Abstract
SusB, an 84-kDa α-glucoside hydrolase involved in the starch utilization system (sus) of Bacteroides thetaiotaomicron, belongs to glycoside hydrolase (GH) family 97. We have determined the enzymatic characteristics and the crystal structures in free and acarbose-bound form at 1.6Å resolution. SusB hydrolyzes the α-glucosidic linkage, with inversion of anomeric configuration liberating the β-anomer of glucose as the reaction product. The substrate specificity of SusB, hydrolyzing not only α-1,4-glucosidic linkages but also α-1,6-, α-1,3-, and α-1,2-glucosidic linkages, is clearly different from other well known glucoamylases belonging to GH15. The structure of SusB was solved by the single-wavelength anomalous diffraction method with sulfur atoms as anomalous scatterers using an in-house x-ray source. SusB includes three domains as follows: the N-terminal, catalytic, and C-terminal domains. The structure of the SusB-acarbose complex shows a constellation of carboxyl groups at the catalytic center; Glu532 is positioned to provide protonic assistance to leaving group departure, with Glu439 and Glu508 both positioned to provide base-catalyzed assistance for inverting nucleophilic attack by water. A structural comparison with other glycoside hydrolases revealed significant similarity between the catalytic domain of SusB and those of α-retaining glycoside hydrolases belonging to GH27, -36, and -31 despite the differences in catalytic mechanism. SusB and the other retaining enzymes appear to have diverged from a common ancestor and individually acquired the functional carboxyl groups during the process of evolution. Furthermore, sequence comparison of the active site based on the structure of SusB indicated that GH97 included both retaining and inverting enzymes.
Bacteroides thetaiotaomicron, the genome of which has been fully sequenced (1), is a bacterial symbiont that is a dominant member of the intestinal microbiota of humans and other mammals. The number of glycoside hydrolases encoded by B. thetaiotaomicron is much greater than reported for any other sequenced bacterium (2), in accordance with the fact that this bacterium is known to salvage energy from nutrients, particularly carbohydrates, which are otherwise nondigestible by the host. According to CAZy (3–5), the genome of B. thetaiotaomicron encodes 230 glycoside hydrolases. The B. thetaiotaomicron proteome includes 172 glycoside hydrolases, 163 outer membrane polysaccharide-binding proteins, and 20 sugar-specific transporters (2). Therefore, studies regarding these enzymes may lead to the discovery of new enzymatic specificities and contribute to the development of enzyme chemistry.
The well known polysaccharide utilization system of B. thetaiotaomicron is the starch utilization system (sus) operon (6). The sus operon contains transcriptional regulator (SusR) (7, 8) and seven genes, the products of which are involved in binding (SusC–F) (9) and hydrolyzing (SusA, -B, and -G) starch (10). Genetic and biochemical analyses indicated that SusC and SusD are required for starch binding in the outer membrane of the bacterium. SusE and SusF are dispensable for starch utilization, but they contribute to stabilization of the starch-binding complex. SusA and SusG show similarity with neopullulanases, which are typical starch-hydrolyzing enzymes. Disruption of susA and susG markedly influences the growth rate of B. thetaiotaomicron (10, 11). Neopullulanase is a member of the α-amylase family, glycoside hydrolase (GH)2 family 13, and both this and related enzymes have been studied extensively (12–16). These enzymes hydrolyze α-1,4- and α-1,6-glucosidic linkages, of the substrates such as amylose, pullulan, and cyclomaltodextrins, and also catalyze transglycosylation to form α-1,4- and α-1,6-glucosidic linkages.
Much less is known about the biochemical characteristics of SusB, i.e. neither its three-dimensional structure nor its catalytic properties are known. SusB has been reported to function in the breakdown of oligosaccharides released by SusA and SusG into glucose units (10). Disruption of susB showed no residual α-glucoside hydrolase activity in the cell-free extract (10). The molecular mass (80 kDa) and pI (5.7) of a partially purified enzyme with α-glucoside hydrolase activity from B. thetaiotaomicron was close to the molecular mass of 84 kDa and pI of 6.0 estimated from the amino acid sequence of SusB. Therefore, SusB has been regarded as an α-glucosidase (10). However, α-glucoside hydrolases are definitively divided into two groups of enzymes, α-glucosidases (EC 3.2.1.20) and glucoamylases (EC 3.2.1.3), which have different catalytic mechanisms; α-glucosidases hydrolyze α-glucosidic linkages with a retaining mechanism, whereas glucoamylases hydrolyze these linkages with an inverting mechanism. Moreover, SusB has no significant sequence similarity to any known α-glucosidases belonging to GH4, -13, or -31, or to glucoamylases belonging to GH15 or -63. The catalytic mechanism of action of SusB is still unclear. Recently, it has been experimentally clarified that a sequence homolog of SusB from Tannerella forsythensis (Bacteroides forsythus), with 83% similarity and 71% identity, has the ability to hydrolyze 4-methylumbelliferyl α-d-glucoside (17). Based on this study, a new glycoside hydrolase family, GH97, was created by CAZy, and 77 sequences including SusB are now categorized as members.
In this study, we clarified the enzymatic characteristics of SusB from B. thetaiotaomicron, which indicated it to be an inverting α-glucoside hydrolase. The three-dimensional structures of free and acarbose-bound SusB have also been determined at 1.6 Å resolution as a first structure of GH97 family. The structure revealed not only the catalytic mechanism but also molecular evolution of this enzyme; SusB shows considerable structural similarity with α-retaining glycosidases even though it catalyzes hydrolysis by an inverting mechanism. Furthermore, we discovered that GH97 family includes both retaining and inverting enzymes.
EXPERIMENTAL PROCEDURES
Materials—B. thetaiotaomicron ATCC 29148 was obtained from American Type Culture Collection (Manassas, VA). Isomaltose, kojibiose, and nigerose were obtained from Wako Pure Chemicals Co. (Osaka, Japan). Malto-oligosaccharides were purchased from Nihon Shokuhin Kako Co. (Tokyo, Japan). p-Nitrophenyl α-glucopyranoside was obtained from Nacalai Tesque (Kyoto, Japan).
Expression of SusB and Mutants—The gene encoding susB (GenBank™ accession number NC_004663) was amplified from the genomic DNA of B. thetaiotaomicron by PCR using the following oligonucleotides: 5′ forward primer 5′-ATA AAT AGA ATG AAA AAG AGA AAG ATT T-3′ and 3′ reverse primer 5′-GTT ATT TCT TTT CCT TAT TAT AAT CTT TTC-3′. PCR amplification was performed for 25 cycles using following conditions: 94 °C, 30 s; 50 °C, 30 s; 74 °C, 2 min. PCR product was used as a template for the next PCR introducing NdeI and XhoI recognition sites. PCR was performed for 25 cycles using following conditions: 98 °C, 10 s; 50 °C, 2 s; 74 °C, 2 min, using the primers listed in Table 1. The resultant DNA fragment was digested with NdeI and XhoI and ligated with pET28a (Novagen, Madison, WI), prior to digestion with NdeI and XhoI. After sequence analysis, a clone without any PCR errors was designated as psusBET28a encoding the SusB protein with His6 and linker sequence at its N terminus (MGSSHHHHHHSSGLVPRGSHM) instead of the predicted signal sequence (1MKKRKILSLIAFLCISFIANA21). The constructed expression vector psusBET28a was transformed into Escherichia coli Rosetta (DE3) (Novagen), and positive colonies were selected. The transformant was first inoculated into 30 ml of LB media containing 30 μgml-1 kanamycin and 50 μgml-1 chloramphenicol at 37 °C with shaking. This overnight culture was diluted in 300 ml of LB media containing 30 μgml-1 kanamycin and further grown at 37 °C up to an A600 of ∼0.7, and then protein production was induced with 0.2 mm isopropyl β-d-thiogalactopyranoside for 14 h at 37 °C. The induced cells were centrifuged at 10,000 × g, and the cell pellet was resuspended in buffer A (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm imidazole), including 0.2 mg/ml lysozyme, and incubated at 4 °C for 30 min, followed by sonication. The sonicated lysates were cloned and amplified by overlap extension PCR using the primers listed in Table 1.
