Abstract
Streptococcus pneumoniae is a major causative agent of mortality throughout the world. The initial event in invasive pneumococcal disease is the attachment of pneumococci to epithelial cells in the upper respiratory tract. Several bacterial proteins can bind to host extracellular matrix proteins and function as adhesins and invasins. To identify adhesins or invasins on the pneumococcal cell surface, we searched for several proteins with an LPXTG anchoring motif in the whole-genome sequence of Streptococcus pneumoniae and identified one, which we called PfbA (plasmin- and fibronectin-binding protein A), that bound to human serum proteins. Immunofluorescence microscopy and fluorescence-activated cell sorter analysis revealed that PfbA was expressed on the pneumococcal cell surface. A ΔpfbA mutant strain was only half as competent as the wild-type strain at adhering to and invading lung and laryngeal epithelial cells. In addition, epithelial cells infected with ΔpfbA showed morphological changes, including cell flattening and a loss of microvilli, that did not occur in cells infected with the wild-type strain. The mutant strain also exhibited a weaker antiphagocytotic activity than wild type in human peripheral blood. Moreover, the growth of wild-type bacteria in human whole blood containing anti-PfbA antibodies was reduced by ∼50% after 3 h compared with its growth without the antibody. These results suggest that PfbA is an important factor in the development of pneumococcal infections.
Streptococcus pneumoniae, which is also simply called pneumococcus, is a Gram-positive Diplococcus and the major pathogen of community-acquired pneumonia. It also causes meningitis, otitis, and septicemia, with a high incidence of morbidity and mortality throughout the world (1, 2). The initial phase of infection involves colonization followed by intimate contact with the host cells, which promotes bacterial uptake by the host cell. Successful conversion from a commensal to an invasive microorganism is accompanied by pathogen transmigration through tissue barriers and its subsequent adaptation to different host niches, which is a multifunctional and highly regulated process (3). Thus, to find ways to prevent pathogenic bacteria from initiating local inflammation or becoming invasive, it is important to understand the mechanisms underlying bacterial adhesion to host cells. Pathogenic microorganisms, particularly Gram-positive bacteria, recognize various components of extracellular matrices to obtain access to host cells in their target tissues. The bacteria can remain in the extracellular environment, which allows them to persist in the host niche, or they can enter host cells and spread into subcellular compartments (4).
Avoiding phagocytosis is also important for bacterial survival, and a variety of mechanisms related to streptococcal survival strategies for avoiding host immunity have been reported. In particular, S. pyogenes expresses a number of proteins that are postulated to play a crucial role in its colonization and antiphagocytic activities (5–8). However, the mechanisms used by S. pneumoniae to escape phagocytosis remain unclear.
A polysaccharide-based vaccine against S. pneumoniae is presently used; however, it is ineffective in children younger than 2 years of age and only 60% effective in older children and adults (9, 10). A newer, conjugate vaccine consisting of a protein linked to the saccharides of seven major disease-causing serotypes has been licensed for use in infants. This vaccine is effective in preventing invasive diseases caused by pneumococci expressing the capsular serotypes contained in the vaccine. Nevertheless, it cannot be expected to provide protection against other serotypes. In addition, antibiotic-resistant strains are on the rise (9, 11, 12). Therefore, there is an urgent need to improve the characterization of S. pneumoniae surface proteins that could serve as candidate targets for protein-based vaccines and the development of new antibiotics (13–15). One of the most promising avenues for creating effective vaccines or drugs is the targeting of adhesins and invasins that promote the adhesion of pathogens to human tissues.
In the present study, we found that inactivation of the pfbA gene in S. pneumoniae significantly reduced the bacterial ability to bind human epithelial cells. We also showed that pfbA expression was involved in protecting pneumococci against phagocytosis. Finally, we found that anti-PfbA antibodies reduced pneumococcal growth in human whole blood by ∼50% compared with its growth without antibodies.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Cells, and Reagents—S. pneumoniae strain R6 was kindly provided by Dr. Shinichi Yokota (Sapporo Medical University). The organism was grown in tryptic-soy (TS)3 broth (Difco) with spectinomycin (500 μg/ml) added to the medium to select an isogenic mutant strain. Escherichia coli strains XL-10 Gold (Stratagene) and BL21 (DE3) pLysE (Novagen) were grown in Luria-Bertani broth (Sigma) or on Luria-Bertani agar plates supplemented with 100 μg/ml of ampicillin and spectinomycin. Human laryngeal and alveolar cell lines HEp-2 (ATCC CCL-23) and A549 (ATCC CCL-185) were purchased from RIKEN Cell Bank (Japan).
Preparation of Pneumococcal Recombinant Proteins—The expression vectors were constructed as follows with the primers listed in Table 1. The eno, spr1345, spr1652, and spr1806 genes were amplified by PCR, and the resultant PCR fragments were cloned into pQE-30 vector (Qiagen). Recombinant proteins which eliminated an N-terminal signal peptide sequence and a C-terminal cell-anchoring region were purified using a QIAexpress protein purification system (Qiagen) according to the manufacturer's instructions.
TABLE 1.
