Abstract
Posttranslational histone modifications serve to store epigenetic information and control both nucleosome assembly and recruitment of non-histone proteins. Histone methylation occurs on arginine and lysine residues and is involved in the regulation of gene transcription. A dynamic control of these modifications is exerted by histone methyltransferases and the recently discovered histone demethylases. Here we show that the hypoxia-inducible factor HIF-1α binds to specific recognition sites in the genes encoding the jumonji family histone demethylases JMJD1A and JMJD2B and induces their expression. Accordingly, hypoxic cells express elevated levels of JMJD1A and JMJD2B mRNA and protein. Furthermore, we find increased expression of JMJD1A and JMJD2B in renal cancer cells that have lost the von Hippel Lindau tumor suppressor protein VHL and therefore display a deregulated expression of hypoxia-inducible factor. Studies on ectopically expressed JMJD1A and JMJD2B indicate that both proteins retain their histone lysine demethylase activity in hypoxia and thereby might impact the hypoxic gene expression program.
Methylation of histones contributes to dynamic changes in chromatin structure (1-5) and thereby influences gene expression, DNA replication, and repair (6, 7). Mono- (me1), di- (me2), or tri- (me3) methylation has been described for five lysines in histone H3 (Lys-4, Lys-9, Lys-27, Lys-36, and Lys-79) as well as for one lysine within histone H4 (Lys-20). In general, di- and trimethylation of histone H3K4, H3K36, and H3K79 appear as hallmarks of active regions of chromatin, whereas the same modifications on H3K9, H3K27, and H4K20 are enriched in condensed, heterochromatic regions. However, it has proven difficult to classify histone marks as simply activating or repressing (6).
Recently, several members of the Jumonji protein family, which is characterized by the catalytic Jumonji C (JmjC)2 domain, have been identified as histone demethylases (reviewed in Refs. 8 and 9). JmjC domain proteins demethylate histone lysine and arginine residues in an oxidative reaction that requires Fe(II) and α-ketoglutarate as cofactors (10-15). Depending on their target specificity, JmjC domain proteins promote transcriptional repression or activation, and thereby impact important processes such as hormone response, stem cell renewal, germ cell development, and cellular proliferation and differentiation.
Interestingly, a range of JmjC proteins is induced in different cancers and has been linked to cell proliferation (10, 16-18) and suppression of senescence (19). Members of the JMJD2 family that target H3K9me3/me2 and H3K36me3/me2 are highly expressed in prostate cancer. JMJD2C/GASC1 associates with the androgen receptor and promotes both transcriptional activation of androgen receptor target genes and proliferation of prostate cancer cells (20). The gene encoding JMJD2C is amplified in esophageal cancer cell lines and required for their proliferation (10, 21). Therefore, some JmjC proteins may have roles in tumorigenesis, in line with the finding that mice lacking the Suv39h H3K9me3 methyltransferases show chromosomal instabilities and a tumor predisposition phenotype (22).
All human tumors display genomic instability, aberrant transcriptional programs, and very often contain areas that are insufficiently perfused, resulting in a local shortage of nutrients and oxygen (hypoxia). This leads to an activation of the transcription factor hypoxia-inducible factor (HIF), the master regulator of oxygen homeostasis (23). HIF is an α/β-heterodimeric DNA binding complex that directs an extensive transcriptional response (24), which involves genes with important roles in angiogenesis (e.g. Ref. 25), glucose/energy metabolism (26-30), and cellular growth and apoptosis (31).
HIF is regulated by the activity and particularly the abundance of the α-subunits HIF-1α (32), HIF-2α (33), and the less studied HIF-3α (34). HIF-1α and HIF-2α are hydroxylated at two proline residues by a family of prolyl hydroxylase domain enzymes in normoxia (35-38), which triggers their binding to the von Hippel Lindau tumor suppressor protein VHL. VHL recruits an E3 ubiquitin ligase complex and thereby initiates the degradation of HIF-α via the ubiquitin/proteasome pathway (39-42). As molecular oxygen is a limiting factor in this reaction, HIF-1α is not efficiently hydroxylated in hypoxic conditions and escapes degradation. In tumors associated with an inactivation of the VHL gene, e.g. in clear-cell renal cell carcinoma (RCC, reviewed in Ref. 43), cellular HIF-α accumulates irrespective of oxygen levels and induces a constitutive expression of hypoxia-responsive genes.