TABLE 1.
Primers used for cloning and mutagenesis of SusB NdeI and XhoI sites are underlined.
| Compounds | Primers |
|---|---|
| Wild type | Forward, ATTGCGCATATGCAACAGAAATTAACCTCA |
| Reverse, TTTTCCCTCGAGTTATAATCTTTTCAA | |
| E508Q | Forward, ATGGTGAATGCACACCAAGCAACCCGCCCTACC |
| Reverse, GGTAGGGCGGGTTGCTTGGTGTGCATTCACCAT | |
| E532Q | Forward, TCCGCCCGCGGTACACAATATGAATCATTCGGA |
| Reverse, TCCGAATGATTCATATTGTGTACCGCGGGCGGA | |
| E439Q | Forward, ATGATGATGCATCACCAAACTTCCGCTTCTGTA |
| Reverse, TACAGAAGCGGAAGTTTGGTGATGCATCATCAT |
Purification of SusB and Mutants—The cell-free extract was applied to 7.5 ml of a nickel-chelating Sepharose fast flow column (Amersham Biosciences), prepared in accordance with the supplier's manual, and equilibrated with buffer A. After the column was washed with buffer B (50 mm Tris-HCl, pH 8.0, 300 mm NaCl) containing 20 mm imidazole, the absorbed fractions were eluted by a linear gradient of 20–500 mm imidazole in buffer B. Next, the fraction containing the SusB protein was loaded onto a Sephadex G-25 column equilibrated with 50 mm sodium acetate, pH 6.0, and 300 mm NaCl to remove imidazole. The protein for crystallization was further purified by DEAE-Sepharose FF (Amersham Biosciences) equilibrated with 20 mm Tris-HCl, pH 8.0. All purification steps were performed at 4 °C.
Enzyme Assay—The hydrolytic activity of wild type and mutants was measured in the standard reaction mixture containing 50 mm sodium acetate, pH 6.5, and 2 mm p-nitrophenyl α-d-glucopyranoside (pNPG), and enzyme was diluted in 50 mm sodium acetate, pH 6.5, containing 0.05 mg ml-1 bovine serum albumin at 37 °C. After incubation, the reaction was stopped by adding 2 volumes of 1 m sodium carbonate. The amount of p-nitrophenol released was measured by determining the absorption at 400 nm in a 1-cm cuvette, considering a molar extinction coefficient of 5,560 m-1 cm-1. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of reducing sugar in 1 min under these reaction conditions.
The kinetic constants were calculated by fitting to the Michaelis-Menten equation by nonlinear regression using the computer program Kaleidagraph version 3.6 (Synergy Software, Reading, PA), in which initial velocities (v) were measured under 10 substrate concentrations (s) in 40 mm sodium acetate buffer, pH 6.5, at 37 °C using 2.4–12 nm enzyme. The glucose released was quantified by the glucose oxidase method.
Ki values for the competitive inhibitors acarbose and Tris(2-aminoethyl)amine were determined with pNPG as the substrate using the equation v = [E]0[S]kcat/([S] + Km(1 + [I]/Ki)), where v is the initial rate of hydrolysis, [E]0 is the total enzyme concentration, and [S] and [I] are the substrate and inhibitor concentrations, respectively.
The pH activity dependence was determined using sodium barbital buffer, pH 4.0–9.0, and 2 mm pNPG as the substrate. For pH stability, aliquots of 20 μl of enzyme (6 nm) were kept at 4 °C for 24 h in sodium barbital buffer, pH 2.5–9.0, or 85 mm glycine-NaOH buffer, pH 9.0–12.8. After incubation, 40 μl of 200 mm MES-NaOH buffer, pH 6.5, was added, and remaining pNPG hydrolysis activity was examined under standard conditions, except 80 mm MES-NaOH buffer was used. The effects of temperature on pNPG hydrolysis were examined under standard conditions (0.49 nm enzyme) but at 25–70 °C for 10 min. For determination of thermal stability, the enzyme (4 nm) was kept at various temperatures for 15 min in 60 mm sodium acetate buffer, pH 6.5, containing 0.02 mg/ml bovine serum albumin, and residual activity was measured in the standard assay. To examine the effects of calcium ions, the apo-form of SusB was prepared by gel filtration through a Bio-Gel P-6 column equilibrated with 100 mm EDTA, pH 6.5, and residual activity was measured under the standard conditions.
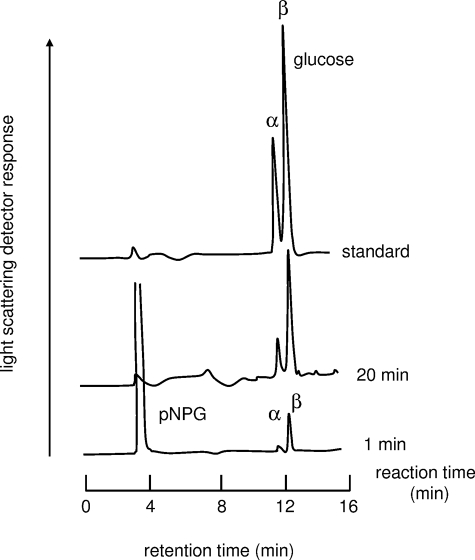
Analysis of the Anomeric Form of the Product—The anomeric form of the hydrolytic product from pNPG was determined by HPLC. The enzyme reaction was performed in 5 mm sodium phosphate buffer, pH 7.0, at 25 °C with an enzyme concentration of 12.5 μm. After incubation for 1 and 20 min, aliquots (10 μl) were loaded onto a TSK-GEL amide-80 column (4.6 × 250 mm; Tosoh, Tokyo, Japan) and eluted with 80% (v/v) acetonitrile at a flow rate of 1.2 ml/min at room temperature, separating the glucose anomers. The products were detected using a light-scattering detector (ELSD model 400; SofTA, Brighton, CO). The retention times of α- and β-glucose were confirmed by loading a solution of α-glucose (Sigma) and β-glucose (Sigma) onto the column.
Sequence Alignment—The amino acid sequences of GH97 members were obtained by BLAST search in sequence cluster UniRef50 using SusB as the query sequence. Among the hit sequences, the top 20 scoring sequences were aligned using ClustalW.