PCR primers used in this study
| Designation | Sequence (5′ to 3′) | References |
|---|---|---|
| For expression of recombinant proteins | ||
| SB20 | GGATCCTTGTCAATTATTACTGATGTTTACGC | 42 |
| SB22 | AAGCTTTTATTTTTTAAGGTTGTAGAATGATTTC | 42 |
| spr1345/BamF | CGGGATCCCGTACAAAAACTGAGAACAAGGTC | This study |
| spr1345/PstR | AACTGCAGCTCATAAGTCGGAACTTCTGGG | This study |
| spr1652/BamF | CGGGATCCGAAGTTGTTACTAGTTCTTCACCG | This study |
| spr1652/HindR | CCCAAGCTTTTTTTGTTTTACATCTACTATTTCTTT | This study |
| spr1806/BamF | CGGGATCCGAAAATACACCAGGTGGAGATAAG | This study |
| spr1806/XmaR | TCCCCCCGGGATTGTTTGCCAGCAGGTGACTGGT | This study |
| For deletional mutagenesis | ||
| aad9 BamF | CGGGATCCTTGATTTTCGTTCGTGAATAC | This study |
| aad9 XbaR | GCTCTAGATTATAATTTTTTTAATCTGTTATTTAAATAG | This study |
| 1652KOuEcoF | GGAATTCGTGTCTTGTTCTAGTTTTCAATTCACCC | This study |
| 1652KOuBamR | CGGGATCCTTGATTATTACTTTTCTTAAAATAGATAG | This study |
| 1652KOdXbaF | GCTCTAGAATGGTAAAGATAAATAAAATTTGTTCG | This study |
| 1652KOdPstR | AACTGCAGTCAAACATCAATAACTACAATACTTGCATC | This study |
| For reverse transcription-PCR | ||
| spr1652D/BamF | GGATCCAGTGTAAAAGATTATGGTGCGGTAGG | This study |
| spr1652D/XmaR | CCCGGGGGAATTTTGAATAGTCACATTTTCAG | This study |
| Spn9802-143F | CAAGTCGTTCCAAGGTAACAAGTCT | 64 |
| Spn9802-304R | CTAAACCAACTCGACCACCTCTTT | 64 |
| Spn9828-19F | GGCATTGTGAATGGATTGATTG | 64 |
| Spn9828-245R | TCATGTGCATCCCAAGCTACA | 64 |
Mutant Construction—Inactivation of the spr1652 gene in S. pneumoniae was performed as described previously (16). pYT339 was constructed by inserting the aad9 gene into the BamHI and XbaI sites of pUC19. To construct the Δspr1652 mutant strain MY7, PCR products from the upstream and downstream regions of spr1652 were ligated into the pYT339 vector, and the resultant plasmids were linearized with HindIII and used to transform competent cells of the S. pneumoniae strain R6. To prepare competent cells, 0.5 ml of exponentialphase organisms in TS broth were added to prewarmed TS broth (9.5 ml) and incubated at 37 °C for 30 min. A portion (1 ml) of the culture was then removed and placed in a tube containing 100 ng of competence-stimulating peptide (17). After further incubation at 37 °C for 15 min, 0.2-ml portions were removed, placed in fresh tubes containing ∼0.1-μg of linearized plasmid (10 μl), and incubated at 37 °C for 2 h. Thereafter, each culture was plated onto TS blood agar and incubated at 37 °C for 24 h. Inactivation of the spr1652 gene in the mutant strain MY7 was confirmed by reverse transcription-PCR amplification using the spr1652D, Spn9802, or Spn9828 primer pairs (Table 1).
RNA Isolation and Reverse Transcription-PCR—Pneumococcal RNA was extracted from exponentially growing cultures in TS broth. Total RNA was prepared from cells using a TRIzol Max Bacterial RNA Isolation kit (Invitrogen), and then 0.8 μg of the total RNA was reverse-transcribed in the presence of random hexamers using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer's instructions.
Preparation of Rabbit Anti-PfbA Serum—All animal procedures were conducted in compliance with the Osaka University Graduate School of Dentistry guidelines and approved by the institutional Animal Care and Ethics Committee (accession number 05-018). On days 0, 21, 42, and 63, New Zealand White rabbits (Charles River Laboratories) were anesthetized with pentobarbital, as previously described (18), then given an injection of 500 μg of recombinant protein in Freund's complete or incomplete adjuvant. Serum was collected on day 84 and stored at -80 °C until use.
Ligand Blotting—Ligand blot analysis was performed as described previously (18). Briefly, human fibronectin (Fn, Sigma), human plasmin (Sigma), human plasminogen (Sigma), and human serum albumin (HSA, Sigma) were biotinylated using an ECL protein biotinylation kit (GE Healthcare), and the concentrations were adjusted to 10 μg/ml. Recombinant proteins were separated by electrophoresis on 10% SDS-PAGE gels, then transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 10% membrane blocking agent (GE Healthcare) at 4 °C for 18 h and incubated with biotinylated proteins for 1 h and with horseradish peroxidase-labeled streptavidin at room temperature for another hour. Bands were detected by using ECL Western-blotting detection reagents (GE Healthcare) and exposing the membrane to x-ray film (Fuji photo film) at room temperature for 5 s.
Biacore Analysis—Biacore analysis was performed as described previously (19). Briefly, Fn, plasmin, plasminogen, or HSA was diluted to 100 μg/ml in 10 mm sodium acetate (pH 4.0) and immobilized on the surface of a CM5 sensor chip using an Amine-coupling kit (GE Healthcare). Lyophilized recombinant PfbA was suspended in HBS-EP buffer (10 mm HEPES (pH 7.4), 150 mm NaCl, 3.4 mm EDTA, and 0.005% Surfactant P20) and adjusted to 0.625, 1.25, 2.5, 5, and 10 μm. For binding analysis the recombinant PfbA was injected at a flow rate of 20 μl/min at 25 °C. A blank was used as the reference, the results of which were subtracted from all raw data. To fit the binding kinetics data to a model, BIA evaluation Version 3.0.2 software (GE Healthcare) was applied, and the 1:1 Langmuir binding model was chosen.
Immunofluorescence and Confocal Microscopic Analysis—Immunofluorescent staining was performed as described previously (20, 21). Briefly, streptococcal cells were washed with phosphate-buffered saline (PBS) and blocked with 10% goat serum (Tissue Culture Biologicals). To observe the localization of PfbA, the bacterial cells were stained with SYBR green I, and surface PfbA was visualized using rabbit anti-PfbA serum followed by Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen). The stained bacteria were analyzed using an LSM 510 confocal laser scanning microscope (Carl Zeiss).
Flow-cytometric Analysis of S. pneumoniae—S. pneumoniae cells were cultured to the mid-log phase and harvested by centrifugation then washed twice with PBS and blocked with PBS containing 10% goat serum and 5% bovine serum albumin (BSA) for 1 h at 4 °C. Next, antisera were diluted 1:100 and incubated with the bacterial cells on ice for 1 h, then FITC-conjugated goat anti-rabbit IgG (Invitrogen) was added, and the mixture was incubated on ice for another 1 h. After washing the samples were analyzed with a CyFlow flow cytometer (Partec) using FlowJo 8.3.2 software (Tree Star, Inc.).
The Fn labeling procedure was performed using a FluoReporter® FITC protein labeling kit (Invitrogen) according to the manufacturer's instructions. S. pneumoniae cells were cultured to the mid-log phase and adjusted to 107 colony-forming units (CFU)/ml, then incubated with 0 or 10 μg/ml of FITC-labeled Fn for 30 min at 37 °C. The Fn binding activity on the surface of live bacterial cells was analyzed with a CyFlow flow cytometer.
Preparation of Streptococci for Adhesion and Invasion Assays—Pneumococci were cultured until the absorbance at 600 nm (A600) reached 0.6–0.7. The cells were then harvested by centrifugation and washed twice with PBS then resuspended in 1 ml of PBS. Biscarboxyethyl-carboxyfluorescein-pentaacetoxymethyl ester (Invitrogen), a nonvital intracellular dye, was added to the bacterial suspension to a final concentration of 1 mm. After a 30-min incubation at 37 °C, the fluorescent streptococci were washed 3 times with PBS and then used immediately in adhesion and invasion assays.
