysis by FANCJ is stimulated in the presence of DNA (13). We next compared the efficiency of FANCJ ATPase activity in the presence of G4 and forked DNAs. We found that whereas both DNAs enhanced the ATPase activity of FANCJ, it was stimulated most in the presence of G4 DNA (Fig. 4C). Nevertheless, the fact that forked DNA stimulates hydrolysis of ATP but is not unwound by FANCJ suggests that the ATPase function of FANCJ is not solely coupled to DNA unwinding.
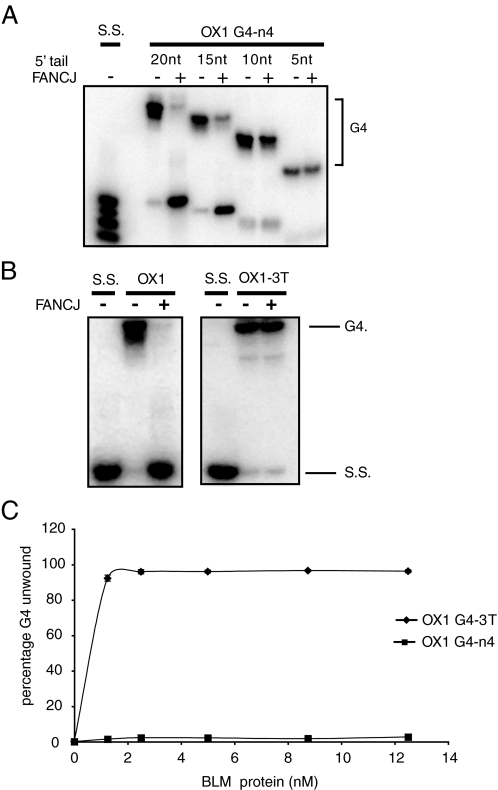
FANCJ Unwinds G4 DNA in a 5′–3′ Direction—We have demonstrated that FANCJ specifically recognizes G4 DNA and that single-stranded DNA does not compete for G4 DNA in an unwinding assay. Nevertheless, we observed that a region of single-stranded DNA immediately 5′ to the G4 structure is required by FANCJ to efficiently unwind OX1-n4 substrate. Substrates with 5′-ssDNA tails of 15 or 20 nucleotides (nt) were completely dissociated by FANCJ, whereas G4 DNA with 5′-ssDNA tails of 5 or 10 nt was not unwound by FANCJ (Fig. 5A). Furthermore, G4 substrate with a 3′-ssDNA tail (OX1–3T) was not unwound by FANCJ, indicating that strand displacement occurs 5′ to 3′ (Fig. 5B). Together these data suggest a mechanism in which FANCJ binds to a region of single-stranded DNA immediately adjacent to the G4 structure and in the presence of ATP, unwinds it in a 5′–3′ direction.
FIGURE 5.
FANCJ unwinds G4 DNA with 5′–3′ polarity. A, FANCJ-mediated unwinding of G4 DNA requires a single-stranded tail of ∼15 nt. FANCJ was incubated with 32P-end-labeled OX1 G4 DNA substrates that have ssDNA tails of different lengths (as indicated). Reactions were carried out as described in the legend to Fig. 1 and under “Experimental Procedures.” Whereas OX1 DNA substrates with tails of 15 or 20 nt were efficiently unwound, those with ssDNA tails of 5 or 10 nt were not unwound by FANCJ. Denatured substrates are shown as markers (S.S.) B, FANCJ unwinds G4 DNA only when the ssDNA tail is positioned 5′ to the G4 structure (OX1), indicating a 5′–3′ polarity. OX1 substrate with a 3′-ssDNA tail (OX1–3T) was not unwound. All reactions were performed under standard reaction conditions described under “Experimental Procedures.” C, BLM efficiently unwinds G4 DNA in the 3′ to 5′ direction. Reactions contained 2.5 nm OX1–3T and the indicated concentrations of BLM and were carried out for 30 min at 37 °C before products were resolved by SDS-PAGE and visualized using a PhosphorImager.
Previous studies identified members of the RecQ family of DNA helicases, including BLM protein, as G4 DNA helicases (18). Therefore, we next compared the G4 DNA unwinding activities of BLM and FANCJ. As expected, BLM unwound G4 substrate with a 3′-ssDNA tail but not that with a 5′-tail, confirming the 3′–5′ directionality of this enzyme (data not shown). Moreover, unwinding of OX-3T by BLM was greater than that of FANCJ on OX-1 under identical conditions (Fig. 5C).