Crystallization—Initial crystallization trials were performed by the sitting-drop vapor diffusion method in 96-well plates using a series of crystallization kits produced by Hampton Research (Laguna Niguel, CA) and Emerald BioSystems (Bainbridge Island, WA). The crystals appeared under condition number 34 of Wizard II (100 mm imidazole, pH 8.0, 10% (w/v) polyethylene glycol 8000). After optimization of the crystallization conditions (pH, precipitant content and concentration, and protein concentration) using the hanging-drop vapor diffusion method in 24-well plates, well ordered crystals were obtained with 100 mm imidazole, pH 7.6–8.0 and 6–12% (v/v), polyethylene glycol 6000, or 2–8% (v/v) polyethylene glycol 20,000 and enzyme concentrations of 10 mg/ml. These triangle-shaped crystals appeared in 1 day and reached maximum size (0.2 × 0.2 × 0.05 mm) in 4 weeks at room temperature. The crystal of SusB in complex with acarbose (SusB-acarbose complex) was obtained by soaking a crystal in reservoir solution containing 10 mm acarbose for 24 h.
Data Collection and Processing—A SAD data set was collected to 2.2 Å resolution on an R-AXIS VII imaging-plate detector using an in-house CrKα (2.29 Å) source (Rigaku FR-E SuperBright with a Cu/Cr dual target). The crystal was transferred to reservoir solution supplemented with 35% (v/v) glycerol as a cryoprotectant, and then mounted using a loop- and buffer-less mount method (18) and flash-cooled under a stream of nitrogen gas at 100 K.
Although the high resolution data set (1.6 Å) was collected at beam line AR-NW12 of the Photon Factory (Tsukuba, Japan), the SusB-acarbose complex data were measured on an R-AXIS IV imaging plate diffractometer using in-house CuKα (1.54 Å) source (Rigaku MicroMax 007). For data collection, the native and complex crystals were soaked in reservoir solution supplemented with 30% (v/v) glycerol and 25% (v/v) 2-methyl-2,4-pentanediol, respectively. Subsequently the crystals were mounted in CryoLoop and flash-cooled under a stream of nitrogen gas at 100 K. All data sets were indexed, integrated, scaled, and merged using the HKL2000 program package (19). The crystals of SusB belong to the space group P21 with unit cell parameters a = 75.6 Å, b = 112.3 Å, c = 102.5 Å, and β = 100.7° (native data). The Vm value was estimated to be 2.54 Å3/Da with two molecules in an asymmetric unit, which corresponded to a solvent content of 51.5% (20). The statistics of data collection are summarized in Table 2.
TABLE 2.
Statistics of data collection Values in parentheses refer to the highest resolution shell.
| Data | Native | SAD | SusB-acarbose complex |
|---|---|---|---|
| Beam line/diffractometer | AR NW12, PF | FR-Ea | MicroMax 007a |
| Wavelength) | 1.0033 Å | 2.2909 Å (CrKα) | 1.5418 Å (CuKα) |
| Space group | P21 | ||
| Unit cell | a = 75.6 Å, b = 112.3 Å, c = 102.6 Å | a = 75.7 Å, b = 112.4 Å, c = 102.5 Å | a = 75.7 Å, b = 112.3 Å, c = 102.5 Å |
| β = 100.7° | β = 100.6° | β = 100.6° | |
| Resolution | 50.0-1.60 Å (1.66-1.60 Å) | 50.0-2.20 Å (2.28-2.20 Å) | 50.0-1.60 Å (1.66-1.60 Å) |
| No. of reflections | 220,597 | 85,123 | 217,504 |
| Completeness | 99.7% (97.5%) | 99.5% (98.5%) | 98.2% (95.8%) |
| Average redundancy | 5.5 (4.4) | 17.4 (15.5) | 7.4 (7.2) |
| I/σ (I) | 19.8 (3.0) | 33.7 (10.8) | 49.9 (7.9) |
| Rsymb | 6.8% (41.7%) | 6.8% (19.0%) | 4.6% (32.8%) |
FR-E and MicroMax 007 are in-house diffractometers (Rigaku)
Rsym = ΣhΣj|〈I〉h – Ih,j|/ΣhΣjIh,j, where 〈I〉h is the mean intensity of symmetry equivalent reflection h
Structure Solution and Refinement—The structure was solved by the SAD method using the anomalous signal of sulfur atoms that exist in natural SusB. The positions of anomalous scatterer atoms were located with the SnB program (21) using the in-house CrKα (2.29 Å) data set. Initial phases were calculated and improved with the SOLVE/RESOLVE program (22) at 2.2 Å resolution, and then extended to 1.6 Å resolution using the native data set during density modification with the DM program (23). The initial model was built automatically to 90% using the ARP/wARP program (24). Then the structure was refined automatically using the LAFIRE program (25) running with the refinement program CNS (26) to a final R-factor of 17.6% and Rfree factor of 19.3%. The structure of SusB-acarbose complex was determined by rigid-body refinement with CNS using the structure of native SusB as the initial model. The model of acarbose in the complex was built based on Fo - Fc difference Fourier map. The final structure of SusB-acarbose complex was obtained by automatic refinement using LAFIRE with CNS. The refinement statistics are given in Table 3.
TABLE 3.
Statistics of structure refinement
| SusB | SusB-acarbose complex | |
|---|---|---|
| Resolution range | 15 to 1.6 Å | 20 to 1.6 Å |
| No. of used reflections | 220,497 | 217,407 |
| Completeness | 99.8% | 98.3% |
| R-factora | 17.6% | 17.1% |
| Rfree factorb | 19.3% | 18.6% |
| Total no. of non-hydrogen atoms | ||
| Protein | 11,566 (5,783 × 2 molecules) | 11,339 (5666, 5673) |
| Water | 1,422 | 1,523 |
| Others | 2 (1 × 2 molecules) | 90 (45 × 2 molecules) |
| Averaged B factors (Å2) | ||
| Protein | 19.0 | 17.4 |
| Water | 24.3 | 24.8 |
| Others | 9.8 | 14.1 |
| Ramachandran plotc | ||
| Most favored regions | 86.85% | 85.9% |
| In addition, allowed regions | 13.07% | 13.9% |
| Generously allowed regions | 0.08 | 0.2 |
R-factor = Σ|Fobs – Fcal|/ΣFobs, where Fobs and Fcal are observed and calculated structure factor amplitudes, respectively
Rfree-factor value was calculated for R-factor, using a subset (10%) of reflections that were not used for refinement
Ramachandran plot was calculated using PROCHECK (27)
RESULTS AND DISCUSSION
Biochemical Characteristics of Recombinant SusB—The recombinant SusB was produced in E. coli and purified by His affinity chromatography. The purified enzyme was migrated as a single band, the Mr of which was estimated to be 83,000 on SDS-PAGE. The Mr of 84,380 calculated from the deduced amino acid sequence was consistent with that of 83,000 seen on SDS-PAGE.
The effects of pH and temperature on the hydrolytic activities toward pNPG were examined. The pH optimum was 6.5, and the enzyme was stable between pH 4.0 and 12.0 at 4 °C for 24 h. The enzyme was stable at temperatures up to 45 °C for 15 min of heat treatment, and maximum hydrolysis rate was obtained at 45 °C. As described below, the crystal structure of SusB included a calcium ion at the active center. The dependence of the enzyme activity on the calcium ion was examined. The reaction rate was reduced from 80 to 3.8 μmol/min/mg (4.8%) for the apoenzyme compared with calcium-bound enzyme.