Streptococcal Adhesion and Invasion Assays—The bacterial adhesion to and invasion of human cells were quantified by standard procedures with minor modifications as described previously (22, 23). Briefly, HEp-2 and A549 cells were cultured in 24-well plates at a density of 1 × 105 cells per well and infected with 3 × 106 CFU of bacteria per well (multiplicity of infection, 1:30) for 1 h. To determine bacterial adhesion, the infected cells were washed 3 times with PBS and harvested with a trypsin solution. The fluorescence intensity in the cell lysates was measured in 96-well plates using a Wallac 1420 ARVO multilabel counter (PerkinElmer Life Sciences) to determine the number of S. pneumoniae. To measure bacterial invasion, the cells were washed 3 times and incubated for 1 h in Dulbecco's modified Eagle's medium (Sigma) containing gentamicin (100 μg/ml) and penicillin (100 units/ml). The cells were washed and lysed, and then the fluorescence intensity was measured as described above to determine the number of invaded S. pneumoniae.
To determine the Fn-dependent adhesion activity, 24 h before the addition of bacteria the cells were rinsed with Dulbecco's modified Eagle's medium and incubated with or without human Fn (10 μg/ml). Collagen-coated plates were used in the Fn-dependent adhesion experiment. The results are expressed as the mean ± S.D. of the percentage of S. pneumoniae recovered per well from six independent determinations. The assays were repeated three times, and representative results are shown. Statistical analysis was performed using a nonparametric Mann-Whitney U test. All tests were carried out using StatView J-5.0 software (SAS Institute Inc.).
Adhesion of PfbA-coated Fluorescent Beads to A549 Cells—Recombinant PfbA in BSA was covalently linked to 0.5-μm-diameter fluorescent beads (Invitrogen) according to the manufacturer's instructions. A549 cells were grown to confluence in 24-well plates and then washed twice with PBS. Next, 1 ml of Dulbecco's modified Eagle's medium containing 15 μl of PfbA-, BSA-, or non-coated fluorescent beads was added to the washed cells, and the plates were incubated at 37 °C, 5% CO2 for 1 h. The cells were washed 3 times with PBS and lysed with 200 μl of trypsin solution. To determine the number of fluorescent beads, the fluorescence intensity of the cell lysates was measured in 96-well plates using the Wallac 1420 ARVO multilabel counter with excitation at 485 nm and emission at 535 nm.
Scanning Electron Microscopic Analysis—A549 cells were grown to semi-confluence on coverslips and infected with the streptococcal strains as described above. The infected cells were incubated for 1 h, fixed with 2% glutaraldehyde for 1 h at room temperature, and washed with distilled water. The samples were dehydrated with 100% t-butyl alcohol and freeze-dried. Finally, the samples were coated with platinum and examined with an emission-scanning electron microscope (JSM-6390LVZ, JEOL Ltd.).
Bactericidal Assay—Lancefield bactericidal assays were performed as described previously (23, 24). Strains R6 and MY7 were grown, washed, and resuspended in 1 ml of PBS as previously described. Diluted cultures (10 μl) were combined with fresh human blood with or without rabbit anti-PfbA IgG or preimmune IgG (final 100 μg/ml, 90 μl), and the mixtures were rotated at 37 °C for 1, 2, or 3 h. The IgG (2.0 mg/ml) from rabbit serum was purified using a monoclonal antibody Trap kit (GE Healthcare) according to the manufacturer's instructions. Viable cell counts were determined by plating diluted samples onto blood agar.
RESULTS
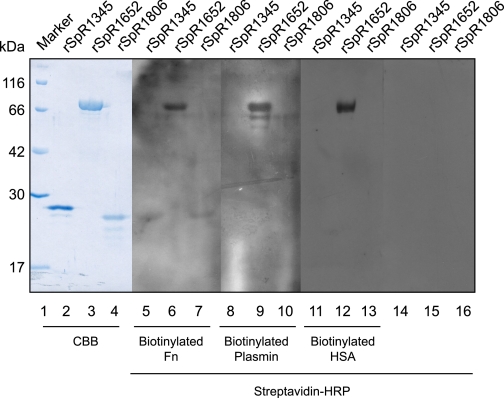
PfbA Protein Binds to Fibronectin, Plasmin(ogen), and Human Serum Albumin—An LPXTG motif is the cell-anchoring sequence in Gram-positive bacteria, and most of the LPXTG-containing proteins work as virulence factors. Gram-positive bacterial adhesins/invasins commonly possess the motif. In the present study we selected three different LPXTG motif-containing proteins, SpR1345, SpR1652, and SpR1806, from the S. pneumoniae strain R6 genome data base because of their recombinant proteins were easily water-soluble. SpR1345 (SP1492) is a mucin-binding protein (25), and SpR1652 (SP1833), annotated as a right-handed β-helical protein, is a probable polysaccharide-modifying enzyme (14). SpR1806 (SP1992) is a hypothetical protein. The 92–160-amino acid region of SpR1806 contained a domain of unknown function DUF1542 (E value 6.3 × 10-6), which is found in several cell-surface proteins. It has been suggested that some of these molecules function in antibiotic resistance and/or cellular adhesion.
Using ligand-blot analysis, we next investigated whether these recombinant proteins bound to host proteins. Our results showed that recombinant SpR1652 bound to Fn, plasmin, plasminogen, and HSA (Fig. 1, Table 2). We, therefore, focused our study on this protein, which we called PfbA (plasmin- and fibronectin-binding protein A). We next analyzed the binding constants using the surface plasmon resonance method with the Biacore system. The KD values of PfbA binding to Fn, plasmin, plasminogen, and HSA indicated moderate affinities and specific interactions (Table 3) that were close to those of other Fn-binding proteins (26).
FIGURE 1.
Ligand blotting among host proteins and pneumococcal LPXTG motif-containing proteins. Recombinant (r) SpR1345, rSpR1652, and rSpR1806 from S. pneumoniae were separated by SDS-PAGE then transferred to polyvinylidene difluoride membranes. Lanes 1–4 were stained with Coomassie Brilliant Blue (CBB). Lanes 5–7, 8–10, and 11–13 were incubated with biotinylated Fn, biotinylated plasmin, and biotinylated HSA, respectively, followed by horseradish peroxidase-labeled streptavidin. Lanes 14–16 were incubated with horseradish peroxidase (HRP)-labeled streptavidin alone. Bound biotinylated molecules were detected with ECL reagents.
TABLE 2.
