Abstract
The eukaryotic translation initiation factor (eIF) 4B promotes the RNA-dependent ATP hydrolysis activity and ATP-dependent RNA helicase activity of eIF4A and eIF4F during translation initiation. eIF4B also helps to organize the assembly of the translational machinery through its interactions with eIF4A, eIF4G, eIF3, the poly(A)-binding protein (PABP), and RNA. Although the function of eIF4B is conserved among plants, animals, and yeast, eIF4B is one of the least conserved of initiation factors at the sequence level. Mammalian eIF4B is a constitutive dimer; however, conflicting reports have suggested that plant eIF4B may exist as a monomer or a dimer. In this study, we show that eIF4B from wheat can form a dimer and we identify the region responsible for its dimerization. Zinc stimulated homodimerization of eIF4B and bound eIF4B with a Kd of 19.7 nm. Zinc increased the activity of the eIF4B C-terminal RNA-binding domain specifically. Zinc promoted the interaction between eIF4B and PABP but not the interaction between eIF4B and eIF4A or eIFiso4G, demonstrating that the effect of zinc was highly specific. The interaction between PABP and eIFiso4G was also stimulated by zinc but required significantly higher levels of zinc. Interestingly zinc abolished the ability of eIFiso4G to compete with eIF4B in binding to their overlapping binding sites in PABP by preferentially promoting the interaction between eIF4B and PABP. Our observations suggest that wheat eIF4B can dimerize but requires zinc. Moreover zinc controls the partner protein selection of PABP such that the interaction with eIF4B is preferred over eIFiso4G.
Protein synthesis is an essential step in gene expression, and several global and specific regulatory mechanisms targeting translation have evolved to control the level of proteins. As the recruitment of a ribosome to an mRNA is typically the rate-limiting step in translation (1), regulation of protein synthesis occurs primarily during translation initiation. Several eukaryotic translation initiation factors (eIFs)2 are required to promote the binding of the 40 S ribosomal subunit to an mRNA and to assemble the 80 S ribosome at the appropriate initiation codon (2, 3). The eIF4 group of initiation factors is particularly important for the recruitment of the ribosome. eIF4F binds to the 5′-cap structure of an mRNA and is composed of eIF4E, which is responsible for cap binding, and eIF4G, a modular scaffolding protein that interacts with eIF4E, eIF4A (an RNA helicase), eIF3 (required for 40 S binding to an mRNA), and the poly(A)-binding protein (PABP) (4). The interaction between PABP and eIF4G results in the circularization of an mRNA and stimulates its translation by promoting 40 S recruitment (5, 6). In addition to eIF4F, plants contain an isoform, called eIFiso4F, which is composed of eIFiso4E and eIFiso4G (7). In its function as a DEA(D/H) helicase, eIF4A uses the energy from ATP hydrolysis to unwind any secondary structure present in the 5′-leader of an mRNA that would otherwise inhibit 40 S subunit scanning (2). eIF4B enhances the helicase activity of eIF4A and eIF4F (8, 9) and mediates mRNA binding to 40 S subunit likely through its interaction with eIF3 (10). The two RNA-binding domains in mammalian eIF4B have been proposed to bind mRNA and ribosomal RNA simultaneously, which also may contribute to 40 S subunit recruitment to an mRNA (11). Like its animal ortholog, plant eIF4B can bind two RNA molecules simultaneously but differs from mammalian eIF4B in that it contains three RNA-binding domains (12). In addition to their interaction with eIF4G, mammalian and plant PABP also interact with eIF4B (12–14), an interaction that increases the affinity of PABP for poly(A) RNA (13). A single interaction domain for eIF4B is present within a 32-amino acid region representing the C-terminal end of RNA recognition motif (RRM) 1 of PABP (15). Two eIFiso4G interaction domains are present in PABP: the first maps to a 36-amino acid region encompassing the C-terminal end of RRM1, the linker region, and the N-terminal end of RRM2 to which eIFiso4G binds strongly, whereas the second site maps to RRM3–4 to which eIFiso4G binds weakly (15). The interaction domain for eIF4B substantially overlaps the N-proximal eIFiso4G interaction domain in PABP. Consequently eIF4B and eIFiso4G exhibit competitive binding to PABP (15), supporting the overlapping nature of their interaction domains.
In addition to its interaction with PABP, eIF4B interacts with eIF4G (or eIFiso4G), eIF4A, and, as mentioned previously, eIF3 and RNA. Thus, eIF4B helps to organize the assembly of the translation initiation machinery. Mammalian eIF4B is known to be a dimer in which the central DRYG-rich domain (16) mediates eIF4B self-association as well as its interaction with eIF3a (10). Although wheat eIF4B has been reported to be a dimer (17), no self-association was detected in a direct pulldown assay (12). Interestingly the RNA binding activity of each of the three wheat eIF4B RNA-binding domains depends on the dimerization of the protein (12). The observation that this aspect of eIF4B function is at variance with the observed lack of self-association has left unresolved the issue of whether or not plant eIF4B can dimerize.
In this study, we present evidence that homodimerization of wheat eIF4B is dependent on a physiological concentration of zinc. The domain responsible for homodimerization mapped to a region encompassing the C-terminal RNA-binding domain. Zinc also stimulated the activity of the C-terminal RNA-binding domain as well as the interaction of PABP to its binding sites on either side of the C-terminal RNA-binding domain in eIF4B. Zinc did not stimulate the activity of the two other RNA-binding domains or the interaction of eIF4B with eIF4A or eIFiso4G, observations suggesting that the effect of zinc was specific to promoting the self-association of eIF4B, its interaction with PABP, and the activity of its C-terminal RNA-binding domain. Because zinc preferentially promoted the interaction between eIF4B and PABP, eIFiso4G was unable to compete with eIF4B in binding to their overlapping binding sites in PABP in the presence of zinc. These results suggest that zinc controls homodimerization of wheat eIF4B as well as the selection of partner proteins of PABP.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Protein Expression—Constructs for the expression of full-length or truncated eIFiso4G, eIF4B, and PABP were described previously (12). Additional constructs expressing eIF4B polypeptides were generated by PCR amplification of the appropriate coding region that was introduced into the BamHI/EcoRI sites of pGEX-2TK (GE Healthcare) to generate GST-tagged protein, into the NdeI/BamHI sites of pET19b (Novagen) to generate His-tagged protein, or into the NcoI/XhoI sites of pET19b to generate non-tagged protein. Point mutations were generated by site-directed PCR mutagenesis or using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega). All constructs were confirmed by sequencing.
RNA Binding Assay—Truncated or full-length GST or His-tagged eIF4B was expressed in Escherichia coli BL21 cells following induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and crude extract was made in Buffer B-100 (200 mm HEPES, pH 7.2, 100 mm KCl, 10% glycerol, 2 mm dithiothreitol) without EDTA but with protease inhibitor mixture (Sigma). Poly(A)-Sepharose 4B (Sigma) was equilibrated in washing buffer (Buffer B-100 without EDTA but with 0.1% Triton X-100). The crude extract was incubated with the resin at 4 °C for 20 min with shaking. The resin was washed extensively with washing buffer, and the bound protein was resuspended in SDS sample buffer. The supernatant was analyzed by SDS-PAGE, and the gel was stained with Coomassie.
To test the metal ion requirement for RNA binding, divalent metal ions were tested by their addition as the chloride salt at the indicated concentration to the standard RNA binding assay reaction containing GST- or His-tagged eIF4B. Following incubation for 20 min at 4 °C, the poly(A)-Sepharose 4B resin was washed, and the extent of RNA binding was determined by SDS-PAGE. Dithiothreitol was omitted in reactions containing CoCl2, FeCl2, or NiCl2 to avoid reduction of the metal.
Protein Interaction—Protein interactions were analyzed as described previously (12) with some modification. GST fusion protein was added to glutathione-Sepharose 4B resin (Amersham Biosciences) washed three times with precooled Buffer B-100 (supplemented with 0.1% Triton X-100). Following incubation with shaking at 4 °C for 1 h, the resin was collected by centrifugation, and the supernatant was removed. Recombinant prey protein from crude extract was added to the resin and incubated for 2 h. The resin was washed four times with Tween phosphate-buffered saline (TPBS; 0.1% Tween 20, 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.4 mm KH2PO4) supplemented with 1 mm dithiothreitol. Bound protein was released following the addition of SDS sample buffer to the resin. Following heating and centrifugation, the protein was resolved by SDS-PAGE, and the gel was stained or used for Western analysis.
To eliminate apparent protein interaction resulting from RNA tethering, RNase A was added to the binding reaction assay. RNase A was added to the resin containing the bait protein and incubated at 37 °C for 20 min. The prey protein was treated similarly. The prey protein and metal ions were then added, and the reaction was incubated at 4 °C for an additional 2 h. After extensive washing, bound protein was released following the addition of SDS sample buffer to the resin. The protein was resolved by SDS-PAGE, transferred to membrane, and used for Western analysis.
Western Analysis—Protein was transferred to 0.22-mm polyvinylidene difluoride membrane by electroblotting. The membranes were blocked in 5% milk, 1% NaN3 in TPBS followed by incubation with antiserum in TPBS with 1% milk for 1.5 h. Membranes were washed twice with TPBS and incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies (Southern Biotechnology Associates, Birmingham, AL) diluted to 1:10,000 for 1 h. The blots were washed twice with TPBS, and the signal was detected typically between 1 and 15 min using chemiluminescence (Pierce). Anti-His antiserum was purchased from Santa Cruz Biotechnology Inc. eIF4A and eIFiso4G antisera (a generous gift of K. Browning) were used at 1:1000 dilution.