Inhibition constants, Ki, for acarbose and Tris were measured using pNPG as the substrate. Inhibition was strictly competitive in both compounds, and Ki values were 0.15 ± 0.03 and 37.5E3 ± 1.1E3 μm for acarbose and Tris, respectively. Strong inhibition by acarbose indicated that the reaction pathway of SusB involves an oxocarbenium ion-like transition state similar to standard glycoside hydrolases as acarbose, which possesses the valienamine unit at the nonreducing terminal end, is widely believed to be an oxocarbenium ion-like transition state analog. The kinetic parameters for various substrates with α-glucosidic linkage were investigated. Michaelis constant (Km) and the molecular activity (kcat) are summarized in Table 4. SusB had a wide specificity on various types of α-glucosidic linkage of not only α-1,4- but also α-1,2-, α-1,3-, and α-1,6-linkage in glucobioses. pNPG is the best substrate with a kcat/Km value 6-fold greater than that of maltose. Among the malto-oligosaccharides with various degrees of polymerization (DP), maltotriose was the best substrate. The kcat/Km value for panose (α-d-glucopyranosyl-(1→6)-α-d-glucopyranosyl-(1→4)-d-glucopyranose) was also high even though the value for isomaltose was low, suggesting that SusB shows higher specificity on trisaccharides. The activity was low on longer malto-oligosaccharides (DP ≥ 5). The role of SusB is to hydrolyze disaccharides and trisaccharides produced by neopullulanase, encoded on susA, into glucose units (10), and thus the preference for trisaccharides agrees with the role of SusB in vivo. The Km value for soluble starch was almost the same as that for maltotetraose, whereas the Km value for amylase DP17 was higher, suggesting that SusB should prefer amylopectin to amylose. The relationship between substrate specificity and structure of the active site is discussed below.
TABLE 4.
Kinetic constants for hydrolysis of various substrates by SusB
| Components | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s–1 | s–1 mm–1 | |
| Maltose | 1.05 ± 0.12 | 182 ± 0.58 | 172 |
| Maltotriose | 0.29 ± 0.01 | 270 ± 5.5 | 943 |
| Maltotetraose | 0.64 ± 0.07 | 321 ± 15 | 504 |
| Maltopentaose | 1.29 ± 0.07 | 434 ± 4.9 | 337 |
| Maltohexaose | 2.71 ± 0.51 | 346 ± 36.5 | 128 |
| Maltoheptaose | 4.58 ± 0.52 | 381 ± 13.5 | 83.3 |
| Amylose DP17 | 6.89 ± 0.45 | 321 ± 9.71 | 46.6 |
| Soluble starch | 0.67 ± 0.06 | 115 ± 2.08 | 172 |
| Kojibiose (α-1,2) | 2.39 ± 0.21 | 141 ± 7.6 | 58.9 |
| Nigerose (α-1,3) | 3.14 ± 0.16 | 207 ± 7.6 | 66.1 |
| Isomaltose (α-1,6) | 6.34 ± 0.35 | 105 ± 4.6 | 16.6 |
| Panose (α-1,6→α-1,4) | 0.31 ± 0.03 | 160 ± 6.5 | 513 |
| p-Nitrophenyl α-glucoside | 0.16 ± 0.01 | 161 ± 4.5 | 1,023 |
Anomeric Configuration of Product—SusB hydrolyzes the α-glucosidic linkage in the nonreducing terminal end of the substrate. Such exo-type hydrolases can be divided into two enzymes, glucoamylase (EC 3.2.1.3, 1,4-α-d-glucan glucohydrolase) and α-glucosidase (EC 3.2.1.20, α-d-glucoside glucohydrolase), by anomeric configuration of product, α-glucose or β-glucose. α-Glucosidase produces α-glucose by a retaining mechanism, whereas glucoamylase produces β-glucose by an inverting mechanism. The anomeric composition of the degradation products of pNPG by SusB was examined by HPLC. As shown in Fig. 1, the enzyme produced β-anomer of glucose in the reaction for 1 min, and α-glucose was detected with increasing reaction time. The α-glucose formation could be explained to occur through spontaneous mutarotation of the hydrolytic product, β-glucose. This result strongly suggests that SusB hydrolyzes the nonreducing terminal side of the α-glucosidic linkage of the substrate with an inverting mechanism, i.e. SusB is not an α-glucosidase but a glucoamylase.
FIGURE 1.
Anomeric analysis of the hydrolytic products of SusB. See text for experimental methods.
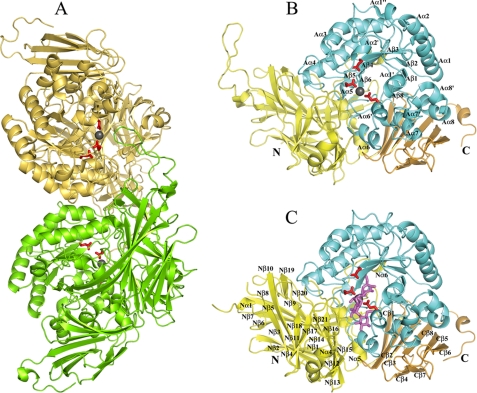
Overall Structure—The crystal structure of SusB was solved by the SAD method at 2.2 Å resolution. Fifty eight of 116 sulfur atoms plus two calcium atoms, of which the anomalous signal was estimated as twice that of sulfur atoms in the native protein, were found and subsequently used for phasing. After phase extension using native data, 90% initial model was automatically built and refined to 1.6 Å resolution. Fig. 2A shows two monomers (Mol-A and Mol-B) in an asymmetric unit. All of the 738 residues were built based on the electron density with the exception of residues 1–21 in the native structure, residues 1–20 and 71–84 of Mol-A, and residues 1–18 and 69–84 of Mol-B in the acarbose-bound complex structure. The two cis-peptides, Thr229-cis-Ala230 and Ile475-cis-Pro476, were found in each monomer. Each monomer of SusB is composed of three domains as follows: the N-terminal domain (domain N), catalytic domain (domain A), and the C-terminal domain (domain C). The three domains associate tightly and form a compact entity (Fig. 2B).
FIGURE 2.
Overall structure of SusB in apo and acrabose complex form. Three catalytic residues, Glu439, Glu508, and Glu532, are shown as stick models in red, and a bound calcium ion is shown as a gray sphere. A, dimer structure of SusB. Mol-A and Mol-B are colored green and khaki, respectively. B, monomer structure of SusB. Domains N, A, and C are shown in yellow, cyan, and gold, respectively (same in C). The secondary structure elements of domain A are labeled following the order of typical (β/α)8 barrel structures, and the prime and double prime refer to the atypical elements. C, monomer structure of SusB with bound acarbose molecule shown as stick model in magenta at the active site pocket. The secondary structure elements of domain N and C are labeled.