Ligand blotting among host proteins and pneumococcal proteins harboring LPXTG motif
|
Host proteins
|
Recombinant surface proteins
|
||
|---|---|---|---|
| rSpR1345 | rSpR1652 | rSpR1806 | |
| Fibronectin | – | + | – |
| Laminin | – | – | – |
| Plasmin | – | + | – |
| Plasminogen | – | + | – |
| Human serum albumin | – | + | – |
| Complement C3a | – | – | – |
| Complement C3b | – | – | – |
| Collagen type I | – | – | – |
| Collagen type IV | – | – | – |
TABLE 3.
Kinetics and affinity constants between PfbA and the binding host proteins determined with Biacore rPfbA was employed as an analyte.
| Ligands | ka | kd | KD |
|---|---|---|---|
| m–1 s–1 | s–1 | μm | |
| Fibronectin | 7.42 × 103 | 0.0302 | 4.07 |
| Plasmin | 2.59 × 104 | 0.0607 | 2.35 |
| Plasminogen | 5.57 × 103 | 0.0327 | 5.88 |
| Human serum albumin | 1.28 × 104 | 0.0908 | 7.12 |
Sequence Analysis of PfbA—In the original genome annotation of S. pneumoniae strain R6 (27), PfbA (SpR1652) was characterized as a cell-wall surface anchor family protein. Indeed, PfbA possessed a typical LPXTG motif in its C terminus followed by a stretch of positively charged amino acid residues. In the N-terminal region, a deduced signal-peptidase cleavage site was located between amino acids 47 and 48. A BLAST search showed an Fn type III repeat in the 115–133-amino acid region of PfbA. In Fn itself, the first of several type III repeats binds to the type I repeat of another Fn molecule, and this interaction has been proposed as a point of regulation in Fn-Fn interactions (28).
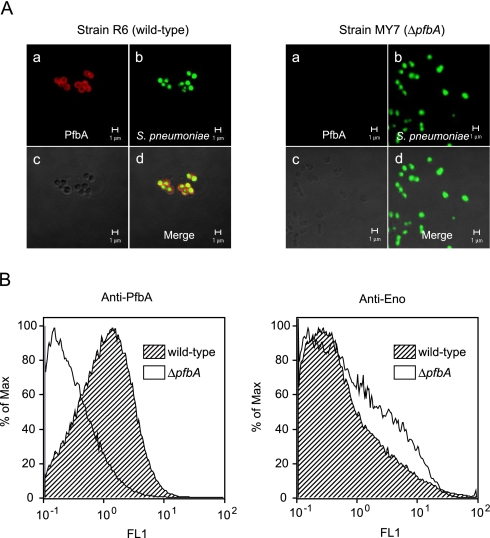
PfbA Is a Pneumococcal Cell-surface Protein—Immunofluorescence microscopic analysis revealed that PfbA was localized to the pneumococcal cell surface (Fig. 2A; red in image). In addition, flow cytometric analysis showed that S. pneumoniae strain R6 was stained positively by the anti-PfbA serum, whereas ΔpfbA strain MY7 did not react with it (Fig. 2B). These findings indicated that PfbA is localized to the cell surface of live pneumococci. There was no significant difference in the surface expression of pneumococcal α-enolase, used as a negative control, between strains R6 and MY7 (Fig. 2B).
FIGURE 2.
Localization of PfbA on S. pneumoniae. A, stained S. pneumoniae surface structures were observed using a confocal laser scanning microscope (Zeiss LSM 510). Panel a, PfbA was visualized with rabbit anti-PfbA serum and Alexa Fluor 594-conjugated goat anti-rabbit IgG (red). Panel b, S. pneumoniae was stained with SYBR Green I (green). Panel c, differential interference micrographs. Panel d, merged image. B, flow cytometric measurement of the binding of PfbA antiserum to the pneumococcal surface. The percent of maximum is the number of cells in each bin divided by the number of cells in the bin that contained the largest number of cells. The bin is a numerical range for the parameter on the x axis. Anti-Enolase was used as a control for R6 (wild-type) and MY7 (ΔpfbA) strains.
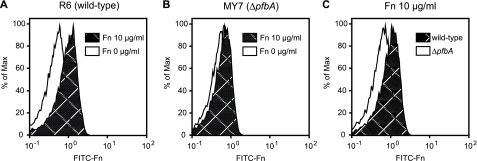
In addition, we examined the differences between strains R6 and MY7 using a fluorescence-activated cell sorter system and FITC-labeled Fn (Fig. 3). The fluorescence intensity was substantially lower for strain R6, incubated without FITC-labeled Fn, as compared with R6, incubated with FITC-labeled Fn, whereas the histograms for strain MY7 showed similar patterns regardless of the existence of FITC-labeled Fn. However, fluorescence intensity was substantially lower for strain MY7 than R6 when the bacteria were incubated with FITC-labeled Fn. These results suggest that PfbA is expressed and functions as an Fn-binding protein on the surface of S. pneumoniae.
FIGURE 3.
Binding activities of live S. pneumoniae with FITC-Fn. Fn binding activity on the bacterial surface was analyzed using a fluorescence-activated cell sorter system. Data are represented by histograms. Presented results are representative of three different experiments with similar histograms. A and B, S. pneumoniae strains were preparedat107 CFU/ml and incubated with (shaded) or without (white) 10 μg/ml of FITC-labeled Fn. C, the results are shown as histograms. FITC-labeled Fn was incubated with the wild-type (shaded) or ΔpfbA (white) strain.
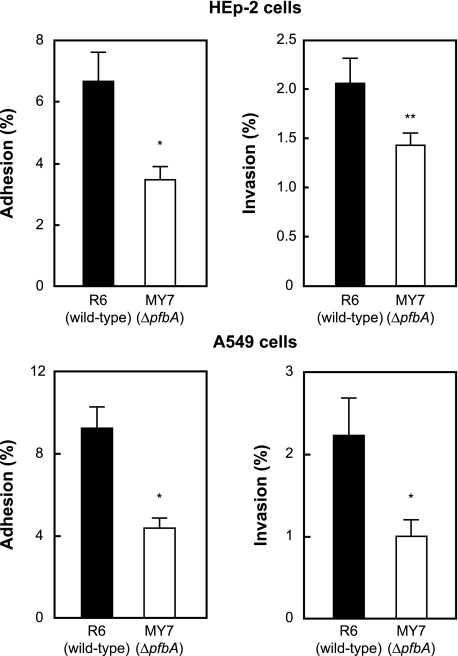
PfbA Facilitates Pneumococcal Adhesion to and Invasion of Epithelial Cells—To investigate the function of PfbA, the adhesion and the invasion efficiencies of strain R6 and its mutant, MY7, were compared using HEp-2 and A549 cells, which are human laryngeal and alveolar lung epithelial cell lines, respectively. Strain MY7 showed half the adhesion and invasion efficiencies of R6 for these epithelial cells (Fig. 4).