Fluorescence Titration Measurements—Fluorescence measurements were performed on a SPEX Fluorolog-τ-2 spectrofluorometer equipped with a high intensity (450-watt) xenon arc lamp at 22 °C. Excitation and emission slit widths of 2.0 and 3 mm were used, and a path length of 1.0 cm was used. All chemicals were molecular biology grade. Binding experiments were performed using 50 and 150 nm full-length eIF4B protein to which increasing amounts of ZnCl2 and MgCl2 were added individually in separate experiments. To study the competition between Zn2+ and Mg2+ metal ion for the protein binding site, eIF4B was titrated with MgCl2 in the presence of 1000 nm ZnCl2. The background emission was eliminated by subtracting the signal from buffer containing an appropriate quantity of substrate. Data were averages of three or more titrations. Experiments using eIF4B were performed using an excitation wavelength of 280 nm to monitor the emission at 332 nm. The binding buffer used for all fluorescence studies consisted of 20 mm HEPES, 100 mm KCl (pH 7.6).
The fluorescence was normalized by using the formula fb = (F0 – Fobs)/(Fobs – F∞)(F0/F∞) + (F0 – Fobs), which is directly related to the fraction of protein bound (fb), where Fobs is the observed fluorescence, F0 is the fluorescence observed in the absence of any metal ion (that is the fluorescence of the protein), and F∞ is the fluorescence at saturation. The normalized fluorescence was used to determine the equilibrium dissociation constant (Kd). The details of the data fitting are described elsewhere (9, 18, 20). Nonlinear least squares fitting of the data was performed using KaleidaGraph software Version 4.
ATPase Activity Assay—20-μl assays containing 15 mm HEPES-KOH (pH 7.0), 100 mm KCl, 0.1 mm MgCl2, 1 mm dithiothreitol, 100 μm non-labeled and [γ-32P]ATP (specific activity, 3000–5000 cpm/pmol), and 0.5 μg of poly(U) RNA as described previously (21) were performed in the presence or absence of 0.1 mm ZnCl2 and the indicated amounts of purified initiation factors. Reactions were incubated at 37 °C for 40 min and stopped by the sequential addition at 4 °C of 0.5 ml of a mixture of 20 mm silicotung-state and 20 mm sulfuric acid, 1.2 ml of 1 mm potassium phosphate (pH 7.0), 0.5 ml of 5% ammonium molybdate in 4 m sulfuric acid, 300 μl of a 1:l mixture of 5% trichloroacetic acid and 100% acetone, and 2 ml of a 1:l mixture of isobutyl alcohol and benzene. The tubes were then mixed and centrifuged at 1500 rpm for 3 min to separate the phases. Free phosphate from 0.5 ml of the organic phase was measured using scintillation counting from which the background was subtracted, and the picomoles of ATP hydrolyzed were quantitated.
RESULTS
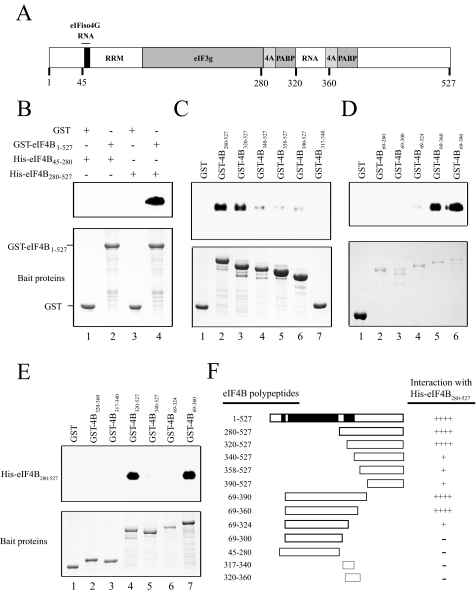
Identification of the eIF4B Dimerization Domain—Homodimerization of animal eIF4B requires a DRYG-rich domain (10), whereas the corresponding region in wheat eIF4B is rich only in glycine and aspartic acid, and its sequence is poorly conserved among plant homologs. To determine whether plant eIF4B can dimerize, full-length GST-tagged wheat eIF4B was used to pull down His-tagged eIF4B polypeptides, which were then detected using anti-His antiserum. GST-eIF4B did not interact with His-eIF4B45–280 (Fig. 1B, lane 2), which contains the region corresponding to the animal DRYG-rich dimerization domain, but did interact with His-eIF4B280–527 (Fig. 1B, lane 4). Neither His-eIF4B45–280 nor His-eIF4B280–527 bound to GST alone (Fig. 1B, lanes 1 and 3, respectively). These data indicated that the region corresponding to the animal DRYG-rich dimerization domain was not involved in the self-association of wheat eIF4B, but rather a region within the C-terminal half of eIF4B was required.
FIGURE 1.
Self-association of wheat eIF4B. In A, the organization of the protein- and RNA-binding domains in wheat eIF4B is shown. In B–E, full-length eIF4B (i.e. residues 1–527) or eIF4B polypeptides were expressed as GST fusion proteins in E. coli (bottom panels) and used in pulldown assays. The region of eIF4B included in each polypeptide is indicated numerically by the residues included. In B, either the N-terminal half of eIF4B (His-eIF4B45–280)(lanes 1 and 2) or the C-terminal half of eIF4B (His-eIF4B280–527)(lanes 3 and 4) was used as the prey protein, whereas in C–E, His-eIF4B280–527 was used as the prey protein. Binding of each His-eIF4B polypeptide to the indicated GST fusion eIF4B polypeptides was determined following binding of each GST fusion construct to glutathione-Sepharose resin. Bound His-eIF4B was detected using SDS-PAGE and Western analysis with anti-His antiserum (top panel). GST was used as a negative control. In F, a summary of eIF4B self-association is shown. RNA-binding domains are indicated by black boxes. Strength of binding of His-eIF4B280–527 to the eIF4B polypeptides is indicated by the number of pluses. –, no detectable binding. 4A, eIF4A; 4B, eIF4B.
To more precisely map the region responsible for homodimerization, a series of N-terminal deletions were tested for their ability to bind His-eIF4B280–527. His-eIF4B280–527 was able to bind to GST-eIF4B280–527 and GST-eIF4B320–527 (Fig. 1C, lanes 2 and 3, respectively) but bound further N-terminal truncations significantly less, e.g. GST-eIF4B340–527, GST-eIF4B358–527, and GST-eIF4B390–527 (Fig. 1C, lanes 4–6, respectively). This suggested that the N-terminal boundary of the dimerization domain is located between residues 320 and 340. This is similar to the N-terminal border of the C-terminal RNA-binding domain (12). A construct containing a region sufficient for RNA binding, i.e. GST-eIF4B317–340 (see below), however, failed to bind His-eIF4B280–527 (Fig. 1C, lane 7), suggesting that this region was not sufficient for homodimerization.
C-terminal truncations of eIF4B were also tested for their ability to bind His-eIF4B280–527. His-eIF4B280–527 bound GST-eIF4B69–390 (Fig. 1D, lane 6) and GST-eIF4B69–360 (Fig. 1D, lane 5), whereas binding to GST-eIF4B69–324 was markedly reduced (Fig. 1D, lane 4). Further C-terminal truncation abolished binding (Fig. 1D, lanes 2 and 3). Together these data suggested that the region between residues 320 and 360 contains the domain required for eIF4B self-association (Fig. 1F). The fact that eIF4B requires a specific region for dimerization indicates that its self-association, although weak under these conditions, is not because of nonspecific interactions. The region between amino acids 320 and 360 includes the C-terminal RNA-binding domain. RNA binding activity, however, does not explain the self-association of eIF4B as other eIF4B constructs containing the N-terminal or central RNA-binding domains did not exhibit eIF4B self-association in the absence of the region between residues 320 and 360 (Fig. 1; also see below). To test whether this region alone is sufficient for homodimerization, a GST fusion eIF4B fragment containing residues 320 to 360 (i.e. GST-eIF4B320–360) was tested for its ability to interact with His-eIF4B280–527. Although His-eIF4B280–527 bound GST-eIF4B320–527 and GST-eIF4B69–360 (Fig. 1E, lanes 4 and 7, respectively) as observed above, no binding to GST-eIF4B320–360 was detected (Fig. 1E, lane 2), suggesting that this region is necessary but may require additional sequence for proper folding to support the interaction.