Domain N (residues 322) consists of five short helices and a bent helix and a distorted β-sandwich with 21 antiparallel β-strands (Fig. 2B). Four loops (residues 185–198, 213–231, 246–254, and 269–278) interact with domain A, and two (residues 185–198 and 213–231) of them form a part of the active site pocket. The helix (Nα2) and a loop (residues 110–119) interact with domain C. Two loops (residues 65–96 and 157–164) and a short β-strand (residues 279–281) contribute to contact with the other molecule in the asymmetric unit. The long loop of residues 65–96, which exists only in SusB in GH97, extends to the active site of the counterpart monomer (Fig. 2B). A DALI search showed that domain N has high degrees of structural similarity with galactose mutarotase (28) (PDB 1L7J; Z score, 12.3; r.m.s.d., 3.9 with 213 C-α atoms), the central domain of copper amine oxidase (29) (PDB 1OAC; Z score, 11.8; r.m.s.d., 4.0 with 220 C-α atoms), and the N-terminal domain of bacterial glucoamylase (30) (PDB 1LF6; Z score, 11.5; r.m.s.d., 3.4 with 180 C-α atoms). The β-sandwich structures of galactose mutarotase and copper amino oxidase are catalytically active by themselves, but the role of the N-terminal domain of bacterial glucoamylase remains unexplored. The structure of SusB revealed that domain N contributes to the formation of the active site.
The catalytic domain A (residues 323–622) forms a (β/α)8 barrel with a long loop (residues 551–566) and seven parallel β-strands packed around a central axis and surrounded by eight helices (Fig. 2B). Although the typical (β/α)8 barrel structure is composed of eight β-strands, the region corresponding to β-strand 7 of the typical (β/α)8 was formed as a loop in SusB. Among the helices of the (β/α)8 barrel, helix 5 is a 310 helix, helix 6 is a disordered 310 helix, and helix 8 was bent close to 90°. In addition, a short α-helix (Aα6′) and five 310 helices (Aα1′, Aα1″, Aα2′, Aα7′, and Aα8′) are located between the β-strands and α-helices of (β/α)8 barrel. Ten residues located in loop between Aβ1 and Aα1, and loop between Aβ2 and Aα2, Aα3 and Aα4 contribute to contact with the other molecule in an asymmetric unit. In a DALI structural similarity search of domain A, the top 10 hits were all glycoside hydrolases included in GH27, -36, -13, -31, -84, and -5 (GH27, α-N-acetylgalactosaminidase (31) (PDB 1KTB) with Z score of 15.7, r.m.s.d. of 3.0 Å for 228 C-α atoms; GH36, α-galactosidase (PDB 1ZY9) with Z score of 16.8, r.m.s.d. of 3.2 Å for 234 C-α atoms; GH13, α-amylase (32) (PDB 2D0H) with Z score of 14.1, r.m.s.d. of 3.3 Å for 221 C-α atoms; GH31, α-glucosidase (33) (PDB 1XSK) with Z score of 13.4, r.m.s.d. of 3.2 Å for 224 C-α atoms; GH84, hyaluronidase (PDB 2J62) with Z score of 13.0, r.m.s.d. of 3.5 Å for 215 C-α atoms, GH5, endoglucanases (34) (PDB 1CEO) with Z score of 11.8, r.m.s.d. of 4.0 Å for 214 C-α atoms), implying that these proteins have a common ancestor, although these enzymes possess different substrate specificity and stereochemistry. Intriguingly, although all members of the above families function by a retaining mechanism, the catalytic mechanism of action of SusB is of the inverting type.
Domain C (residues 623–738) is a β-sandwich formed by eight antiparallel β-strands (Fig. 2B). One β-sheet is composed of six strands (Cβ1, Cβ3, Cβ4, Cβ6, Cβ7, and Cβ8), whereas the second small β-sheet has only two strands (Cβ1 and Cβ5). The former β-sheet interacts with Aα6, Aα7, and Aα8 of domain A and keeps these helices out of the solvent. Although many carbohydrate-binding domains found in glycosidases contain a β-sandwich fold as seen in SusB, a DALI structural comparison of domain C showed that the C-terminal β-sandwich domain of SusB is most similar to β-xylosidases (PDB 1PX8; Z score, 8.9, r.m.s.d. 2.3 with 86 C-α atoms; and PDB 1W91; Z score, 8.7, r.m.s.d. 2.6 with 87 C-α atoms), which belong to GH39 family and do not possess carbohydrate-binding module (35). The interaction between domain C and α-helices of domain A suggests that domain C is involved in stabilizing the catalytic domain.
Native PAGE shows that SusB exists as a dimer in solution. Consistent with this observation, two SusB molecules in an asymmetric unit contact each other to form a dimer. These two molecules are related by a noncrystallographic 2-fold axis, and they are very similar with r.m.s.d. of 0.12 Å for 717 C-α atoms. The total buried surface area per monomer upon dimerization was calculated using the program CNS to be 2,101 Å2, which was 8% of the total molecular accessible surface (26,998 Å2). The dimer contact region is at domains N and A involving 26 residues of each monomer. Although the long loop (residues 65–96) of each monomer is extended into the active pocket of the other in the native structure (Fig. 2A), participation of this loop in the catalysis is not clear as this loop is disordered in the SusB-acarbose complex.
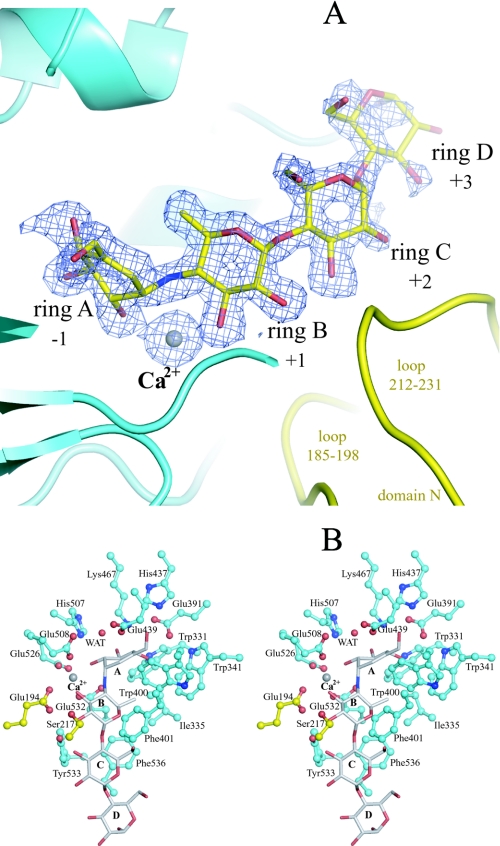
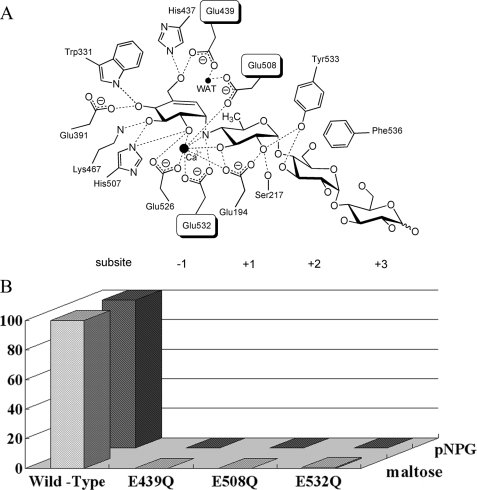
Active Center and Catalytic Mechanism of Action of SusB—To clarify the catalytic mechanism of action of SusB, the structure of the complex of SusB with acarbose that is a pseudotetrasaccharide inhibitor (36–38) has been elucidated. Both electron density maps of 2Fo - Fc and Fo - Fc clearly showed the shape of the acarbose molecule per monomer (Fig. 3A). The active site pocket is positioned at the C-terminal end of the barrel similarly to other (β/α)8 barrel proteins (Fig. 2C). The loops from the (β/α)8 barrel contribute the major part of the active site pocket, and two loops from domain N (residues 185–198 and 212–231) additionally construct a narrow plus-subsite groove (Figs. 2C and 3A).