FIGURE 4.
Effects of PfbA on bacterial adhesion to and invasion of human epithelial cells. S. pneumoniae strain R6 and its isogenic ΔpfbA mutant strain MY7 were examined for their adhesion and invasion activities. Pneumococcal infection of epithelial cell lines A549 and HEp-2 was performed for 1 h at 37 °C, 5% CO2. The number of adherent and invaded bacteria was determined as described under “Experimental Procedures.” Three experiments were performed, and the data show the mean of six wells from a representative experiment. S.D. are represented by vertical lines.*, p < 0.005; **, p < 0.05.
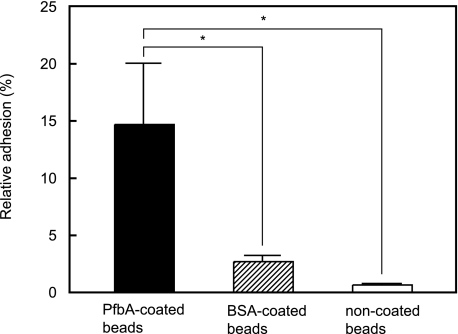
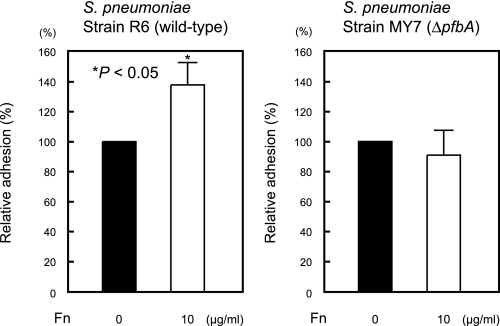
Next, we confirmed the cell binding activity of PfbA by using PfbA-coated fluorescent beads. The relative rates of adhesion of PfbA-, BSA-, and non-coated fluorescent beads to A549 cells are shown in Fig. 5. The PfbA-coated fluorescent beads showed at least 5-fold greater adhesion than the BSA-coated beads and 20-fold greater adhesion than the non-coated samples, indicating that PfbA functions directly as an adhesin. Furthermore, strain R6 showed increased adhesion to A549 cells after the addition of Fn, but strain MY7 did not (Fig. 6). These results indicate that PfbA serves, at least in part, as an Fn-dependent adhesin and invasin.
FIGURE 5.
PfbA-coated fluorescent beads bind to A549 cells. BSA-coated and non-coated fluorescent beads were used as negative controls. Three experiments were performed, and the data presented show the mean of six wells from a representative experiment. S.D. are represented by vertical lines. *, p < 0.005 (Mann-Whitney U test.)
FIGURE 6.
PfbA is an Fn-dependent adhesin. Experiments were performed in the absence or presence of 10 μg/ml Fn. Bacterial adhesion to A549 cells in the absence of Fn was considered to be 100%. *, significant difference (p < 0.05) between the means, determined using a Mann-Whitney U test.
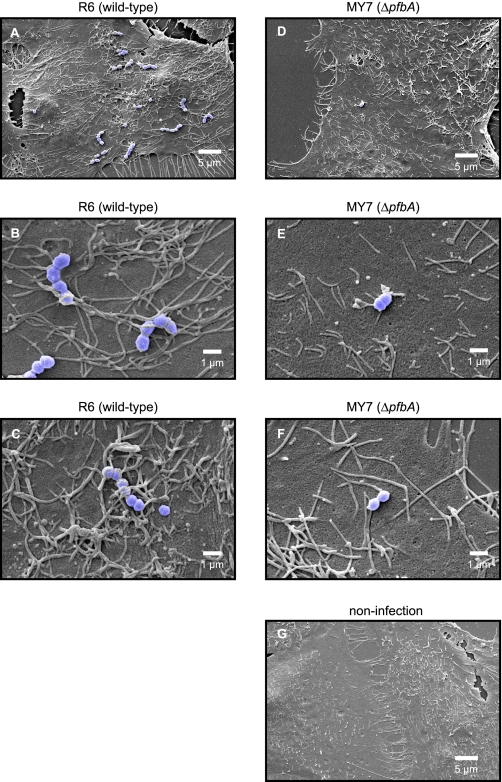
Scanning Electron Microscopic Analysis of Morphologic Changes of A549 Cells Infected by Pneumococci—Scanning electron microscopic analysis showed that only a few microvilli had passive contact with strain MY7 cells at the point of their adhesion to the cell surface (Fig. 7, D–F), whereas the point of contact of strain R6 cells with the cellular membrane showed a massive recruitment of neighboring microvilli (Fig. 7, A and B). At the attachment point, elongated microvilli adhered to the bacterial surface and formed a tight network (Fig. 7B), which then fused to cover the streptococcal chains (Fig. 7C).
FIGURE 7.
Scanning electron microscopic analysis of pneumococcal adhesion to A549 cells. Pneumococcal infection of A549 epithelial cells was performed for 1 h at 37 °C, 5% CO2. A549 cells were infected with pneumococci at a multiplicity of infection of 1:30. S. pneumoniae strain R6 (wild-type) triggered a massive recruitment of microvilli (A–C), whereas MY7 (ΔpfbA) triggered a lower level of recruitment (D–F). The microvilli surrounded the R6 pneumococcal cells (B and C). A549 cells infected with no pneumococci appeared similar to those infected with MY7 with regard to the recruitment of microvilli (G).
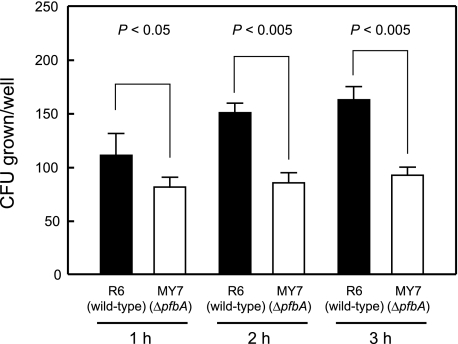
PfbA Possesses Antiphagocytotic Activity—We performed bactericidal assays to investigate the function of PfbA during the early phase of infection. A low number of bacteria (100 CFU) was incubated with 100 μl of human whole blood, and then antiphagocytotic activities were determined based on the viability of the wild-type and ΔpfbA mutant strains in human blood. The bacterial growth activity of the ΔpfbA mutant strain MY7 in human whole blood was ∼73, 57, and 57% after 1, 2, and 3 h, respectively, that of the wild-type strain R6 (Fig. 8).
FIGURE 8.