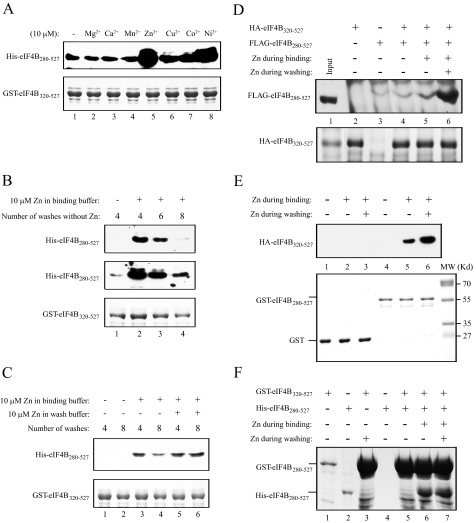
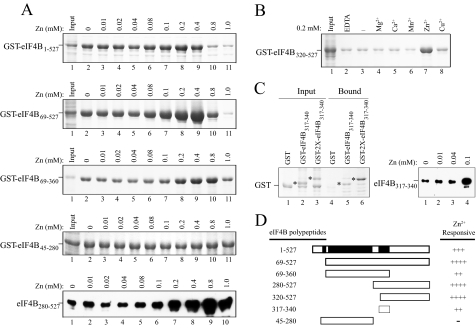
Dimerization of eIF4B Is Stimulated by Zinc—eIF4B, either as a GST fusion or when non-tagged, bound cobalt-based affinity resin (data not shown), suggesting that it could be a metal-loprotein. Cobalt is a borderline metal as defined by Pearson (22). This group of metal ions, which includes Zn2+, Cu2+, and Ni2+, has a preference for the imidazole ring of histidine residues in proteins (23, 24); seven histidine residues are present in wheat eIF4B. To examine whether metal ions may stimulate eIF4B self-association, several divalent metal ions were tested for their ability to stimulate the interaction between GST-eIF4B320–527 and His-eIF4B280–527 using the pulldown assay. The presence of 10 μm Mg2+, Ca2+, or Mn2+ had no effect on eIF4B self-association (Fig. 2A, top panel, compare lanes 2–4 with lane 1). In contrast, 10 μm Zn2+ substantially stimulated eIF4B self-association (Fig. 2A, top panel, lane 5). Ni2+, Cu2+, and Co2+ also stimulated eIF4B self-association (Fig. 2A, top panel, lanes 6–8) but to a smaller extent than did Zn2+.
FIGURE 2.
eIF4B self-association is increased by zinc. In A–E, the bait proteins used in the pulldown assay are shown in the Coomassie-stained gels (bottom panels). In A, binding of His-eIF4B280–527 to GST-eIF4B320–527 in the presence of the indicated metal ions was determined following binding of GST-eIF4B320–527 to glutathione-Sepharose resin (top panel). Bound His-eIF4B280–527 was detected using SDS-PAGE and Western analysis with anti-His antiserum. In B, the requirement for zinc during binding and washing of the GST-eIF4B320–527·His-eIF4B280–527 complex was analyzed using wash buffer without 10 μm zinc present. Bound His-eIF4B280–527 was detected using Western analysis in a short exposure (top panel) or long exposure (middle panel). In C, the requirement for zinc during binding and washing of the GST-eIF4B320–527·His-eIF4B280–527 complex was analyzed using binding or wash buffer without zinc present. Bound His-eIF4B280–527 was detected using Western analysis (top panel). In D, the effect of zinc on the interaction between HA-eIF4B320–527 and FLAG-eIF4B280–527 was examined in the presence or absence of 10 μm zinc during binding and washing of the complex. Bound FLAG-eIF4B280–527 was detected by Western analysis using anti-FLAG antiserum (top panel). In E, the effect of zinc on the interaction between HA-eIF4B320–527 and GST-eIF4B280–527 was examined in the presence or absence of 10 μm zinc during binding and washing of the complex. Bound HA-eIF4B320–527 was detected by Western analysis using anti-HA antiserum (top panel). In F, quantitation of the effect of zinc on the eIF4B self-association was determined using GST-eIF4B320–527 to pull down His-eIF4B280–527 in the presence or absence of 10 μm zinc during binding and washing of the complex. Lane 1, 4% of the GST-eIF4B320–527 used for the binding reactions; lane 2, 4% of the His-eIF4B280–527 used for the binding reactions; lane 3, GST-eIF4B320–527 retained on glutathione-Sepharose resin; lane 4, His-eIF4B280–527 retained on glutathione-Sepharose resin in the absence of GST-eIF4B320–527; lane 5, His-eIF4B280–527 pulled down by GST-eIF4B320–527 in the absence of zinc; lane 6, His-eIF4B280–527 pulled down by GST-eIF4B320–527 in the presence of zinc during binding but not during the four subsequent washes; lane 7, His-eIF4B280–527 pulled down by GST-eIF4B320–527 in the presence of zinc during binding and washing. GST-eIF4B320–527 and His-eIF4B280–527 were detected by Coomassie staining of the SDS-PAGE gel.
In the pulldown assay, zinc is included during the incubation of the prey protein with the bait protein as well as in the subsequent washes of the bait·prey protein complex. To determine how quickly the enhancement of eIF4B self-association by Zn2+ is lost when the latter is absent in the wash buffer, the protein interaction assay was performed in the presence of Zn2+ but washed with buffer lacking Zn2+. In the absence of Zn2+, little to no self-association of eIF4B was observed at a normal exposure of the Western blot (Fig. 2B, top panel, lane 1), although a low level of interaction was observed with a longer exposure (Fig. 2B, middle panel, lane 1). Following formation of the bait·prey protein complex in the presence of Zn2+, a substantial stimulation of self-association was observed even after four washes in the absence of Zn2+ (Fig. 2B, top panel, lane 2). Additional washes of the complex in the absence of Zn2+, however, resulted in a decrease in the level of self-association detected (Fig. 2B, top panel, lanes 3 and 4). To determine whether the presence of Zn2+ during the interaction and wash steps maintains the self-association, the effect of wash buffer containing Zn2+ was compared with wash buffer lacking Zn2+. When Zn2+ was absent from both the binding and wash buffer, little self-association of eIF4B was detected (Fig. 2C, top panel, lanes 1 and 2). The presence of Zn2+ during the incubation of the prey protein with the bait protein but not during the subsequent four washes of the bait·prey protein complex resulted in a level of self-association similar to that when Zn2+ was present in both (Fig. 2C, top panel, compare lane 3 with lane 5). The presence of Zn2+ during eight washes of the bait·prey protein complex maintained the self-association compared with the decrease in self-association when Zn2+ was absent from the wash buffer (Fig. 2C, top panel, compare lane 6 with lane 4). These results indicate that binding of Zn2+ is relatively stable but can be removed from eIF4B by extensive washing in the absence of Zn2+.
As GST can dimerize (26), GST-eIF4B constructs may be expected to dimerize preferentially and reduce their interaction with His-eIF4B constructs. Moreover, it was formally possible that the His tag in the His-eIF4B constructs might bind zinc and facilitate an interaction between His-eIF4B constructs, thus reducing their interaction with GST-eIF4B constructs. To eliminate any effect that the GST or His tags may have had on the observed self-association of eIF4B, the HA and FLAG tags were substituted for the GST and His tags, respectively. HA-tagged eIF4B320–527 was used to pull down FLAG-tagged eIF4B280–527, and the presence of FLAG-eIF4B280–527 was detected using anti-FLAG antiserum. In the absence of HA-eIF4B320–527, only limited binding of FLAG-eIF4B280–527 to the resin was detected (Fig. 2D, top panel, lane 3). In the absence of Zn2+, little self-association was detected above the background binding (Fig. 2D, top panel, lane 4). As observed when the GST- and His-tagged proteins were used, however, the presence of Zn2+ during the pulldown and subsequent washes resulted in a substantial increase in self-association (Fig. 2D, top panel, lane 6). The increase in eIF4B self-association by zinc was similar to that observed between the GST- and His-tagged constructs (Fig. 2B). As observed above, the absence of Zn2+ from the wash buffer resulted in reduced self-association (Fig. 2D, top panel, lane 5). Similar results were obtained for the effect of Zn2+ on the interaction between GST-eIF4B280–527 and HA-eIF4B320–527. In the absence of Zn2+, little self-association was detected (Fig. 2E, top panel, lane 4). Inclusion of Zn2+ in the binding and wash buffers resulted in a substantial increase in self-association (Fig. 2E, top panel, lane 6), whereas the absence of Zn2+ from the wash buffer resulted in reduced self-association (Fig. 2E, top panel, lane 5). GST alone did not interact with HA-eIF4B320–527 in the presence or absence of Zn2+ (Fig. 2E, top panel, lanes 1–3). These results demonstrate that the effect of Zn2+ on eIF4B self-association is independent of the tag used and that eIF4B self-association is not obscured by the use of the GST and His tags.
To determine the degree of eIF4B self-association, GST-eIF4B320–527 was used to pull down His-eIF4B280–527 in the presence or absence of zinc, and the amount of His-eIF4B280–527 bound to GST-eIF4B320–527 was determined by Coomassie staining following resolution of the proteins by SDS-PAGE. Following quantitation of the proteins and normalization for the difference in their molecular weights, the relative molar amount of GST-eIF4B320–527 and His-eIF4B280–527 in a complex in the absence of zinc was 1:0.006 (Fig. 2F, lane 5), suggesting a level of association less than 1%. In the presence of zinc during binding but not in the subsequent wash steps, the relative molar amount of GST-eIF4B320–527 and His-eIF4B280–527 in a complex was 1:0.16 (Fig. 2F, lane 6), an increase in association of 26-fold over that observed in the absence of zinc. When zinc was present during binding and in the subsequent wash steps, the relative molar amount of GST-eIF4B320–527 to His-eIF4B280–527 in a complex was 1:0.28 (Fig. 2F, lane 7), an increase in association of 46-fold over that observed in the absence of zinc. These results indicate that a significant fraction of the eIF4B is present as a complex in the presence of zinc.