HIF activation contributes to the classical tumor phenotypes of up-regulated glycolysis and angiogenesis (27, 44, 45). Moreover, HIF induce genes that promote invasive cancer growth and metastasis (46-48) and a de-differentiated phenotype in tumor cells (49-54). Accordingly, the expression of HIF and its target genes can serve as markers for an unfavorable prognosis in cancer patients (e.g. Refs. 55-57).
We hypothesized that expression of members of the JmjC protein family might be controlled by oxygen tension. Here we show that hypoxia and HIF induce transcription of the JMJD1A and JMJD2B genes leading to increased protein expression. In addition, we provide evidence that the demethylase activity of both enzymes is reduced but still present even in severe hypoxia, indicating that a HIF-mediated induction of their expression might provide a compensatory mechanism to maintain a dynamic regulation of H3K9 methylation under low oxygen stress.
EXPERIMENTAL PROCEDURES
Cell Culture—Human renal proximal tubule epithelial cells (obtained from Lonza) were maintained in REGM with supplements and growth factors (Lonza) according to the provider's instructions. HeLa and HEK293 cells were grown in high-glucose Dulbecco's modified Eagle's medium (Invitrogen) containing Glutamax. Human prostate cancer LNCaP cells were cultured in RPMI (Invitrogen). 786-0 cells stably transfected with pRcCMV vector or pRcCMV-HA-VHL (58) were maintained in Dulbecco's modified Eagle's medium with 0.1 mg/ml G418 (Calbiochem). All media were supplemented with 10% fetal bovine serum, penicillin, and streptomycin and the cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air (20% O2). For hypoxia treatment, cells were incubated in an atmosphere containing 1, 0.5, or 0.2% oxygen and 5% CO2 at 37 °C in a hypoxia work station (Ruskinn), for the indicated time periods. Cells were treated, as indicated, with 100 μm desferrioxamine mesylate (DFO; Sigma).
Microarray Analysis—Low passage renal proximal tubule epithelial cell cultures were exposed to normoxia or 1% oxygen for 24 h and total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Three independent biological replicates were performed. RNA was re-purified using an RNeasy mini kit (Qiagen). Analysis was performed with HG_U133A GeneChips (Affymetrix) as described previously (46). Data have been deposited in the NCBI Gene Expression Omnibus and are accessible through accession number GSE12792.
siRNA Silencing—HeLa cells were transfected with oligonucleotides (Dharmacon) at a final concentration of 25-50 nm using Oligofectamine (Invitrogen) according to the manufacturer's protocol. siRNA probe sequences are described in the supplemental information. Cells were incubated for 32 h in normoxia and an additional 16 h in hypoxia before harvesting.
Quantitative Real-time Reverse Transcription PCR—Total RNA was isolated from cells using an RNeasy mini kit (Qiagen) and treated with DNase I. cDNA was synthesized with a Taq-Man Reverse Transcription kit (Applied Biosystems). Quantitative PCR was performed with SYBR Green 2× PCR master mix (Applied Biosystems) in an ABI Prism 7300 Real Time PCR system (Applied Biosystems). Melting temperature profiles of final products and gel electrophoresis of test PCR were used to ensure amplicon specificity. The relative fold change in expression of each mRNA was calculated using the ΔΔCt method relative to 18S rRNA or human large ribosomal protein (hRPLPO) mRNA.
Specific primer sets were designed using the PrimerDesign software (Applied Biosystems). Sequence information is described under supplemental information.
Plasmids—pCMV-HA-hJMJD2B was kindly provided by Jesper Christensen (Biotech Research and Innovation Centre, Copenhagen). The cDNA encoding hJMJD1A was amplified by PCR from a human KIAA clone (KIAA0742). The PCR product was inserted into the XhoI and SalI sites of the pENTR1A vector (Invitrogen) and verified by sequencing. To generate an expression vector, the entry clone was transferred into a Gateway-compatible pCMV-HA (hemagglutinin) vector.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to create JMJD2B(H189G/E191Q) and JMJD1A(H1120G/D1122N). Both PCR products were verified by sequencing and transferred into a Gateway-compatible pCMV-HA expression vector. Primer sequences are described under supplemental information.