Role of PfbA in antiphagocytosis. Bacteria (10 μl, ∼100 CFU) were added to heparinized whole blood (90 μl) and then gently mixed for 1, 2, or 3 h at 37 °C. Next the mixture was serially diluted and plated on TS agar. After incubation the number of CFU was determined. Three experiments were performed, and the data are from a representative experiment. S.D. are represented by vertical lines.
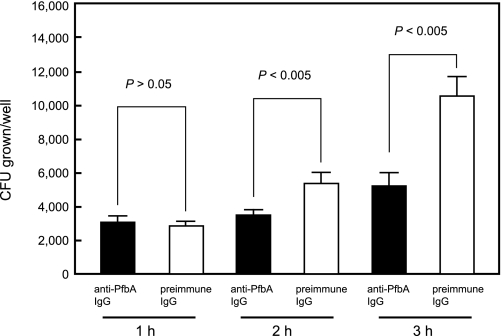
Opsonic Activity of the Anti-PfbA Antibody—We also examined the efficacy of the anti-PfbA antibody for passive immunotherapy during the period in which the bacteria grew in the tissues. Briefly, a large number of S. pneumoniae (1000 CFU) was mixed with anti-PfbA or anti-PfbA IgG antibodies, after which the mixture was adjusted to 100 μl with human blood. Bacterial growth in human whole blood containing the anti-PfbA antibodies was ∼107, 65, and 50% after 1, 2, and 3 h, respectively, of bacterial growth in the blood containing the preimmune IgG (Fig. 9).
FIGURE 9.
Survival of S. pneumoniae in the presence of rabbit anti-PfbA IgG. Bacteria (10 μl, ∼1000 CFU) were added to heparinized whole blood (85 μl), mixed with rabbit anti-PfbA IgG or preimmune IgG (5 μl), and gently rotated for 1, 2, or 3 h at 37 °C. Next, the mixture was serially diluted and plated on TS agar. After incubation the number of CFU was determined. Three experiments were performed, and the data are from a representative experiment. S.D. are represented by vertical lines.
DISCUSSION
Colonization and invasive infection by S. pneumoniae involve its expression of adhesins that target host components, and these proteins have been proposed to contribute to pathogen colonization or resistance to both the innate and adaptive immune systems (29). Several bacterial surface proteins, such as the choline binding proteins SpsA (also referred to as CbpA and PspC) (30–33) and PspA (34, 35), the ABC metal permeases Adc and PsaA (36–38), and the plasmin(ogen) binding proteins (39) PavA (16, 40) and SlrA (41), are crucial for pneumococcal pathogenesis. These pneumococcal proteins adhere to a variety of extracellular matrix (42, 43) and serum proteins, such as factor H (44–46), immobilized Fn (16, 47), plasmin(ogen) (42, 48), and lactoferrin (34, 43), and these interactions are involved in pneumococcal pathogenesis (30, 32, 34, 39, 48). In the present study we demonstrated that PfbA possesses Fn-, plasmin(ogen)-, and human serum albumin-binding activities. Interestingly, our results also indicated that PfbA expression is important for pneumococcal colonization in cell culture and for antiphagocytotic activity in bactericidal assays.
Bacterial adhesion to and internalization into human epithelial cells were reduced in the absence of PfbA. The ability to bind Fn either in the fluid phase or when immobilized onto a surface is a common property of streptococci and has been postulated to accelerate the adhesion of S. pyogenes to epithelial cells (49) and promote the binding of viridans streptococci in the thrombotic vegetations associated with infective endocarditis (50). Moreover, Fn binding activity is associated with the invasive properties of S. pyogenes. The matrix form of Fn enhances the binding of S. pyogenes to host cells (51), and PrtF1, a major Fn-binding protein, mediates this organism's invasion of human epithelial cells (22, 52, 53). Bacterial surface proteins bound to Fn form a bridge to α5β1 integrins, which leads to the rearrangement of the cytoskeletal actin in the host cells and the uptake of the bacteria (54, 55). S. pneumoniae strain TIGR4 is reported to possess two Fn-binding proteins, PavA and SP0082 (which is equal to SpR0075 in strain R6) (56). Although PavA seems not to function directly as an adhesin, it is probably involved in modulating other as yet unidentified virulence determinants of pneumococci (40). It remains to be determined whether SP0082 contributes to pneumococcal adhesion and invasion. The present results indicate that PfbA has a direct role as an adhesin and that it acts at least in part in the Fn-dependent adhesion to and invasion of epithelial cells. Scanning electron micrographs of A549 cells infected with pneumococci showed that infection with the ΔpfbA mutant strain resulted in morphological changes, including cell flattening and loss of microvilli, which were not observed in cells infected with the wild-type pneumococcal strain. These findings indicate that PfbA might mediate microvilli elongation or recruitment through binding to fibronectin.
Several Fn binding proteins are reported to play a role in phagocytosis resistance (57, 58). In the present experiments we found that the wild-type pneumococcal strain was more resistant to phagocytosis in human peripheral blood than the ΔpfbA mutant strain. Several hypotheses could explain the antiphagocytosis effect of PfbA. A previous study demonstrated that plasmin, formed by the conversion of plasminogen by staphylokinase, cleaves human IgG as well as human C3b from the bacterial surface, leading to impaired phagocytosis by human neutrophils (59). PfbA has plasmin(ogen)-binding activity and, therefore, its antiphagocytosis function might be related to the activity of plasmin. Additional possible mechanisms include the avoidance of phagocytosis by the molecular mimicry of host-protein-bound PfbA or by forming bacterial aggregates via an Fn-collagen complex, as previously reported for SfbI-expressing S. pyogenes (57). Although the exact molecular mechanism is not yet understood, PfbA apparently represents a pneumococcal antiphagocytic effector molecule.
Studies have demonstrated that a protective immune response against S. pyogenes can develop after immunization with the Fn-binding proteins SfbI (PrtF1) (60), Fbp54 (61), or FbaA (19). In addition, PfbA is predicted to belong to the β-helix group of proteins, which are rare and generally specific to the microbial world, and drugs fashioned to target them may benefit from their specificity toward infectious agents (62).
There has been growing concern about pneumococcal infection in the elderly, because the number of elderly has been increasing worldwide over the last century (63) and because aging is associated with the development of pneumococcal pneumonia (2). Therefore, the global morbidity and the mortality from pneumococcal infections may greatly increase in the near future. The identification of an antigenic protein that localizes to the pneumococcal cell surface would be useful for the development of vaccines and the prevention of pneumococcal infections. The present findings demonstrated that pneumococcal growth in human whole blood containing PfbA antibodies was reduced by 50% compared with its growth without PfbA antibodies. Thus, PfbA may be an attractive candidate for a component of a protein-based pneumococcal vaccine.