FIGURE 3.
A, composite of the acarbose molecule and a calcium ion, and their omit map. The contour level of the Fo - Fc map is 3σ. Domains N and A are shown in yellow and cyan, respectively (the same colors are used in B). B, stereo views of the active pocket with bound acarbose (gray) and a calcium ion (gray). All oxygen and nitrogen atoms are shown in red and blue, respectively.
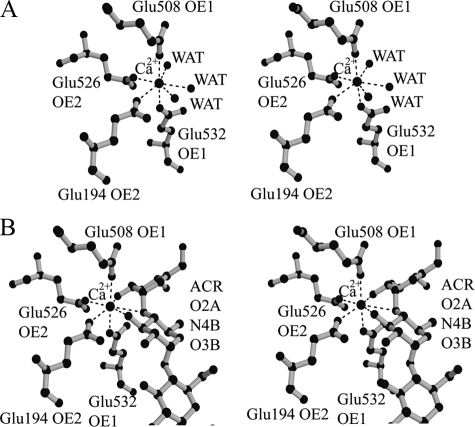
As shown in Fig. 4, one calcium ion is located at the active pocket in the structures of both native and acarbose complexes based on both electron density maps (2Fo - Fc and Fo - Fc) and its coordination manner in which all binding atoms are oxygen with a pentagonal bipyramid geometry. The O-ε1 of Glu508 and O-ε1 of Glu532 are positioned on the tops of the pentagonal bipyramid, whereas the O-ε2 of Glu194, O-ε2 of Glu526, and three water molecules form the plane of the pentagonal bipyramid (Fig. 4A). The distances between the calcium ion and the above atoms in the native form (Mol-A) are 2.26–2.55 Å. In the SusB-acarbose complex, the binding pentagonal bipyramid was twisted and the three water molecules were replaced by acarbose (O2A of ring A, O3B of ring B, and N4B) (Fig. 4B). As no reagents containing calcium ions had been added throughout the purification and crystallization procedures, these observations indicated that the calcium ion was derived from the E. coli host cell and bound tightly to the active center of SusB. The assay experiment of apoenzyme shows that calcium ion plays an important role to the catalytic activity; for example, the calcium ion accumulates functional groups and substrate and serves as a general Lewis acid, polarizing the anomeric carbon and rendering it a better electrophile.
FIGURE 4.
Stereo view of a calcium binding site in native (A) and acarbose complex (B) structures. The calcium ion is shown as a large black sphere, and water molecules are shown as small black spheres. All residues coordinated to the calcium ion are indicated.
The valienamine unit (ring A, subsite -1) of acarbose is located at the deepest end of the active site pocket formed by domain A and is held dominantly by hydrogen bonds of numerous residues (Figs. 3B and 5A). A significant number of hydrogen bonds are evidently responsible for recognition of the glucose moiety at subsite -1. The 4-amino-4,6-dideoxy-α-d-glucose unit (ring B, subsite +1) and two glucose units (ring C and D, subsite +2 and +3) bind a long loop (508–542 residues) containing a 310 helix Aα6′ between Aβ6 and Aα6, and additionally loops from domain N (residues 185–198 and 212–231) cover rings B and C from the opposite face (Figs. 2C and 3B). Tyr533 on the long loop (residues 508–542) hydrogen bonds to O-2 of ring B. Glu194 and Ser217 on loops from domain N make hydrogen bonds to O-2 and O-3, and O-2 of ring B, respectively (Figs. 3B and 5A). The ring B at subsite +1 is also surrounded by hydrophobic residues Ile335, Trp341, Trp400, and Phe401. Nevertheless, these aromatic residues are not involved in typical hydrophobic stacking interactions with ring B. However, in the case of pNPG as a substrate, which is the best substrate (i.e. with the highest kcat/Km value) of SusB, the aromatic nitrophenyl ring may be located against the aromatic rings of Trp341, Trp400, or Phe401 and thus resulting in fixed torsion angles of the glucopyranoside-phenyl bond in the transition state of pNPG and decreased activation energy of the transition state. SusB shows relatively loose fixation of ring B compared with ring A, resulting in the wide substrate specificity of SusB, which hydrolyzes not only α-1,4- but also α-1,6-, α-1,3-, and α-1,2-glucosidic linkages. Although ring C (subsite +2) forms only one hydrogen bond with Tyr533, the moiety is juxtaposed to Phe536 (Figs. 3B and 5A). The substrate specificity with a preference for trisaccharides of SusB is probably because of the tight hydrophobic stacking interaction of Phe536. Ring D (subsite +3) is situated in the active site pocket close to the surface of the protein. No hydrogen bonding or other interactions between hydroxyl groups of ring D and SusB were found, and substrates longer than tetrasaccharide seem to protrude out into the solvent. These observations agree with the kinetic parameters that the kcat/Km values of SusB decrease with longer malto-oligosaccharides.
FIGURE 5.
Catalysis by SusB. A, schematic drawing of the hydrolytic center of SusB with the acarbose binding. Hydrogen bonds are shown as dotted lines, and Ca2+ and water molecules as black spheres. B, hydrolysis activities of 3 SusB mutants. The saturated activities relative to those of the wild type SusB are shown. The activity measurements were performed three times, and the experimental values were averaged.
SusB is an inverting glycoside hydrolase, which cleaves the nonreducing α-glucosidic bond via inversion of anomeric configuration at C-1 and formation of β-glucose. It is generally accepted that the catalytic mechanism of action of inverting glycosidases involves two carboxylic acids, one acting as general catalytic acid donating protons to the O-glycosidic linkage and thus enhancing leaving group departure and another acting as a general catalytic base assisting the nucleophilic attack of water at C-1. The spatial position of SusB and acarbose molecule shows that O-ε2 of Glu532, which is on the 310 helix in the long loop (residues 508–542), is closest to N4B of acarbose (2.8 Å) (Fig. 3B). In addition, Glu532 is absolutely conserved in members of this family (Fig. 6). Therefore, Glu532 is the most likely candidate for the catalytic acid. Comparing the native structure with the acarbose complex structure revealed that the side chain of Glu532 rotates around 30° and comes closer to N4B of acarbose (2.8 Å) resulting in formation of a hydrogen bond. In inverting glycoside hydrolases, a catalytic water molecule, the proton of which is abstracted by the action of a general base catalyst, must be positioned to attack an anomeric carbon from the opposite side of the general acid catalyst across the glycosidic bond (β-face of α-glucoside). In the active site of SusB, a water molecule is located near the anomeric carbon (3.37 Å) and pinched by the side chains of Glu508 and Glu439, which are located at the C termini of β3 and β5, respectively (Fig. 3B). This is the most likely candidate for the catalytic water molecule, and these glutamate residues could play a role as the catalytic base and facilitate nucleophilic attack of the catalytic water molecule. The importance of these residues (Glu439, Glu508, and Glu532) was verified by mutational experiments; point mutations of these residues to Gln resulted in the complete loss of the hydrolysis activity (Fig. 5B).
FIGURE 6.