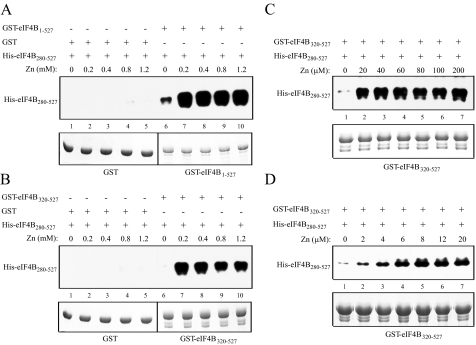
The concentration of zinc required for eIF4B self-association was next examined. Addition of zinc stimulated the binding of His-eIF4B280–527 to full-length GST-eIF4B as it did for GST-eIF4B320–527 and did so over a range of zinc concentrations (Fig. 3, A and B, respectively). Stimulation of eIF4B self-association by zinc was nearly as great at 20 μm as it was at 200 μm (Fig. 3C). To determine the lowest concentration needed for the stimulation of eIF4B self-association, zinc concentrations below 20 μm were also tested. Stimulation by zinc was detected at a concentration as low as 2 μm, but 6–8 μm was necessary for near full stimulation (Fig. 3D). Because 8 μm eIF4B was used in the interaction assay, near full stimulation at 8 μm zinc suggests a 1:1 molar ratio of zinc to eIF4B.
FIGURE 3.
Determination of the optimum zinc concentration for eIF4B dimerization. In A–D, GST, GST-eIF4B1–527, and GST-eIF4B320–527 used in the pulldown assay are shown in the Coomassie-stained gel (bottom panels). Binding of His-eIF4B280–527 to each GST fusion construct in the presence of the indicated amount of zinc was determined following binding of the latter to glutathione-Sepharose resin (top panels). Bound His-eIF4B280–527 was detected by SDS-PAGE and Western analysis.
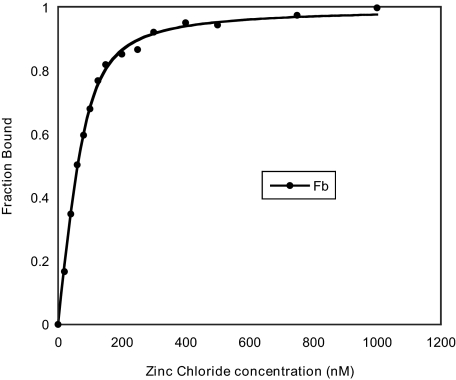
The strength and specificity of zinc binding to eIF4B was measured by determining its equilibrium dissociation constant (Kd) using fluorescence titration (Fig. 4). Zn2+ bound eIF4B with a Kd of 19.7 nm (Table 1). In contrast, Mg2+ bound eIF4B with a Kd of 42.4 nm, suggesting that eIF4B binds Zn2+ more strongly than it does Mg2+. The Kd for Mg2+ was virtually unchanged when 1000 nm Zn2+ was included in the analysis (Table 1), suggesting that Zn2+ binds to eIF4B independently of Mg2+.
FIGURE 4.
Fluorescence intensity measurements on binding of full-length eIF4B with zinc chloride at 22 °C. For the titrations the excitation wavelength was 280 nm, and the emission was 332 nm. The fluorescence is normalized by using the formula fb = (F0 – Fobs)/(Fobs – F∞)(F0/F∞) + (F0 – Fobs), which is directly related to the fraction of protein bound (fb). The solid line is the fitted curve.
TABLE 1.
Equilibrium dissociation constants for the interaction of full-length eIF4B with ZnCl2 and MgCl2 determined by fluorescence titration at 22 °C
| Complex | Kd |
|---|---|
| nm | |
| eIF4B1-527 + ZnCl2 | 19.68 ± 1.58 |
| eIF4B1-527 + MgCl2 | 42.37 ± 5.27 |
| eIF4B1-527 + 1 mm ZnCl2 + MgCl2 | 40.09 ± 5.07 |
The effect that Zn2+ has on the activity of eIF4B was examined next. eIF4B promotes the ATPase activity of the eIFiso4G·eIF4A complex in wheat (8, 9). Consequently the effect of Zn2+ on the ATPase activity of the eIFiso4G·eIF4A·eIF4B complex was investigated. The presence of Zn2+ had little to no effect on the basal level of ATPase activity of eIFiso4G, eIF4A, or eIF4B alone or in any combination of two of the factors (Table 2). However, Zn2+ enhanced the ATPase activity of the eIFiso4G·eIF4A·eIF4B complex by ∼2-fold, suggesting that Zn2+ promotes the ATPase activity of the eIFiso4G·eIF4A·eIF4B complex specifically.
TABLE 2.
Zinc stimulates the ATPase activity of the eIF4A·eIF4B·eIFiso4G complex
|
Amount
|
ATP hydrolyzed
|
|||
|---|---|---|---|---|
| 0 μm zinc | 10 μm zinc | |||
| μg | pmol | |||
| eIF4A | 0.8 | <10 | <10 | |
| eIF4B | 1.0 | 22.8 | 28.7 | |
| eIFiso4G | 1.3 | <10 | 16.9 | |
| eIF4A·eIF4B | 0.8/1.0 | 28.0 | 15.8 | |
| eIF4B·eIFiso4G | 1.0/1.3 | 32.6 | 32.0 | |
| eIF4A·eIFiso4G | 0.8/1.3 | 11.7 | <10 | |
| eIF4A·eIF4B·eIFiso4G | 0.8/1.0/1.3 | 29.7 | 61.9 | |
To demonstrate the requirement for zinc in eIF4B dimerization directly, phenanthroline, a zinc chelator, was used in the binding reaction. In the absence of zinc, the addition of phenanthroline to the binding reaction did not inhibit eIF4B self-association (supplemental Fig. 1A, top panel, compare lanes 2–4 with lane 1). As observed above, the addition of 10 μm zinc substantially stimulated eIF4B self-association (supplemental Fig. 1A, top panel, lane 5). The addition of phenanthroline reversed the stimulatory effect of zinc on eIF4B dimerization but did so only at a concentration greater than 10 μm (supplemental Fig. 1A, top panel, lane 8).
To demonstrate that it was zinc that was chelated by the phenanthroline, additional zinc was included in a reaction where phenanthroline had reversed the stimulatory effect of zinc on eIF4B dimerization. As observed in supplemental Fig. 1A, the stimulation of eIF4B self-association afforded by 10 μm zinc was inhibited by phenanthroline (supplemental Fig. 1B, top panel, compare lanes 4–7 with lane 3). Significant inhibition was observed at a 3:1 ratio of phenanthroline to zinc, and full inhibition of the zinc-induced stimulation was achieved at a 6:1 ratio. Stimulation of eIF4B self-association was restored when additional zinc was added to the binding reaction containing 90 μm phenanthroline. Full stimulation of eIF4B dimerization was observed when an additional 30 μm zinc (for a total zinc concentration of 40 μm) was added (supplemental Fig. 1B, top panel, lane 10). Although the stoichiometric ratio of zinc binding to phenanthroline is unknown, phenanthroline binds iron in a 3:1 ratio. These results suggest that zinc may bind phenanthroline in a ratio from 3:1 to 6:1 consistent with the chemistry of Zn2+ in that it is typically coordinated by four ligands (25).
To examine whether other metal ions could reverse the inhibitory effect of phenanthroline on eIF4B self-association to the same extent as Zn2+, binding reactions containing 90 μm phenanthroline were supplemented with various metal ions. Although some stimulation of eIF4B self-association was observed, particularly by Cu2+ and Co2+ (supplemental Fig. 1C), none exhibited a stimulatory effect on eIF4B self-association nearly as great as that observed for Zn2+.
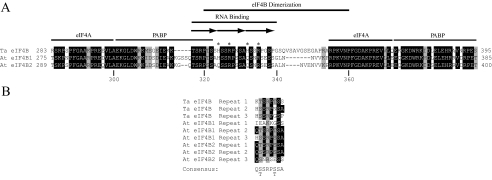
Two Histidine Residues within the eIF4B Dimerization Domain Affect the Strength of Self-association in the Absence of Zinc—Because borderline metal ions can bind histidine residues (23, 24), it was possible that one or more of the histidine residues within wheat eIF4B were involved in the zinc-mediated stimulation of eIF4B self-association. Of the seven histidine residues in wheat eIF4B, six are located in the C-terminal half of the protein (i.e. His-324, His-332, His-387, His-475, His-480, and His-504) and two lie within the C-terminal RNA-binding domain (i.e. His-324 and His-332) (Ref. 12 and Fig. 5A). Each of these two histidines is present within two of the three copies of the octapeptide repeat that makes up the C-terminal RNA-binding domain (Fig. 5B). The observation that the C-terminal half of eIF4B had higher affinity to cobalt resin than did the N-terminal half (data not shown) and that GST-eIF4B69–360, which contains His-324 and His-332 within the eIF4B dimerization domain, also bound to cobalt resin (data not shown) suggested that binding zinc may involve one or more of these histidine residues. Consequently the contribution that His-324 and His-332 may make to the zinc-mediated stimulation of eIF4B homodimerization was examined.
FIGURE 5.