FA-J Cells Accumulate Deletions at Genomic Sequences with G4 DNA Signature—In C. elegans defects in the FANCJ homolog Dog-1 correlate with the accumulation of deletions at G-tracts with the G4 DNA signature. We reasoned that this might also be true in human cells from FA-J patients.
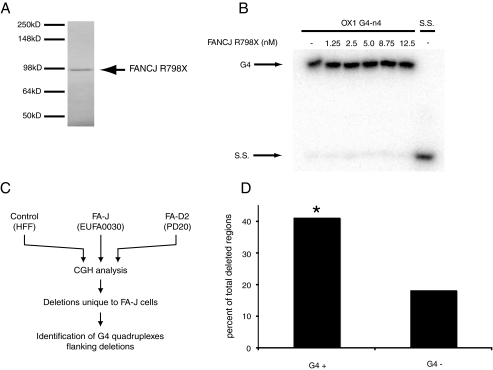
The FA-J patient cell line, EUFA0030, encodes a truncated form of FANCJ due to the substitution of a stop codon in place of arginine at residue 798. This truncation results in loss of helicase domain VII. Patients with this mutation have characteristic FA symptoms. To analyze this mutant protein we expressed and purified FANCJR798X to homogeneity in Sf9 cells (Fig. 6A). As expected, FANCJR798X was defective in ATP hydrolysis and unable to dissociate G4 DNA substrates in vitro (Fig. 6B).
FIGURE 6.
FA-J cells accumulate deletions in regions containing G4 DNA. A, mutant FANCJR798X protein was expressed and purified from Sf9 cells. Protein was resolved by SDS-PAGE and visualized using simply blue stain (Invitrogen). B, FANCJR798X does not unwind G4 DNA substrate in vitro. 32P-End-labeled OX1 G4 DNA was incubated with increasing concentrations of FANCJ (as indicated) in the presence of ATP. Migration of the expected single-stranded oligonucleotide DNA product is indicated (S.S.). Reaction products were analyzed by polyacrylamide gel electrophoresis and visualized using a PhosphorImager (GE Healthcare). C, schematic of array CGH analysis of control, FA-J, and FA-D2 cell lines and subsequent analysis of FA-J-specific deletion breakpoint regions for sequences predicted to form G4 quadruplexes. D, G4 quadruplex DNA flanks deletions in FA-J cells, in an orientation specific manner. 6 kb of sequence up- and downstream of each deletion observed by CGH as specific to FA-J cells was analyzed using the Quadparser algorithm to identify putative G4 DNA signatures. Deletions that associate with G4 DNA signatures were then classified by strand orientation. G4+ designates G4 DNA signatures that lies 3′ to a deleted region, and G4– are those regions that are associated with a G4 DNA signatures situated 5′ to the deleted region. Statistical analysis was performed using a Hidden Markov model to show significance (*, p = 0.04) for the correlation between G4+ DNA signatures and FA-J-specific deleted regions.
We next used CGH to identify regions of the genome that are deleted in the FA-J patient cell lines EUFA0030 but not in either PD20 (deficient in FANCD2) or human fetal fibroblast cells (wild-type; Fig. 6C). To minimize the risk of false positives, only regions that were deleted for two or more consecutive CGH probes were considered for further analysis. The Quadparser algorithm (19) was used to investigate whether the sequence immediately up- or downstream of these deleted regions had the potential to form G4 quadruplexes. This algorithm was designed to identify sequences of the form d(G3+N1–7-G3+N1–7G3+N1–7G3+), which are predicted to fold into stable G-tetrad stacks under physiological conditions.
From the 213 genomic deletions identified as unique to FA-J, we identified a bias toward breakpoint regions adjacent to DNA sequences with a G4 DNA signature (Fig. 6D). This bias was most pronounced among larger deletions (approximately 36 kb or more), where nearly 75% possess sequences with the G4 signature in the vicinity of the breakpoint.
In C. elegans, dog-1 mutants genomic G-tract deletions associated with G4 signature DNA occur with a distinct polarity, with the 3′-breakpoint within the G4 signature DNA, extending 5′ beyond the signature region (i.e. the deletion has 3′–5′ orientation with respect to the G4 signature) (8). Although in FA-J cells, it was not possible to identify exact breakpoint there was a 4-fold bias in G4 signatures situated 3′ of the deletion compared with 5′ of the deleted region (Fig. 6D).
We conclude that in human cells the expression of a mutated form of FANCJ, which is unable to unwind G4 structures in vitro, is consistent with the accumulation of genomic deletions in the vicinity of sequences with G4 signature with a polarity mirroring that observed in C. elegans dog-1 mutants.