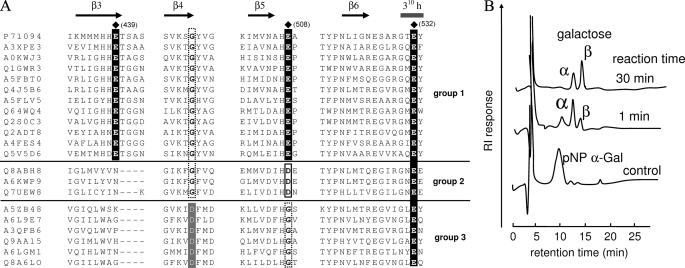
Partial multiple amino acid sequence alignment of GH97 and anomeric analysis. A, cluster names around β3, β4, β5, and β6 are shown as abbreviations, omitting the prefix “UniRef50_”; for example, the accurate name of P71094 is UniRef50_P71094. Secondary structural elements of SusB are indicated as β-strand (→) and helix (-). The catalytic residues of SusB are indicated by (♦) and conserved equivalent residues are indicated by dark shading. Conserved Asp residues in the group 2 are boxed. Conserved Asp residues at β4 in group 3 are indicated by light shading. B, anomeric analysis of the hydrolytic products of protein named Q8A6LO from B. thetaiotaomicron in A. See text for experimental methods.
The carboxylic side chain of Glu526, which is absolutely conserved in members of this family, is positioned close to the cleavage site and hydrogen bonds to the O-2 of ring A and the calcium ion (2.86 and 2.33 Å, respectively) (Fig. 5A). The hydrogen bond to O-2 of the glycan moiety could enhance stabilization of the oxocarbenium-like transition state by donating an electron. Glu526 is also located 3.15 Å from the acid catalyst residue, and is potentially responsible for increasing pKa of the acid catalyst. The acid catalyst should be protonated in the free enzyme, for this purpose, the acid catalyst requires a modulator to raise the pKa, at the optimum pH condition, pH 6.5. The negative electrical charge of the Glu526 side chain probably stabilizes the protonated state of the acid catalyst. These observations suggest that Glu526 is a key residue for forming the reaction environment and substrate binding in the GH97 family.
Structural Comparison with Other (β/α)8 Barrel GH Enzymes—The GH14 β-amylases are a well known group of (α→β) inverting glycoside hydrolases bearing a canonical (β/α)8 barrel, which catalyze release of β-configured maltose from the nonreducing ends of α-glucosyl linkages, such as starch. However, structural analysis by DALI search indicated a lower level of similarity between domain A of SusB and Bacillus cereus β-amylase (39) (PDB 1B9Z; Z score, 9.3; r.m.s.d., 3.9 Å with 203 C-α position) than retaining enzymes. Moreover, the arrangement of the catalytic residues is completely different between SusB and β-amylase. The catalytic acid and two base residues of SusB are located at the ends of β6, β3, and β5, respectively, but those of β-amylase are on β4 (acid) and β7 (base), respectively. Therefore, substrate binding and the catalytic mechanism are quite different between SusB and β-amylase. As mentioned above, analysis using the DALI server indicated that SusB and retaining enzymes, such as members of GH27, -36, -31, -5, -84, and -13, show significant structural similarity. Among them, GH27, -31, and -36 enzymes seem to have a closer evolutionary relationship with SusB than other relatives even though their catalytic mechanisms are different. Structural superimposition of SusB, GH27 α-N-acetylgalactosaminidase, GH31 α-xylosidase, and GH36 α-galactosidase showed that they share not only a similar fold structure but also some amino acid residues important for catalysis, Glu391, Lys467, and Glu532 (residues are numbered with reference to SusB) (Fig. 5A). Glu391 and Lys467 are hydrogen-bonded to O-4 and O-3 of acarbose ring A, respectively, at subsite -1 and Glu532 is the catalytic acid residue in SusB. Equivalent residues in GH27, -31, and -36 enzymes are also hydrogen-bonded to O-4 and O-3 of the sugar at subsite -1 and catalytic acid/base residues (Fig. 5A).
In both inverting and retaining mechanisms, a pair of carboxyl groups function as the catalyst, but the role of one catalyst located in the opposite face of the glucosidic linkage (β-face of α-glucoside) is markedly different. The carboxyl group acts as a general base catalyst in the inverting mechanism, assisting catalytic water in nucleophilic attack on anomeric carbon, whereas in the retaining mechanism a nucleophile catalyst directly attacks the anomeric carbon. The residues at the end of β6, providing protons to the α-glycosidic linkage, are conserved across GH97, -27, -31, and -36, as mentioned above (equivalent to Glu532 of SusB). In contrast, whereas GH27, -31, and -36 enzymes have a residue providing a nucleophilic catalyst at the end of β4, there is no carboxylic acid group around the end of β4 in SusB. Alternatively, SusB possesses Glu439 and Glu508 on the ends of β3 and β5, respectively, and they pinch a catalytic water molecule near β4 (Fig. 5). As GH97, -27, -31, and -36 have a common ancestor, these findings suggest that SusB has acquired the machinery to catch catalytic water near β4, whereas the α-retaining enzymes have acquired a nucleophilic catalyst residue at the end of β4 during the process of evolution, and thus SusB hydrolyzes α-glucosidic linkages with an inverting mechanism.
This is similar to the situation in the well characterized lysozymes. There are two types of lysozyme, those with a retaining mechanism of catalysis, such as hen egg white lysozyme (HEWL), and those with an inverting mechanism, such as T4 phage lysozyme (T4L). Both enzymes share a common “lysozyme fold” even though there is no similarity between their amino acid sequences (HEWL, GH22; T4L, GH24). Each lysozyme has a highly conserved glutamic acid on the β-side of the substrate (Glu35 in HEWL, Glu11 in T4L; acting as a proton donor). However, it shows considerable variability on the α-face. The variability is responsible for the differences in the catalytic mechanism (40). The relation between T4L and HEWL closely resembles that between SusB and GH27, -31, -36, and -13 demonstrated in this study.
Furthermore, multiple amino acid sequence alignment of GH97 family members raised the possibility of a similar relation in GH97. Based on the multiple alignment, GH97 enzymes can be divided into three groups (Fig. 6A) as follows: 1) enzymes possessing three critical residues (Glu439, Glu508, and Glu532), such as SusB; 2) enzymes possessing Asp and Glu residues, equivalent to Glu508 and Glu532 of SusB, respectively; 3) enzymes possessing a Glu residue equivalent to Glu532, but no Glu residue equivalent to Glu508 and Glu439. As the carboxyl group of the Asp residue of the enzymes categorized into the group 2 could function as a base catalyst, the enzymes probably have a catalytic mechanism similar to that of SusB. However, in group 3, the acidic residue equivalent to Glu508 is replaced with Gly, and there is no candidate base catalyst around the β-face. Thus, the enzymes in the group 3 presumably have a different mechanism or catalyze a different reaction. Interestingly, the enzymes in the group 3 show a conserved Asp residue at the end of β4, where conserved Gly is situated in groups 1 and 2 (Fig. 6A). As mentioned above, the enzymes structurally related to SusB show the conserved Asp residue at β4, and the Asp residue functions as a nucleophilic catalyst. Thus, it is possible that the Asp residues at the end of β4 in the group 3 can act as a nucleophilic catalyst, and catalysis is via a retaining mechanism. To confirm this assumption, we chose one (Q8A6LO from B. thetaiotaomicron) of the enzymes in the group 3 (Fig. 6A), and performed the anomeric analysis. The result (Fig. 6B) clearly showed that GH97 includes both inverting and retaining enzymes. The details of Q8A6LO protein will be published elsewhere.