Domain organization of the central region of wheat eIF4B. In A, a comparison of the sequence of the C-terminal RNA-binding domain and dimerization domain from wheat eIF4B (Ta eIF4B) with the corresponding sequence of Arabidopsis eIF4B1 and eIF4B2 (At eIF4B1 and At eIF4B2, respectively) is shown. Conservation of identical or similar residues relative to wheat eIF4B is indicated by shading. Histidine and arginine residues in wheat eIF4B are indicated by asterisks. The portion of eIF4B illustrated in each case is indicated by residue numbers before and after each sequence. In B, a comparison of the three octapeptide repeats within the C-terminal RNA-binding domain of wheat eIF4B and Arabidopsis eIF4B1 and eIF4B2 is shown. Conservation of identical or similar residues relative to wheat eIF4B is indicated by shading.
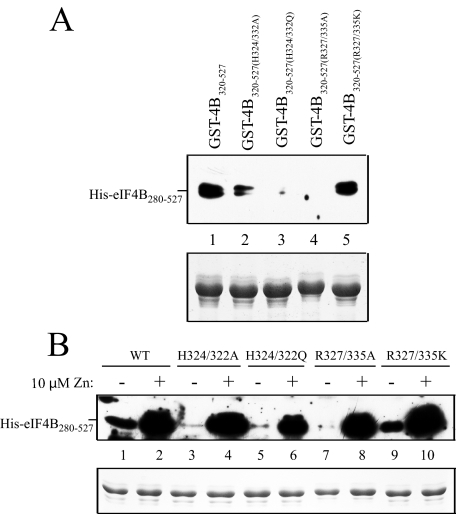
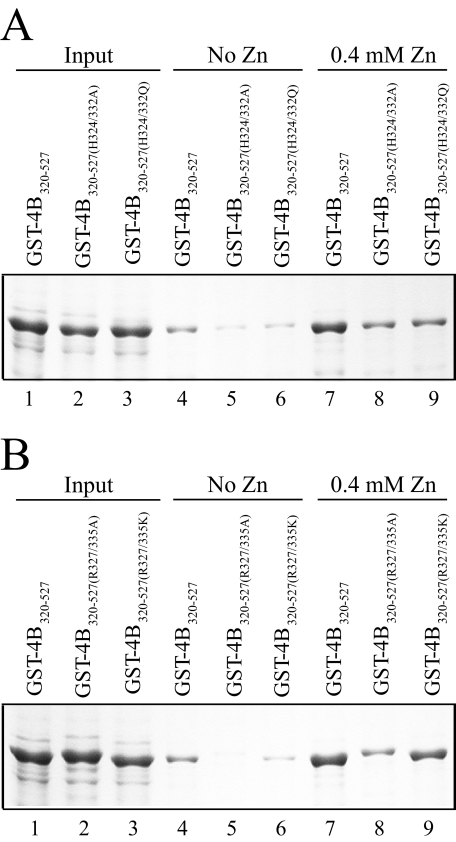
To test this, histidine residues His-324 and His-332 were replaced with alanine to generate GST-eIF4B320–527(H324A/ H332A). Because the corresponding sites in Arabidopsis eIF4B are glutamine residues (i.e. in eIF4B2) or a histidine and glutamine residue, respectively (i.e. in eIF4B1), His-324 and His-332 were also replaced by glutamine residues to generate GST-eIF4B320–527(H324Q/H332Q). The mutant eIF4B proteins were then used in pulldown assays with His-eIF4B280–527. In the absence of zinc, the H324A/H332A double mutation resulted in a moderate reduction in eIF4B self-association (Fig. 6A, compare lane 2 with lane 1), whereas interaction between the H324Q/H332Q double mutation and His-eIF4B280–527 was virtually abolished (Fig. 6A, lane 3). The two arginine residues within the RNA-binding/dimerization domains (i.e. Arg-327 and Arg-335), which are often required for the activity of RNA-binding domains, were mutated to alanine (i.e. GST-eIF4B320–527(R327A/R335A)). Their mutation also virtually abolished eIF4B self-association (Fig. 6A, lane 4). In contrast, mutation of the same two arginine residues to lysine (i.e. GST-eIF4B320–527(R327K/R335K)) did not affect eIF4B self-association (Fig. 6A, lane 5).
FIGURE 6.
Analysis of the contribution of histidine and arginine residues within the dimerization domain to eIF4B self-association. In A, the H324A/H332A, H324Q/H332Q, R327A/R335A, and R327K/R335K mutations were introduced into GST-eIF4B320–527. The wild-type (WT) and mutant GST-eIF4B320–527 (GST-4B320–527) constructs used in the pulldown assay are shown in the Coomassie-stained gel (bottom panel) and were used to pull down His-eIF4B280–527 in the absence of zinc. In B, the same binding reactions were performed with or without 10 μm zinc. In each case, bound protein was resolved by SDS-PAGE and detected using Western analysis (top panels).
To determine the zinc responsiveness of each mutant, the ability of 10 μm zinc to stimulate the association with eIF4B was examined. In the presence of zinc, the H324A/H332A double mutation did not prevent the zinc-mediated stimulation of eIF4B self-association (Fig. 6B, compare lane 4 with lane 2), although a slight reduction was observed for the H324Q/H332Q double mutation (Fig. 6B, compare lane 6 with lane 2). Little to no reduction in the zinc-mediated stimulation of eIF4B self-association was observed for the R327A/R335A mutation (Fig. 6B, lane 8) or the R327K/R335K mutation (Fig. 6B, lane 10). These data suggest that the two histidine and arginine residues within the wheat eIF4B dimerization domain affect the strength of the dimerization of eIF4B in the absence of zinc. That eIF4B self-association was largely maintained in the presence of zinc suggests additional residues are involved for zinc responsiveness.
Zinc Stimulates the Activity of the C-terminal RNA-binding Domain—Because the eIF4B dimerization domain also encompasses the C-terminal RNA-binding domain, we examined whether the zinc-mediated stimulation of eIF4B self-association promoted RNA binding, a prediction based on the previous observation that this RNA-binding domain was active only when present as a dimer (12). RNA binding of full-length GST-eIF4B was moderately stimulated by zinc (Fig. 7A). However, as eIF4B contains three RNA-binding domains, the C-terminal RNA-binding domain was tested separately from the other two domains to determine whether they differ in zinc stimulation. No stimulation of RNA binding was observed for GST-eIF4B45–280 (Fig. 7A), which contains the N-terminal RNA-binding domain and the RRM but not the C-terminal RNA-binding domain. In contrast, stimulation of RNA binding was observed for GST-eIF4B69–527 and GST-eIF4B69–360 (Fig. 7A), both of which contain the C-terminal RNA-binding domain and the RRM. Zinc also stimulated the RNA binding activity of untagged eIF4B280–527, which contains just the C-terminal RNA-binding domain (Fig. 7A), demonstrating that the effect of zinc on RNA binding is independent of any tag. The RNA binding activity of GST-eIF4B320–527, which contains just the C-terminal RNA-binding domain, was also stimulated by zinc but not other metal ions (Fig. 7B). Interestingly greater stimulation by zinc was observed for constructs that included sequence C-terminal to residue 360, e.g. GST-eIF4B69–527, GST-eIF4B320–527, and untagged eIF4B280–527 (Fig. 7A).
FIGURE 7.
Zinc stimulates the RNA binding activity of the eIF4B C-terminal RNA domain. In A, each GST fusion or untagged construct was expressed in E. coli (lane 1). RNA binding activity in the absence or presence of the indicated amount of zinc was determined following the addition of crude extract to poly(A)-Sepharose 4B resin. Bound proteins were resolved by SDS-PAGE and detected using Coomassie staining. In B, the specificity of metal ion requirement was tested by the inclusion of the indicated metal ions in the RNA binding assay. In C, the C-terminal RNA domain from wheat eIF4B is sufficient for RNA binding. The region from residues 317 to 340 was fused to GST as one copy (i.e. GST-eIF4B317–340) or two copies (i.e. GST-2×eIF4B317–340), expressed in E. coli (left panel, lanes 1–3), and tested for binding to poly(A)-Sepharose 4B resin (left panel, lanes 4–6). Asterisks indicate each fusion construct. RNA binding activity of GST-eIF4B317–340 in the absence or presence of the indicated amount of zinc was determined following binding to poly(A)-Sepharose 4B resin (right panel). In D, a summary of the RNA binding activity of eIF4B polypeptides is shown. RNA-binding domains are indicated by black boxes. Strength of eIF4B binding is indicated by the number of pluses. –, lack of stimulation by zinc.
The eIF4B dimerization domain lies between residues 320 and 360, whereas the C-terminal RNA-binding domain lies between residues 317 and 340 (12) (Fig. 5A). To determine whether the sequence from residues 317 to 340 is sufficient for RNA binding, this region was tested for its RNA binding activity when fused to GST (i.e. GST-eIF4B317–340). GST-eIF4B317–340 exhibited RNA binding when present as a single copy or when the same sequence was tandemly repeated (i.e. GST-2×eIF4B317–340) (Fig. 7C, left panel). Although this sequence is sufficient for RNA binding, it is insufficient for dimerization as demonstrated in Fig. 1. Zinc, however, increased the RNA binding activity of this minimum C-terminal RNA-binding domain (Fig. 7C, right panel). These data demonstrate that although the eIF4B dimerization domain and C-terminal RNA-binding domain overlap they differ in the sequences required for each activity.