This study was supported in part by a grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology, grants-in-aid for Scientific Research (B), by the Japan Society for the Promotion of Science (JSPS) Fellows from JSPS, and by grants from the Naito Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TS, tryptic soy; Fn, fibronectin; HSA, human serum albumin; PBS, phosphate-buffered saline; BSA, bovine serum albumin; CFU, colony-forming units; FITC, fluorescein isothiocyanate.
References
- 1.File, T. M. (2003) Lancet 362 1991-2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutiérrez, F., Masiá, M., Mirete, C., Soldán, B., Rodríguez, J. C., Padilla, S., Hernández, I., Royo, G., and Martin-Hidalgo, A. (2006) J. Infect. 53 166-174 [DOI] [PubMed] [Google Scholar]
- 3.Hammerschmidt, S., Wolff, S., Hocke, A., Rosseau, S., Müller, E., and Rohde, M. (2005) Infect. Immun. 73 4653-4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talay, S. R. (2005) Contrib. Microbiol. 12 90-113 [DOI] [PubMed] [Google Scholar]
- 5.Bisno, A. L., Brito, M. O., and Collins, C. M. (2003) Lancet Infect. Dis. 3 191-200 [DOI] [PubMed] [Google Scholar]
- 6.Voyich, J. M., Musser, J. M., and DeLeo, F. R. (2004) Microbes Infect. 6 1117-1123 [DOI] [PubMed] [Google Scholar]
- 7.Urban, C. F., Lourido, S., and Zychlinsky, A. (2006) Cell. Microbiol. 8 1687-1696 [DOI] [PubMed] [Google Scholar]
- 8.Terao, Y., Mori, Y., Yamaguchi, M., Shimizu, Y., Ooe, K., Hamada, S., and Kawabata, S. (2008) J. Biol. Chem. 283 6253-6260 [DOI] [PubMed] [Google Scholar]
- 9.Butler, J. C., Hofmann, J., Cetron, M. S., Elliott, J. A., Facklam, R. R., and Breiman, R. F. (1996) J. Infect. Dis. 174 986-993 [DOI] [PubMed] [Google Scholar]
- 10.Hofmann, J., Cetron, M. S., Farley, M. M., Baughman, W. S., Facklam, R. R., Elliott, J. A., Deaver, K. A., and Breiman, R. F. (1995) N. Engl. J. Med. 333 481-486 [DOI] [PubMed] [Google Scholar]
- 11.Novak, R., Henriques, B., Charpentier, E., Normark, S., and Tuomanen, E. (1999) Nature 399 590-593 [DOI] [PubMed] [Google Scholar]
- 12.Oishi, K., Yoshimine, H., Watanabe, H., Watanabe, K., Tanimura, S., Kawakami, K., Iwagaki, A., Nagai, H., Goto, H., Kudoh, S., Kuriyama, T., Fukuchi, Y., Matsushima, T., Shimada, K., Matsumoto, K., and Nagatake, T. (2006) Respirology 11 429-436 [DOI] [PubMed] [Google Scholar]
- 13.Jedrzejas, M. J. (2001) Microbiol. Mol. Biol. Rev. 65 187-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigden, D. J., Galperin, M. Y., and Jedrzejas, M. J. (2003) Crit. Rev. Biochem. Mol. Biol. 38 143-168 [DOI] [PubMed] [Google Scholar]
- 15.Jedrzejas, M. J. (2004) Front. Biosci. 9 891-914 [DOI] [PubMed] [Google Scholar]
- 16.Holmes, A. R., McNab, R., Millsap, K. W., Rohde, M., Hammerschmidt, S., Mawdsley, J. L., and Jenkinson, H. F. (2001) Mol. Microbiol. 41 1395-1408 [DOI] [PubMed] [Google Scholar]
- 17.Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11140-11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terao, Y., Kawabata, S., Kunitomo, E., Murakami, J., Nakagawa, I., and Hamada, S. (2001) Mol. Microbiol. 42 75-86 [DOI] [PubMed] [Google Scholar]
- 19.Terao, Y., Okamoto, S., Kataoka, K., Hamada, S., and Kawabata, S. (2005) J. Infect. Dis. 192 2081-2091 [DOI] [PubMed] [Google Scholar]
- 20.Terao, Y., Kawabata, S., Nakata, M., Nakagawa, I., and Hamada, S. (2002) J. Biol. Chem. 277 47428-47435 [DOI] [PubMed] [Google Scholar]
- 21.Terao, Y., Yamaguchi, M., Hamada, S., and Kawabata, S. (2006) J. Biol. Chem. 281 14215-14223 [DOI] [PubMed] [Google Scholar]
- 22.Ozeri, V., Rosenshine, I., Mosher, D. F., Fässler, R., and Hanski, E. (1998) Mol. Microbiol. 30 625-637 [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi, M., Terao, Y., Ogawa, T., Takahashi, T., Hamada, S., and Kawabata, S. (2006) Microbes Infect. 8 2791-2796 [DOI] [PubMed] [Google Scholar]
- 24.Wessels, M. R., Goldberg, J. B., Moses, A. E., and DiCesare, T. J. (1994) Infect. Immun. 62 433-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bumbaca, D., Littlejohn, J. E., Nayakanti, H., Lucas, A. H., Rigden, D. J., Galperin, M. Y., and Jedrzejas, M. J. (2007) Proteins 66 547-558 [DOI] [PubMed] [Google Scholar]
- 26.Schwarz-Linek, U., Höök, M., and Potts, J. R. (2006) Microbes Infect. 8 2291-2298 [DOI] [PubMed] [Google Scholar]
- 27.Hoskins, J., Alborn, W. E., Arnold, J., Blaszczak, L. C., Burgett, S., DeHoff, B. S., Estrem, S. T., Fritz, L., Fu, D. J., Fuller, W., Geringer, C., Gilmour, R., Glass, J. S., Khoja, H., Kraft, A. R., Lagace, R. E., LeBlanc, D. J., Lee, L. N., Lefkowitz, E. J., Lu, J., Matsushima, P., McAhren, S. M., McHenney, M., McLeaster, K., Mundy, C. W., Nicas, T. I., Norris, F. H., O'Gara, M., Peery, R. B., Robertson, G. T., Rockey, P., Sun, P. M., Winkler, M. E., Yang, Y., Young-Bellido, M., Zhao, G., Zook, C. A., Baltz, R. H., Jaskunas, S. R., Rosteck, P. R., Jr., Skatrud, P. L., and Glass, J. I. (2001) J. Bacteriol. 183 5709-5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguirre, K. M., McCormick, R. J., and Schwarzbauer, J. E. (1994) J. Biol. Chem. 269 27863-27868 [PubMed] [Google Scholar]
- 29.Pandiripally, V., Wei, L., Skerka, C., Zipfel, P. F., and Cue, D. (2003) Infect. Immun. 71 7119-7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, J. R., Mostov, K. E., Lamm, M. E., Nanno, M., Shimida, S., Ohwaki, M., and Tuomanen, E. (2000) Cell 102 827-837 [DOI] [PubMed] [Google Scholar]