DISCUSSION
Using model oligonucleotide substrates we have demonstrated that FANCJ is a structure-specific DNA helicase with a strong preference for unwinding G4 DNA. We also find that cells from human patients lacking FANCJ exhibit large deletions in regions of the genome matching the G4 DNA signature. Together these findings suggest that FANCJ may function to preserve genome stability through the resolution of G4 secondary structures formed in genomic G/C tracts.
It was reported that FANCJ unwinds forked DNA substrates in vitro (7, 21). However, we find that the helicase activity of FANCJ is highly specific for certain types of G4 DNA. We detected very little unwinding of either forked or duplex DNA substrates by FANCJ over a wide range of protein concentrations, even using substrate and conditions identical to those used in previous reports (7). Furthermore, we found that in competition assays FANCJ exhibits a preference of more than 50-fold for G4 DNA compared with forked DNA substrate. We are currently unable to explain this discrepancy with the earlier reports except to note that the ratio of protein to DNA substrate used in previous studies was higher than used in our assay. It is possible that under these conditions the separation of DNA strands, which occurs upon binding of certain helicases to DNA, may destabilize the substrate sufficiently to cause dissociation of DNA strands.
Although FANCJ unwinds G4 structures specifically and does not stably bind to single-stranded DNA in a competition assay, unwinding of G4 DNA requires a region of ssDNA (>11 nt or more) immediately 5′ of the G4 structure (21). Accordingly, FANCJ unwinds G4 structures with a defined 5′–3′ polarity. We envisage that FANCJ may bind transiently to ssDNA but that this association is stabilized by additional stronger specific interactions with G4 DNA.
Unwinding of G4 DNA by FANCJ is also accompanied by hydrolysis of ATP. However, because forked DNA also stimulates hydrolysis of ATP by FANCJ without being unwound, these functions may not be completely coupled. One explanation is if ATP hydrolysis is required both for translocation of FANCJ along single-stranded DNA and also for destabilization of G4 DNA ahead of the enzyme (22).
What then, is the function of FANCJ in cells? Our work supports genetic studies in C. elegans, suggesting that FANCJ functions to promote the stability of G4 DNA signature sequences in the genome (8, 11). We show clearly that the human FA cell line EUFA0030 encodes a mutated form of FANCJ, which is unable to dissociate G4 DNA in vitro and like dog-1 mutant worms, EUFA0030 cells accumulate deletions at or near sequences matching the G4 DNA signature. Although we have so far been unable to map the breakpoints of the large deletions in FA-J cells it is of note that similar to those observed in nematodes, they exhibit a bias toward deletions terminating to the 5′ side of G-tracts (8).
One notable difference between the deletions found in FA-J cells and those in C. elegans is their size. Although our CGH strategy did not enable us to identify small deletions (less than 6 kb), the deletions we scored as unique in FA-J cells are very much larger (up to 80 kb) than those observed in C. elegans, which are only a few hundred base pairs. One explanation for this phenomenon is that, as a result of self-fertilization in C. elegans, deletions will become homozygous. Consequently it is likely that only small, non-essential, deletions would be tolerated and maintained in viable progeny. Larger deletions would be prone to target essential genes, and therefore would be selected against. In contrast, the deletions identified here in FA-J cells are heterozygous (data not shown), and therefore, are able to persist.
More recently Zhao et al. (23) reported that in addition to small deletions, dog-1 mutant C. elegans exhibit a broader spectrum of mutations including duplications and complex rearrangements. Further investigation of FA-J mutant cells should reveal whether a similar situation exists in humans.
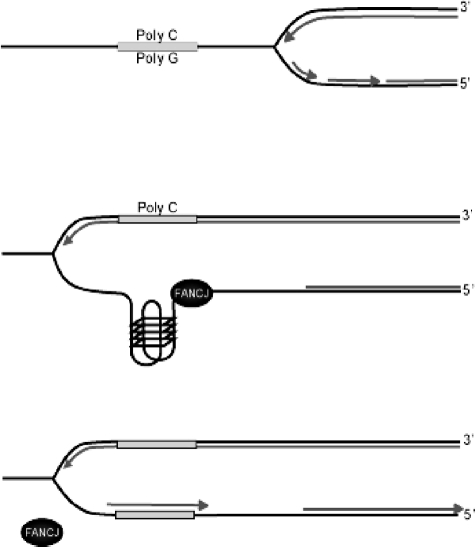
The polarity of the deletions found in dog-1 mutants led Cheung et al. (8) to speculation that DOG-1 unwinds G4 DNA structures that form on the lagging strand during replication. Although currently biochemical evidence for unwinding of G4 DNA by DOG-1 is lacking, our data support a similar model involving FANCJ in human cells (Fig. 7). Moreover, the 5′–3′ polarity of the FANCJ helicase is entirely consistent with resolution of G4 structures targeted to the lagging strand during replication.