The Two Histidine and Arginine Residues within the eIF4B C-terminal RNA-binding Domain Are Required for RNA Binding Activity—To determine whether the two histidine residues (i.e. His-324 and His-332) or two arginine residues (i.e. Arg-327 and Arg-335) within the RNA-binding/dimerization domain are required for RNA binding activity, the RNA binding of the mutants described above was examined. As GST has the ability to dimerize (26) and the formation of a dimer is required for the activity of the C-terminal RNA-binding domain, GST fusions were used to be able to detect RNA binding in the absence of zinc and therefore assess the contribution of the histidine and arginine residues to the RNA binding activity independent of any requirement for protein dimerization. In the absence of zinc, the H324A/H332A double mutant exhibited significantly reduced RNA binding (Fig. 8A, compare lane 5 with lane 4). Similar results were obtained with the H324Q/H332Q double mutant (Fig. 8A, lane 6). Zinc stimulated RNA binding of both mutants, but both remained significantly impaired in RNA binding relative to the wild-type sequence (Fig. 8A, lanes 8 and 9, respectively) in contrast to the minimal effect these same mutations had on eIF4B dimerization in the presence of zinc (Fig. 6B). The R327A/R335A double mutant exhibited substantially lower RNA binding activity (Fig. 8B, compare lane 5 with lane 4). Although the R327K/R335K double mutation also exhibited reduced RNA binding, the reduction was not as great as when the arginine residues were altered to alanines (Fig. 8B, lane 6). Zinc stimulated RNA binding of both mutants, but the RNA binding activity of the R327K/R335K mutant remained greater than that of the R327A/R335A mutant (Fig. 8B, compare lane 9 with lane 7). However, the RNA binding activity of both mutants remained lower than the wild type, which, as with the histidine mutants, is in contrast to the minimal effect these same mutations had on eIF4B dimerization in the presence of zinc (Fig. 6B). These results indicate that the histidine residues and the arginine residues in particular in the C-terminal RNA-binding domain are required for full RNA binding activity.
FIGURE 8.
Analysis of the contribution of histidine and arginine residues within the C-terminal RNA domain to RNA binding activity. In A, binding of wild-type GST-eIF4B320–527 (GST-4B320–527) and the H324A/H332A and H324Q/H332Q mutants to poly(A)-Sepharose 4B resin in the absence or presence of zinc is shown. In B, binding of wild-type GST-eIF4B320–527 and the R327A/R335A and R327K/R335K mutants to poly(A)-Sepharose 4B resin in the absence or presence of zinc is shown. In each case, bound proteins were resolved by SDS-PAGE and detected using Coomassie staining.
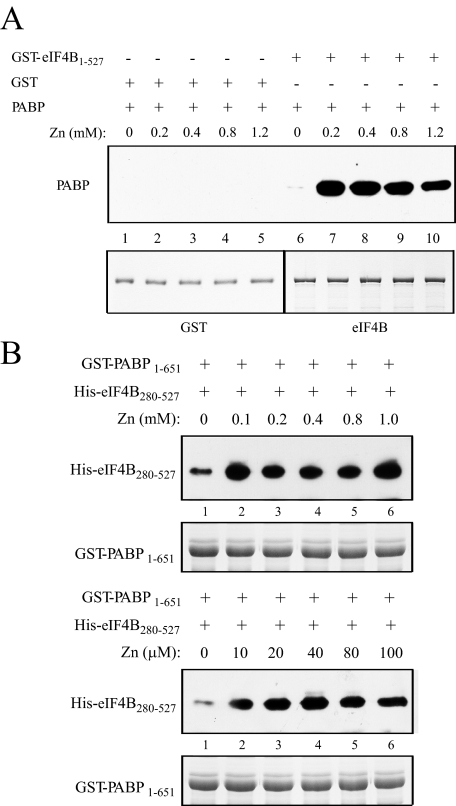
The Interaction between eIF4B and PABP Is Stimulated by Zinc—eIF4B contains two binding sites each for PABP and eIF4A that are present on either side of the C-terminal RNA-binding domain (12) (Fig. 1A). In each case, the eIF4A binding site lies N-proximal to the PABP binding site (Fig. 5A). Because zinc stimulates eIF4B dimerization as well as the activity of the C-terminal RNA-binding domain, the effect of zinc on the interaction of those partner proteins that bind close to the dimerization/C-terminal RNA-binding domain, i.e. PABP and eIF4A, was examined. At concentrations that promote the activity of the C-terminal RNA-binding domain, zinc substantially increased the interaction between eIF4B and PABP (Fig. 9A, compare lanes 7–10 with lane 6). PABP did not bind GST in the presence or absence of zinc (Fig. 9A, compare lanes 2–5 with lane 1), demonstrating that the effect of zinc is specific to the interaction between eIF4B and PABP. To test whether zinc can stimulate the interaction between eIF4B and PABP at even lower concentrations, a GST-PABP fusion was used to pull down His-eIF4B280–527. This approach avoided the use of GST-eIF4B as the presence of the GST may cause dimerization (26) and may thus obscure the effect that a low zinc concentration might have on the eIF4B and PABP interaction if eIF4B dimerization is a prerequisite for its interaction with PABP. 100 μm zinc stimulated binding between eIF4B and PABP to the same level that 1 mm did (Fig. 9B). As observed for eIF4B dimerization, even 10 μm zinc was sufficient to stimulate fully the interaction (Fig. 9B).
FIGURE 9.
Zinc stimulates the interaction between eIF4B and PABP. In A, GST and full-length GST-eIF4B used in each pulldown assay are shown in the Coomassie-stained gel (bottom panel). Binding of full-length His-PABP to GST-eIF4B at the indicated levels of zinc was determined by Western analysis following binding of the latter construct to glutathione-Sepharose resin (top panel). GST was used as a negative control. In B, full-length GST-PABP used in each pulldown assay is shown in the Coomassie-stained gel (bottom panel). Binding of His-eIF4B280–527 to GST-PABP at the indicated levels of zinc was determined by Western analysis following binding of the latter construct to glutathione-Sepharose resin (top panel).
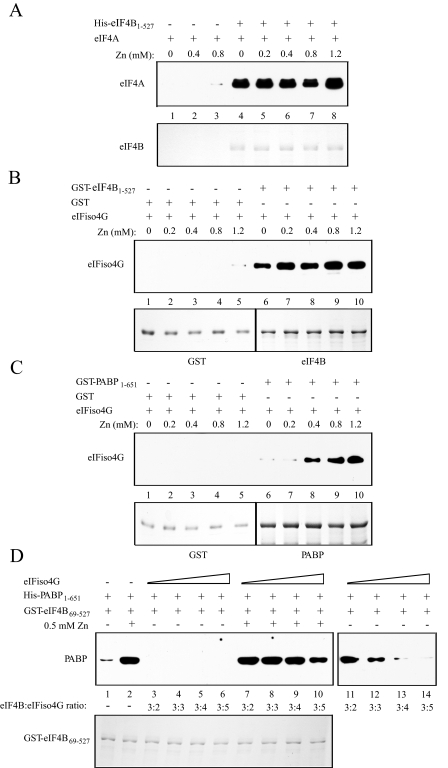
The interaction between eIF4B and eIF4A was readily detected in the absence of zinc (Fig. 10A, lane 4) in good agreement with previous results (12), and zinc did not stimulate their interaction (Fig. 10A, compare lanes 5–8 with lane 4). eIF4B also interacts with eIFiso4G (12). The eIFiso4G-binding domain lies near the N terminus of eIF4B (Fig. 1A) and is C-proximal to the N-terminal RNA-binding domain (12). The interaction between eIF4B and eIFiso4G was readily detected in the absence of zinc (Fig. 10B, lane 6) in good agreement with previous results (12). Little to no stimulation by zinc was observed (Fig. 10B, compare lanes 7–10 with lane 6). These data indicate that zinc stimulates the binding of PABP but not eIF4A or eIFiso4G to eIF4B, thus demonstrating specificity in which partner protein interactions are promoted by zinc.
FIGURE 10.
The effect of zinc on the interaction between eIF4B and eIF4A, eIF4B and eIFiso4G, or PABP and eIFiso4G. In A, full-length His-eIF4B used in each pulldown assay is shown in the Coomassie-stained gel (bottom panel). Binding of full-length eIF4A to His-eIF4B at the indicated levels of zinc was determined following binding of His-eIF4B to TALON metal affinity resin (top panel). Bound eIF4A was detected by SDS-PAGE and Western analysis using anti-eIF4A antiserum. In B, GST and full-length GST-eIF4B used in each pulldown assay are shown in the Coomassie-stained gel (bottom panel). Bound eIFiso4G was detected by SDS-PAGE and Western analysis using anti-eIFiso4G antiserum following binding of GST-eIF4B to glutathione-Sepharose resin (top panel). In C, GST and full-length GST-PABP used in each pulldown assay are shown in the Coomassie-stained gel (bottom panel). Bound eIFiso4G was detected by SDS-PAGE and Western analysis using anti-eIFiso4G antiserum following binding of GST-PABP to glutathione-Sepharose resin (top panel). In D, increasing amounts of eIFiso4G were added to binding reactions containing a constant amount of GST-eIF4B69–527 (bottom panel) and His-PABP1–651 in the absence or presence of zinc. Following binding of GST-eIF4B69–527 to glutathione-Sepharose resin, bound PABP was detected by Western analysis using anti-His antiserum (top panel). GST-eIF4B69–527 was used in the binding reactions (bottom panel) because it lacks the eIFiso4G interaction domain. A longer exposure of lanes 3–6 is shown in lanes 11–14 (right panel).