Acknowledgments
We thank staff of beamline AR-NW12, Photon Factory, Japan, for their help with data collection.
The atomic coordinates and structure factors (code 2D73 and 2ZQ0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) NC_004663.
This work was supported by the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science, and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GH, glycoside hydrolase; pNPG, p-nitrophenyl α-d-glucopyranoside; HPLC, high pressure liquid chromatography; MES, 4-morpholineethanesulfonic acid; PDB, Protein Data Bank.
References
- 1.Xu, J., Bjursell, M., Himrod, J., Deng, S., Carmichael, L., Chiang, H., Hooper, L., and Gordon, J. (2003) Science 299 2074-2076 [DOI] [PubMed] [Google Scholar]
- 2.Comstock, L., and Coyne, M. (2003) BioEssays 25 926-929 [DOI] [PubMed] [Google Scholar]
- 3.Henrissat, B. (1991) Biochem. J. 280 309-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrissat, B., and Bairoch, A. (1993) Biochem. J. 293 781-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat, B., and Bairoch, A. (1996) Biochem. J. 316 695-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper, L., Midtvedt, T., and Gordon, J. (2002) Annu. Rev. Nutr. 22 283-307 [DOI] [PubMed] [Google Scholar]
- 7.Cho, K., Cho, D., Wang, G., and Salyers, A. (2001) J. Bacteriol. 183 7198-7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Elia, J., and Salyers, A. (1996) J. Bacteriol. 178 7180-7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves, A., Wang, G., and Salyers, A. (1997) J. Bacteriol. 179 643-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Elia, J., and Salyers, A. (1996) J. Bacteriol. 178 7173-7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith, K., and Salyers, A. (1991) J. Bacteriol. 173 2962-2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, H., Kim, M., Cho, H., Kim, J., Kim, T., Choi, J., Park, C., Oh, B., and Park, K. (2002) J. Biol. Chem. 277 21891-21897 [DOI] [PubMed] [Google Scholar]
- 13.Park, K., Kim, T., Cheong, T., Kim, J., Oh, B., and Svensson, B. (2000) Biochim. Biophys. Acta 1478 165-185 [DOI] [PubMed] [Google Scholar]
- 14.Kim, J., Cha, S., Kim, H., Kim, T., Ha, N., Oh, S., Cho, H., Cho, M., Kim, M., Lee, H., Kim, J., Choi, K., Park, K., and Oh, B. (1999) J. Biol. Chem. 274 26279-26286 [DOI] [PubMed] [Google Scholar]
- 15.Kamitori, S., Kondo, S., Okuyama, K., Yokota, T., Shimura, Y., Tonozuka, T., and Sakano, Y. (1999) J. Mol. Biol. 287 907-921 [DOI] [PubMed] [Google Scholar]
- 16.Hondoh, H., Kuriki, T., and Matsuura, Y. (2003) J. Mol. Biol. 326 177-188 [DOI] [PubMed] [Google Scholar]
- 17.Hughes, C., Malki, G., Loo, C., Tanner, A., and Ganeshkumar, N. (2003) Oral Microbiol. Immunol. 18 309-312 [DOI] [PubMed] [Google Scholar]
- 18.Kitago, Y., Watanabe, N., and Tanaka, I. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61, 1013-1021 [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 20.Matthews, B. W. (1968) J. Mol. Biol. 33 491-497 [DOI] [PubMed] [Google Scholar]
- 21.Weeks, C. M., and Miller, R. (1999) J. Appl. Crystallogr. 32 120-124 [Google Scholar]
- 22.Terwilliger, T. C. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 17-23 [DOI] [PubMed] [Google Scholar]
- 23.Cowtan, K. D., and Main, P. (1996) Acta Crystallogr. Sect. D Biol. Crystallogr. 52, 43-48 [DOI] [PubMed] [Google Scholar]
- 24.Morris, R. J., Perrakis, A., and Lamzin, V. S. (2003) Methods Enzymol. 374 229-244 [DOI] [PubMed] [Google Scholar]
- 25.Yao, M., Zhou, Y., and Tanaka, I. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62, 189-196 [DOI] [PubMed] [Google Scholar]
- 26.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 27.Laskowski, R. A., Mac Arthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 28.Thoden, J., and Holden, H. (2002) J. Biol. Chem. 277 20854-20861 [DOI] [PubMed] [Google Scholar]
- 29.Parsons, M., Convery, M., Wilmot, C., Yadav, K., Blakeley, V., Corner, A., Phillips, S., McPherson, M., and Knowles, P. (1995) Structure (Lond.) 3 1171-1184 [DOI] [PubMed] [Google Scholar]
- 30.Aleshin, A. E., Feng, P.-H., Honzatko, R. B., and Reilly, P. J. (2003) J. Mol. Biol. 327 61-73 [DOI] [PubMed] [Google Scholar]
- 31.Garman, S., Hannick, L., Zhu, A., and Garboczi, D. (2002) Structure (Lond.) 10 425-434 [DOI] [PubMed] [Google Scholar]
- 32.Abe, A., Yoshida, H., Tonozuka, T., Sakano, Y., and Kamitori, S. (2005) FEBS J. 272 6145-6153 [DOI] [PubMed] [Google Scholar]
- 33.Lovering, A. L., Lee, S. S., Kim, Y. W., Withers, S. G., and Strynadka, N. C. (2005) J. Biol. Chem. 280 2105-2115 [DOI] [PubMed] [Google Scholar]
- 34.Dominguez, R., Souchon, H., Lascombe, M., and Alzari, P. M. (1996) J. Mol. Biol. 257 1042-1051 [DOI] [PubMed] [Google Scholar]
- 35.Yang, J., Yoon, H., Ahn, H., Lee, B., Pedelacq, J., Liong, E., Berendzen, J., Laivenieks, M., Vieille, C., Zeikus, G., Vocadlo, D., Withers, S., and Suh, S. (2004) J. Mol. Biol. 335 155-165 [DOI] [PubMed] [Google Scholar]
- 36.Truscheit, E., Frommer, W., Junge, B., Müller, L., Schmidt, D. D., and Wingender, W. (1981) Angew. Chem. Int. Ed. Engl. 20 744-761 [Google Scholar]
- 37.Svensson, B., and Sierks, M. R. (1992) Carbohydr. Res. 227 29-44 [DOI] [PubMed] [Google Scholar]
- 38.Mosi, R., Sham, H., Uitdehaag, J. C., Ruiterkamp, R., Dijkstra, B. W., and Withers, S. G. (1998) Biochemistry. 37 17192-17198 [DOI] [PubMed] [Google Scholar]
- 39.Mikami, B., Adachi, M., Kage, T., Sarikaya, E., Nanmori, T., Shinke, R., and Utsumi, S. (1999) Biochemistry 38 7050-7061 [DOI] [PubMed] [Google Scholar]
- 40.Kuroki, R., Weaver, L., and Matthews, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8949-8954 [DOI] [PMC free article] [PubMed] [Google Scholar]