FIGURE 7.
Model depicting the role of FANCJ in the maintenance of G-tracts. FANCJ resolves G4 DNA during replication. This model is a modification of that proposed by Cheung et al. (8) for DOG-1 in C. elegans. During replication in regions of the genome with long G/C tracts, the separation of DNA strands reveals stretches of ssDNA with the potential to form stable G4 DNA structures that may cause a block in DNA synthesis. In wild type cells FANCJ resolves this potential replication block by unwinding G4 DNA in a 5′–3′ direction, enabling synthesis to continue. In the absence of FANCJ/DOG-1 the G4 sequence is most likely excised and resected by nucleases and DNA rejoined by cellular DNA repair machinery giving rise to a deletion.
FANCJ is one of several G4 unwinding enzymes identified to date. Most notable are members of the RecQ family of helicases, which includes RecQ in Escherichia coli, Sgs1 of yeast, and the human proteins BLM and WRN (18). These proteins all function in the resolution of DNA structures that arise during DNA replication, recombination, and repair.
Apart from the polarity of G4 unwinding, BLM and FANCJ have rather similar biochemical properties. In our experiments, BLM unwound G4 substrate with opposite (3′–5′) polarity but similar specificity to FANCJ. However, genetic studies in C. elegans demonstrate that the homologs of BLM and WRN contribute to G/C tract stability only as a back-up pathway, when DOG-1 is absent (10). For BLM, this probably involves its function in replication restart at stalled or damaged replication forks (24) or through the resolution of intermediates in homologous recombination (25). Therefore, we think it likely that FANCJ/DOG-1 plays an important role as the first line of defense against potentially hazardous G4 DNA structures.
Although G4 DNA structures have been identified at telomeres (26) there is no strong evidence that FANCJ functions in the maintenance of telomere length. We think it more likely that specialized G4 unwinding enzymes are required for telomere stability during elongation (26). Therefore, it is noteworthy that an ortholog of FANCJ, called Rtel, is required in murine cells for telomere maintenance and therefore maybe a candidate for a telomere-specific G4 helicase (27).
How does the unwinding of G4 DNA by FANCJ relate to its role in the FA pathway? Recent evidence suggests that the role of DOG-1 in the FA pathway is genetically distinct from its function in G/C tract stability (9). Likewise, we have reported previously that, in DT40 FANCJ has a function additional to its role in the repair of DNA cross-links through the FA pathway (28). Consistent with this view G-tract deletions identified here are enriched in FA-J cells compared with FA-D2.
In summary we have identified a novel biochemical activity for FANCJ, as a structure-specific DNA helicase with a preference for G4 DNA that is consistent with its role in an evolutionarily conserved pathway for the maintenance of potentially unstable genomic G/C tracts (Fig. 4C). Furthermore, we show for the first time how a single component of the Fanconi anemia pathway can function to promote the maintenance of genome stability.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FA, Fanconi anemia; ssDNA, single-stranded DNA; CGH, comparative genome hybridization; ATPγS, adenosine 5′-O-(thiotriphosphate); nt, nucleotide.