Zinc Stimulates the Interaction between PABP and eIFiso4G—Because zinc stimulated the interaction between PABP and eIF4B, the ability of zinc to stimulate the interaction between PABP and other partner proteins was examined. Because PABP and eIFiso4G are known to interact directly (13–15), the effect of zinc on this interaction was investigated. Their interaction was detectable in the absence of zinc as reported previously (15), but zinc promoted the interaction (Fig. 10C, compare lanes 7–10 with lane 6). No interaction between eIFiso4G and GST was observed in the presence or absence of zinc (Fig. 10C, lanes 1–5). The lowest zinc concentration observed to stimulate the interaction between eIFiso4G and PABP was 200–400 μm, a concentration higher than that observed for the interaction between eIF4B and PABP (Fig. 9) or for eIF4B dimerization (Fig. 3). Zinc stimulated the interaction of eIFiso4G to each of the two eIFiso4G binding sites within PABP (data not shown).
Zinc Determines PABP Partner Protein Selection—The eIF4B interaction domain at the C-terminal end of RRM1 of PABP overlaps the N-proximal, strong eIFiso4G interaction domain in PABP (12). The overlapping nature of their interaction domains results in competition between eIF4B and eIFiso4G in their binding to PABP (12). For an mRNA undergoing translation, the strength of interaction of PABP for each partner protein will determine the specificity of its interaction with the initiation factors assembled at the 5′-end of the mRNA. Because zinc stimulates the interaction between PABP and eIF4B and the interaction between PABP and eIFiso4G, we examined whether the presence of zinc altered the partner protein selection of PABP.
A competition experiment was performed as described previously (15) in which increasing amounts of eIFiso4G were added to binding reactions containing eIF4B and a limited amount of PABP. The ability of eIFiso4G to compete with eIF4B for binding with PABP was then monitored by the amount of PABP that remained bound to eIF4B. GST-tagged eIF4B was used in the binding reactions to facilitate its isolation on glutathione resin with the associated PABP. To prevent direct binding of eIFiso4G to eIF4B (12), which would obscure the detection of competition between eIFiso4G and eIF4B for PABP, GST-eIF4B69–527 was used in the assay as it lacks the interaction site for eIFiso4G.
In the absence of eIFiso4G, the interaction between eIF4B and PABP was observed (Fig. 10D, top panel, lane 1). The addition of increasing amounts of eIFiso4G abolished the binding of PABP to eIF4B (Fig. 10D, top panel, lanes 3–6) in good agreement with our previous observation (15). These data indicate that the interaction of eIFiso4G with PABP can preclude the interaction of eIF4B with PABP in the absence of zinc. In the presence of 0.5 mm zinc, a concentration high enough to stimulate fully the interaction between PABP and eIFiso4G as well as the interaction between PABP and eIF4B, increasing amounts of eIFiso4G failed to abolish the binding of PABP to eIF4B and, at most, moderately reduced the interaction between PABP and eIF4B and even then only at a high concentration of eIFiso4G (Fig. 10D, top panel, lanes 7–10). These data suggest that zinc increases the strength of the PABP-eIF4B interaction more than the PABP-eIFiso4G interaction, resulting in the preferential interaction of PABP and eIF4B even in the presence of eIFiso4G.
DISCUSSION
Despite the low level of sequence conservation, the interaction of eIF4B with other initiation factors and RNA is largely conserved among species (12). Interestingly the functional domains involved in these interactions show significant differences in their organization and in sequence conservation (12). Like its mammalian ortholog, wheat eIF4B was reported to be a dimer (16, 17) based on gel exclusion analysis, but subsequent analysis indicated an extended shape for the protein, and thus it may actually be a monomer.3 Biochemically, however, eIF4B functions as a dimer in that each of its three RNA-binding domains depends on protein dimerization for activity (12). In this study, we showed that the region in wheat eIF4B corresponding to the central DRYG-rich region of mammalian eIF4B responsible for the dimerization of the animal protein (16) does not support dimerization of the plant protein. Nevertheless wheat eIF4B can dimerize, and the dimerization domain mapped to a region that includes residues 320-360 (Fig. 5A). Less than 1% of eIF4B self-associated in the absence of zinc, suggesting that dimerization is weak. The presence of zinc at physiological concentrations, however, specifically and substantially stimulated eIF4B dimerization as well as the ATPase activity promoted by eIF4B on the eIFiso4G·eIF4A complex. Although zinc varies in concentration among plant species and growth conditions, its foliar concentration can range from 15 to 100 mg kg–1 of fresh tissue (∼0.230–1.54 mm) (27). Stimulation of eIF4B self-association by zinc was detectable at a concentration as low as 2 μm with near maximum stimulation observed between 8 and 20 μm and full stimulation observed by 200 μm. Despite the stimulatory effect that zinc had on eIF4B dimerization, 28% of eIF4B was observed to self-associate in the presence of zinc (Fig. 2F). This may be an underestimate as some of the His-eIF4B280–527 in the pulldown assay would be expected to self-associate and thus would not be available to be pulled down as a complex with GST-eIF4B320–527. Nevertheless the partial dimerization of eIF4B may indicate either that other factors may contribute to its self-association, e.g. interaction with partner proteins or post-translational modification, or that the pool of cellular eIF4B may exist in an equilibrium of monomer and dimer forms.
The effect of zinc on eIF4B was specific in several ways. Zinc promoted the self-association of eIF4B and its interaction with PABP but not its binding to eIF4A or eIFiso4G, demonstrating that not all protein interactions with eIF4B are stimulated by zinc. Zinc promoted the activity of the C-terminal RNA-binding domain but not the activity of the N-terminal RNA-binding domain or the RRM, demonstrating that the effect of zinc was specific to a single RNA-binding domain. Zinc bound eIF4B with a Kd of 19.7 nm, and its binding was independent of Mg2+. Only zinc reversed the inhibition of eIF4B self-association by a divalent metal cation chelator to result in a high level of eIF4B dimerization. Moreover the effect of zinc on eIF4B was independent of the tag present, and zinc even stimulated the RNA binding of eIF4B280–527 containing no tag, results that demonstrated that the effect of zinc was not because of any potential interaction with a specific tag.
The eIF4B dimerization domain overlaps the C-terminal RNA-binding domain and lies between twin eIF4A and PABP interaction domains (Fig. 5A). As mentioned above, zinc stimulated the interaction between eIF4B and PABP but had no effect on the binding between eIF4B and eIF4A. The interaction between eIF4B and eIF4A appears to be strong as it is readily detected, whereas that between eIF4B and PABP appears to be significantly weaker. Therefore, zinc may not be required for eIF4A to bind to eIF4B as it is already a strong interaction. As zinc had no effect on the activity of the RRM or N-terminal RNA-binding domain or on the interaction of eIFiso4G, which binds proximal to the N-terminal RNA-binding domain, the effect of zinc may be limited to the region encompassing the C-terminal RNA-binding and PABP interaction domains. It is possible that zinc induces a conformational change in this region of eIF4B to facilitate its self-association or may act as a bridge between the two proteins in a dimer. The observation that the interaction between eIF4B and PABP is stimulated by zinc at a concentration similar to that needed to promote eIF4B self-association supports the possibility that its interaction with PABP may involve its dimerization. Moreover the dimerization of eIF4B was previously reported as necessary for its RNA binding activity (12). Therefore, the extent to which zinc promotes the self-association of eIF4B may contribute to the RNA binding of the protein.
The eIF4B dimerization domain is characterized by the presence of three tandemly repeated octapeptide sequences, which constitute the entire C-terminal RNA-binding domain (Fig. 5). Mutational analysis of the histidine or arginine residues in these repeats suggested that both contributed to eIF4B self-association in the absence of zinc. Specifically mutation of the two histidines to glutamines abolished eIF4B self-association, whereas their mutation to alanines did not. Similarly mutation of the two arginines to alanines abolished eIF4B self-association, whereas their mutation to lysines did not. The ability of these mutants to bind wild-type eIF4B was retained in the presence of zinc. These results indicate that His-324, His-332, Arg-327, and Arg-335 contribute to the stability of eIF4B dimerization particularly in the absence of zinc. The observation that eIF4B self-association is retained in the presence of zinc may be the result of one wild-type eIF4B molecule in the dimer. The deleterious impact that mutation of these residues had on eIF4B self-association in the absence of zinc suggests that their mutation may affect local protein folding that impairs the already weak self-interaction. Interestingly the mutation of Arg-327 and Arg-335 to lysines had little effect on eIF4B dimerization even in the absence of zinc, supporting the conclusion that arginines at these positions are not specifically required for eIF4B dimerization but rather may play an indirect role in protein folding.