- 31.Elm, C., Braathen, R., Bergmann, S., Frank, R., Vaerman, J. P., Kaetzel, C. S., Chhatwal, G. S., Johansen, F. E., and Hammerschmidt, S. (2004) J. Biol. Chem. 279 6296-6304 [DOI] [PubMed] [Google Scholar]
- 32.Iannelli, F., Chiavolini, D., Ricci, S., Oggioni, M. R., and Pozzi, G. (2004) Infect. Immun. 72 3077-3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenow, C., Ryan, P., Weiser, J. N., Johnson, S., Fontan, P., Ortqvist, A., and Masure, H. R. (1997) Mol. Microbiol. 25 819-829 [DOI] [PubMed] [Google Scholar]
- 34.Shaper, M., Hollingshead, S. K., Benjamin, W. H., Jr., and Briles, D. E. (2004) Infect. Immun. 72 5031-5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yother, J., and Briles, D. E. (1992) J. Bacteriol. 174 601-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry, A. M., and Paton, J. C. (1996) Infect. Immun. 64 5255-5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dintilhac, A., Alloing, G., Granadel, C., and Claverys, J. P. (1997) Mol. Microbiol. 25 727-739 [DOI] [PubMed] [Google Scholar]
- 38.Marra, A., Lawson, S., Asundi, J. S., Brigham, D., and Hromockyj, A. E. (2002) Microbiology 148 1483-1491 [DOI] [PubMed] [Google Scholar]
- 39.Bergmann, S., Rohde, M., Preissner, K. T., and Hammerschmidt, S. (2005) Thromb. Haemostasis 94 304-311 [DOI] [PubMed] [Google Scholar]
- 40.Pracht, D., Elm, C., Gerber, J., Bergmann, S., Rohde, M., Seiler, M., Kim, K. S., Jenkinson, H. F., Nau, R., and Hammerschmidt, S. (2005) Infect. Immun. 73 2680-2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermans, P. W., Adrian, P. V., Albert, C., Estevão, S., Hoogenboezem, T., Luijendijk, I. H., Kamphausen, T., and Hammerschmidt, S. (2006) J. Biol. Chem. 281 968-976 [DOI] [PubMed] [Google Scholar]
- 42.Bergmann, S., Rohde, M., Chhatwal, G. S., and Hammerschmidt, S. (2001) Mol. Microbiol. 40 1273-1287 [DOI] [PubMed] [Google Scholar]
- 43.Hammerschmidt, S., Bethe, G., Remane, P. H., and Chhatwal, G. S. (1999) Infect. Immun. 67 1683-1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dave, S., Brooks-Walter, A., Pangburn, M. K., and McDaniel, L. S. (2001) Infect. Immun. 69 3435-3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duthy, T. G., Ormsby, R. J., Giannakis, E., Ogunniyi, A. D., Stroeher, U. H., Paton, J. C., and Gordon, D. L. (2002) Infect. Immun. 70 5604-5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janulczyk, R., Iannelli, F., Sjöholm, A. G., Pozzi, G., and Björck, L. (2000) J. Biol. Chem. 275 37257-37263 [DOI] [PubMed] [Google Scholar]
- 47.van der Flier, M., Chhun, N., Wizemann, T. M., Min, J., McCarthy, J. B., and Tuomanen, E. I. (1995) Infect. Immun. 63 4317-4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergmann, S., Rohde, M., and Hammerschmidt, S. (2004) Infect. Immun. 72 2416-2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasty, D. L., Ofek, I., Courtney, H. S., and Doyle, R. J. (1992) Infect. Immun. 60 2147-2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baddour, L. M. (1994) Infect. Immun. 62 2143-2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada, N., Watarai, M., Ozeri, V., Hanski, E., Caparon, M., and Sasakawa, C. (1997) J. Biol. Chem. 272 26978-26984 [DOI] [PubMed] [Google Scholar]
- 52.Molinari, G., Talay, S. R., Valentin-Weigand, P., Rohde, M., and Chhatwal, G. S. (1997) Infect. Immun. 65 1357-1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molinari, G., Rohde, M., Guzmán, C. A., and Chhatwal, G. S. (2000) Cell. Microbiol. 2 145-154 [DOI] [PubMed] [Google Scholar]
- 54.Wang, B., Yurecko, R. S., Dedhar, S., and Cleary, P. P. (2006) Cell. Microbiol. 8 257-266 [DOI] [PubMed] [Google Scholar]
- 55.Schröder, A., Schröder, B., Roppenser, B., Linder, S., Sinha, B., Fässler, R., and Aepfelbacher, M. (2006) Mol. Biol. Cell 17 5198-5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bumbaca, D., Littlejohn, J. E., Nayakanti, H., Rigden, D. J., Galperin, M. Y., and Jedrzejas, M. J. (2004) OMICS 8 341-356 [DOI] [PubMed] [Google Scholar]
- 57.Dinkla, K., Rohde, M., Jansen, W. T., Carapetis, J. R., Chhatwal, G. S., and Talay, S. R. (2003) Mol. Microbiol. 47 861-869 [DOI] [PubMed] [Google Scholar]
- 58.Hyland, K. A., Wang, B., and Cleary, P. P. (2007) Infect. Immun. 75 3188-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rooijakkers, S. H., van Wamel, W. J., Ruyken, M., van Kessel, K. P., and van Strijp, J. A. (2005) Microbes Infect. 7 476-484 [DOI] [PubMed] [Google Scholar]
- 60.Guzmán, C. A., Talay, S. R., Molinari, G., Medina, E., and Chhatwal, G. S. (1999) J. Infect. Dis. 179 901-906 [DOI] [PubMed] [Google Scholar]
- 61.Kawabata, S., Kunitomo, E., Terao, Y., Nakagawa, I., Kikuchi, K., Totsuka, K., and Hamada, S. (2001) Infect. Immun. 69 924-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simkovsky, R., and King, J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3575-3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lutz, W., Sanderson, W., and Scherbov, S. (2008) Nature 451 716-719 [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, N., Seki, M., Nakano, Y., Kiyoura, Y., Maeno, M., and Yamashita, Y. (2005) J. Clin. Microbiol. 43 4528-4534 [DOI] [PMC free article] [PubMed] [Google Scholar]