References
- 1.Seal, S., Thompson, D., Renwick, A., Elliott, A., Kelly, P., Barfoot, R., Chagtai, T., Jayatilake, H., Ahmed, M., Spanova, K., North, B., McGuffog, L., Evans, D. G., Eccles, D., Easton, D. F., Stratton, M. R., and Rahman, N. (2006) Nat. Genet. 38 1239–1241 [DOI] [PubMed] [Google Scholar]
- 2.Levitus, M., Waisfisz, Q., Godthelp, B. C., de Vries, Y., Hussain, S., Wiegant, W. W., Elghalbzouri-Maghrani, E., Steltenpool, J., Rooimans, M. A., Pals, G., Arwert, F., Mathew, C. G., Zdzienicka, M. Z., Hiom, K., De Winter, J. P., and Joenje, H. (2005) Nat. Genet. 37 934–935 [DOI] [PubMed] [Google Scholar]
- 3.Litman, R., Peng, M., Jin, Z., Zhang, F., Zhang, J., Powell, S., Andreassen, P. R., and Cantor, S. B. (2005) Cancer Cell 8 255–265 [DOI] [PubMed] [Google Scholar]
- 4.Levran, O., Attwooll, C., Henry, R. T., Milton, K. L., Neveling, K., Rio, P., Batish, S. D., Kalb, R., Velleuer, E., Barral, S., Ott, J., Petrini, J., Schindler, D., Hanenberg, H., and Auerbach, A. D. (2005) Nat. Genet. 37 931–933 [DOI] [PubMed] [Google Scholar]
- 5.Joenje, H., and Patel, K. J. (2001) Nat. Rev. 2 446–457 [DOI] [PubMed] [Google Scholar]
- 6.Cantor, S. B., Bell, D. W., Ganesan, S., Kass, E. M., Drapkin, R., Grossman, S., Wahrer, D. C., Sgroi, D. C., Lane, W. S., Haber, D. A., and Livingston, D. M. (2001) Cell 105 149–160 [DOI] [PubMed] [Google Scholar]
- 7.Gupta, R., Sharma, S., Sommers, J. A., Jin, Z., Cantor, S. B., and Brosh, R. M., Jr. (2005) J. Biol. Chem. 280 25450–25460 [DOI] [PubMed] [Google Scholar]
- 8.Cheung, I., Schertzer, M., Rose, A., and Lansdorp, P. M. (2002) Nat. Genet. 31 405–409 [DOI] [PubMed] [Google Scholar]
- 9.Youds, J. L., Barber, L. J., Ward, J. D., Collis, S. J., O'Neil, N. J., Boulton, S. J., and Rose, A. M. (2008) Mol. Cell. Biol. 28 1470–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youds, J. L., O'Neil, N. J., and Rose, A. M. (2006) Genetics 173 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruisselbrink, E., Guryev, V., Brouwer, K., Pontier, D. B., Cuppen, E., and Tijsterman, M. (2008) Curr. Biol. 18 900–905 [DOI] [PubMed] [Google Scholar]
- 12.Mallery, D. L., Vandenberg, C. J., and Hiom, K. (2002) EMBO J. 21 6755–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantor, S., Drapkin, R., Zhang, F., Lin, Y., Han, J., Pamidi, S., and Livingston, D. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr., Bohr, V. A., and Hickson, I. D. (2001) Nucleic Acids Res. 29 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen, D., and Gilbert, W. (1992) Biochemistry 31 65–70 [DOI] [PubMed] [Google Scholar]
- 16.Sen, D., and Gilbert, W. (1988) Nature 334 364–366 [DOI] [PubMed] [Google Scholar]
- 17.Sen, D., and Gilbert, W. (1990) Nature 344 410–414 [DOI] [PubMed] [Google Scholar]
- 18.Sun, H., Karow, J. K., Hickson, I. D., and Maizels, N. (1998) J. Biol. Chem. 273 27587–27592 [DOI] [PubMed] [Google Scholar]
- 19.Huppert, J. L., and Balasubramanian, S. (2005) Nucleic Acids Res. 33 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, F. W., and Feigon, J. (1992) Nature 356 164–168 [DOI] [PubMed] [Google Scholar]
- 21.Wu, Y., Shin-ya, K., and Brosh, R. M., Jr. (2008) Mol. Cell. Biol. 28 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soultanas, P., Dillingham, M. S., Wiley, P., Webb, M. R., and Wigley, D. B. (2000) EMBO J. 19 3799–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao, Y., Tarailo-Graovac, M., O'Neil, N. J., and Rose, A. M. (2008) DNA Repair 7 1846–1854 [DOI] [PubMed] [Google Scholar]
- 24.Davies, S. L., North, P. S., and Hickson, I. D. (2007) Nat. Struct. Mol. Biol. 14 677–679 [DOI] [PubMed] [Google Scholar]
- 25.Wu, L., and Hickson, I. D. (2003) Nature 426 870–874 [DOI] [PubMed] [Google Scholar]
- 26.Paeschke, K., Simonsson, T., Postberg, J., Rhodes, D., and Lipps, H. J. (2005) Nat. Struct. Mol. Biol. 12 847–854 [DOI] [PubMed] [Google Scholar]
- 27.Ding, H., Schertzer, M., Wu, X., Gertsenstein, M., Selig, S., Kammori, M., Pourvali, R., Poon, S., Vulto, I., Chavez, E., Tam, P. P., Nagy, A., and Lansdorp, P. M. (2004) Cell 117 873–886 [DOI] [PubMed] [Google Scholar]
- 28.Br6192idge, W. L., Vandenberg, C. J., Franklin, R. J., and Hiom, K. (2005) Nat. Genet. 37 953–957 [DOI] [PubMed] [Google Scholar]