The C-terminal RNA-binding domain was mapped to the region between residues 317 and 340. A polypeptide representing this region was sufficient to bind RNA, thus demonstrating its activity (Fig. 7C). The three octapeptide repeats comprising the C-terminal RNA-binding domain are conserved in number and sequence in Arabidopsis eIF4B1 and eIF4B2 (Fig. 5). Although the first copy of these repeats is not essential for RNA binding, deletion of the remaining two abolishes RNA binding activity (12). His-324, His-332, Arg-327, and Arg-335 lie within the second and third repeats. His-324, His-332, Arg-327, and Arg-335 were necessary for the full activity of the C-terminal RNA-binding domain in the presence or absence of zinc. Mutation of His-324 and His-332 to either alanine or glutamine residues impaired RNA binding as did the mutation of Arg-327 and Arg-335 to alanines. Mutation of Arg-327 and Arg-335 to lysine residues also impaired RNA binding but to a lesser extent. Interestingly this same mutation had little effect on eIF4B self-association, suggesting that the presence of lysines at these positions specifically affects RNA binding but not eIF4B self-association. Mutation of the histidine or arginine residues did not abolish the stimulation of RNA binding by zinc perhaps because eIF4B dimerization was not impaired by these mutations in the presence of zinc as observed above and dimerization of eIF4B is required for the full activity of the C-terminal RNA-binding domain (12). Thus, these data suggest that His-324 and/or His-332 as well as Arg-327 and/or Arg-335 are necessary for the activity of the C-terminal RNA-binding domain.
The presence of zinc stimulated the RNA binding activity of constructs containing sequence C-terminal to residue 360, e.g. GST-eIF4B69–527, GST-eIF4B320–527, and untagged eIF4B280–527, more so than constructs that terminated at residue 360, e.g. GST-eIF4B69–360 (Fig. 7A), suggesting that sequence C-terminal to the C-terminal RNA-binding domain itself may be required to fully respond to the stimulatory effect of zinc. Zinc, however, did stimulate the RNA binding activity of residues 317–340 (i.e. GST-eIF4B317–340) when this polypeptide was tested in isolation, indicating that even this minimum RNA-binding domain remains responsive to zinc. Detection of the stimulation of RNA binding by zinc required a somewhat higher level of zinc (80–100 μm) than that needed to observe stimulation of eIF4B dimerization or the interaction between eIF4B and PABP, suggesting that the effect of zinc on RNA binding requires a significant fraction of the eIF4B to undergo dimerization before an increase in RNA binding over that observed in the absence of zinc can be detected. Maximum stimulation of eIF4B dimerization required up to 200 μm zinc. It is also possible that zinc plays a more direct role in RNA binding in that zinc stimulated binding of the minimum RNA-binding domain (residues 317–340), which lacks a portion of the eIF4B dimerization domain, when fused to GST, which itself would have facilitated dimerization.
Zinc also stimulated the interaction between PABP and eIFiso4G (Fig. 10). A concentration of 200–400 μm zinc was required to promote the interaction; this was higher than that for eIF4B dimerization or for the interaction between eIF4B and PABP but still within physiological levels (24). Because of the overlapping nature of the eIFiso4G and eIF4B interaction domains in PABP, eIFiso4G and eIF4B compete for binding PABP (15). Even when the ratio of eIF4B to eIFiso4G was 3:2, eIFiso4G competed effectively with eIF4B in binding PABP (Fig. 10D) (12). In the presence of zinc, however, eIFiso4G was unable to compete effectively with eIF4B. Thus, despite the stimulatory effect that zinc had on the PABP-eIFiso4G interaction as well as the PABP-eIF4B interaction, the results from the competition experiment demonstrate that zinc disproportionately increases the interaction between PABP and eIF4B to favor it over the interaction between PABP and eIFiso4G when both initiation factors are present. As multiple molecules of PABP would be expected to be bound to a typical poly(A) tail, particularly for newly synthesized mRNAs, the competing nature of the binding of eIFiso4G and eIF4B to PABP would likely result in their interaction with separate molecules of PABP, providing additional stability of the PABP·eIF4F·eIF4B complex through multiple protein interactions. The observation that eIFiso4G and eIF4B can interact (12) and that the combination of eIFiso4G and eIF4B synergistically promotes the multimeric binding of PABP to poly(A) RNA (13) supports the notion that they can bind separate molecules of PABP and increase the stability of the complex. Moreover wheat PABP exists as differentially phosphorylated species (19), and its phosphorylation state affects partner protein selection (14), further reducing the potential for competition between eIFiso4G and eIF4B. Thus, from these data, we conclude that zinc can contribute to these interactions by increasing the self-association of eIF4B, the activity of the eIF4B C-terminal RNA-binding domain, and the interaction between eIF4B and PABP. Whether zinc also promotes similar interactions for the animal or yeast orthologs will be of interest to determine to what extent the involvement of zinc is conserved among eukaryotes.
Supplementary Material
Acknowledgments
We thank Dr. Karen Browning for pET3d-eIFiso4G and pET3d-eIF4B and for eIFiso4G and eIF4A antisera.
This work was supported by United States Department of Agriculture Grant 2003-35100-13375 and the University of California Agricultural Experiment Station (to D. R. G.) and by National Science Foundation Grant MCB-0814051 (to D. J. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: eIF, eukaryotic translation initiation factor; PABP, poly(A)-binding protein; RRM, RNA recognition motif; GST, glutathione S-transferase; HA, hemagglutinin.
K. Browning, personal communication.
References
- 1.Preiss, T., and Hentze, M. (2003) BioEssays 10 1201–1211 [DOI] [PubMed] [Google Scholar]
- 2.Kapp, L. D., and Lorsch, J. R. (2004) Annu. Rev. Biochem. 73 657–704 [DOI] [PubMed] [Google Scholar]
- 3.Pestova, T. V., Lorsch, J. R., and Hellen, C. U. T. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J. W. B., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 4.Gallie, D. R. (2002) Plant Mol. Biol. 50 949–970 [DOI] [PubMed] [Google Scholar]
- 5.Wells, S. E., Hillner, P. E., Vale, R. D., and Sachs, A. B. (1998) Mol. Cell 2 135–140 [DOI] [PubMed] [Google Scholar]
- 6.Tarun, S. Z., and Sachs, A. B. (1995) Genes Dev. 9 2997–3007 [DOI] [PubMed] [Google Scholar]
- 7.Browning, K. S. (1996) Plant Mol. Biol. 32 107–144 [DOI] [PubMed] [Google Scholar]
- 8.Rogers, G. W., Jr., Richter, N. J., Lima, W. F., and Merrick, W. C. (2001) J. Biol. Chem. 276 30914–30922 [DOI] [PubMed] [Google Scholar]
- 9.Bi, X. P., Ren, J. H., and Goss, D. J. (2000) Biochemistry 39 5758–5765 [DOI] [PubMed] [Google Scholar]
- 10.Methot, N., Song, M. S., and Sonenberg, N. (1996) Mol. Cell. Biol. 16 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Methot, N., Pickett, G., Keene, J. D., and Sonenberg, N. (1996) RNA 2 38–50 [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, S., and Gallie, D. R. (2006) J. Biol. Chem. 281 24351–24364 [DOI] [PubMed] [Google Scholar]
- 13.Le, H., Tanguay, R. L., Balasta, M. L., Wei, C.-C., Browning, K. S., Metz, A. M., Goss, D. J., and Gallie, D. R. (1997) J. Biol. Chem. 272 16247–16255 [DOI] [PubMed] [Google Scholar]
- 14.Le, H., Browning, K. S., and Gallie, D. R. (2000) J. Biol. Chem. 275 17452–17462 [DOI] [PubMed] [Google Scholar]
- 15.Cheng, S., and Gallie, D. R. (2007) J. Biol. Chem. 282 25247–25258 [DOI] [PubMed] [Google Scholar]
- 16.Milburn, S. C., Hershey, J. W. B., Davies, M. V., Kelleher, K., and Kaufman, R. J. (1990) EMBO J. 9 2783–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metz, A. M., Wong, K. C., Malmstrom, S. A., and Browning, K. S. (1999) Biochem. Biophys. Res. Commun. 266 314–321 [DOI] [PubMed] [Google Scholar]
- 18.Khan, M. A., and Goss, D. J. (2004) Biochemistry 43 9092–9097 [DOI] [PubMed] [Google Scholar]
- 19.Gallie, D. R., Le, H., Caldwell, C., Tanguay, R. L., Hoang, N. X., and Browning, K. S. (1997) J. Biol. Chem. 272 1046–1053 [DOI] [PubMed] [Google Scholar]
- 20.Wei, C. C., Balasta, M. L., Ren, J., and Goss, D. J. (1998) Biochemistry 37 1910–1916 [DOI] [PubMed] [Google Scholar]
- 21.Grifo, J. A., Abramson, R. D., Satler, C. A., and Merrick, W. C. (1984) J. Biol. Chem. 259 8648–8654 [PubMed] [Google Scholar]
- 22.Pearson, R. G. (1990) Coord. Chem. Rev. 100 403–425 [Google Scholar]
- 23.Porath, J., Carlsson, J., Olsson, I., and Belfrage, G. (1975) Nature 258 598–599 [DOI] [PubMed] [Google Scholar]
- 24.Porath, J. (1988) Trends Anal. Chem. 7 254–259 [Google Scholar]
- 25.Barak, P., and Helmke, P. A. (1993) in Zinc in Soil and Plants (Robson, A. D., ed) pp. 1–13, Kluwer Academic Publishers, Dordrecht, Netherlands
- 26.Trakshel, G. M., and Maines, M. D. (1988) Biochem. J. 252 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longnecker, N. E., and Robson, A. D. (1993) in Zinc in Soil and Plants (Robson, A. D., ed) pp. 79–91, Kluwer Academic Publishers, Dordrecht, Netherlands
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.