Chromatin Immunoprecipitation Assay—ChIP assays were performed as described in Bracken et al. (59). Briefly, HeLa cells, incubated for 6 h either in normoxia or hypoxia (0.5% oxygen), were fixed in 1% formaldehyde. The reaction was stopped by addition of 2 m glycine. Cells were lysed in 100 mm NaCl, 50 mm Tris-Cl, pH 8.1, 5 mm EDTA, pH 8.0, 0.02% NaN3 supplemented with leupeptin, aprotinin, and phenylmethylsulfonyl fluoride. Cells were pelleted and resuspended in a mixture of lysis buffer and Triton dilution buffer (100 mm Tris-Cl, pH 8.6, 100 mm NaCl, 5 mm EDTA, pH 8.0, 0.02% NaN3, 5% Triton X-100) supplemented with the inhibitors mentioned above and sonicated to shear the chromatin. Samples were precleared with protein A-Sepharose beads (GE Healthcare), sonicated herring sperm DNA (Sigma), and bovine serum albumin for 20 min at 4 °C. 30 μl were saved as input samples. Immunoprecipitation with anti-HIF-1α (Abcam, ab2185), anti-RNA polymerase II (Santa Cruz Biotechnology, sc899), and anti-HA (Santa Cruz Biotechnology, sc-805) was performed overnight at 4 °C. Immune complexes bound to protein A beads were washed with mixed micelle buffer (150 mm NaCl, 20 mm Tris-Cl, pH 8.1, 5 mm EDTA, pH 8.0, 5.2% sucrose, 0.02% NaN3, 1% Triton X-100, 0.2% SDS), buffer 500 (0.1% deoxycholic acid, 1 mm EDTA, pH 8.0, 50 mm HEPES, pH 7.5, 1% Triton X-100, 0.2% NaN3), LiCl buffer (0.5% deoxycholic acid, 1 mm EDTA, pH 8.0, 250 mm LiCl, 0.5% Nonidet P-40, 10 mm Tris-Cl, pH 8.0, 0.02% NaN3), and TE buffer. Elution was performed in 1% SDS, 0.1 m NaHCO3 for 1 h at 65 °C and cross-links were reversed at 65 °C overnight. Samples were treated with RNase (Roche) for 1 h at 37 °C and Proteinase K (Sigma) for 2 h at 55 °C. DNA was extracted with phenol/chloroform. DNA was precipitated by addition of ethanol, NaOAc, and glycogen overnight at -20 °C. Pelleted material was washed in 70% ethanol, dried, and resuspended in H2O. Analysis of the samples was performed by real time PCR as described above. Sequence information for primer sets is described under supplemental information.
Luciferase Reporter Assays—All promoter fragments were cloned into the KpnI/HindIII sites of the pGL2 basic reporter vector (Promega). Fragments were PCR-amplified from human genomic DNA using the primers described under supplemental information. The QuikChange kit (Stratagene) was used to introduce point mutations into the hypoxia-response elements. Reporter assays were performed as previously described (46). Luciferase activity was determined in a Fluostar Optima microtiter plate reader (BMG Labtech).
Immunoblot Analysis—For immunoblot analysis cells were lysed in TNN buffer (50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40 supplemented with protease and phosphatase inhibitors). Proteins were separated on 8% SDS-PAGE and transferred to a nitrocellulose membrane. Primary antibodies used were anti-HIF-1α (BD Bioscience 610959), anti-HIF-2α (Abcam ab12606), anti-JMJD1A (Abcam ab52002), anti-JMJD2B (Abcam ab27531; Novus 100-74605), anti-vinculin (Sigma V9131), and anti-β-actin (Chemicon MAB1501). Histones were isolated with urea buffer (1% SDS, 9 m urea, 25 mm Tris-HCl, pH 6.8, 1 mm EDTA, 700 mm 2-mercaptoethanol) and separated on 15% SDS-PAGE. Primary antibodies were anti-H3K9me1 (Abcam ab9045), anti-H3K9me2 (Upstate 07-441), and anti-H3K9me3 (Upstate 07-523). Quantification of band intensities was performed in ImageGauge software (Fuji).
Flow Cytometry—Demethylase activity of HA-JMJD1A and HA-JMJD2B was determined using a 4-color FACS-Calibur (BD Biosciences). HeLa cells were transfected using FuGENE (Roche) and immediately transferred into hypoxia for 24 h. Samples were fixed in 70% ethanol, permeabilized (0.1% Triton X-100), blocked (10% goat serum in phosphate-buffered saline), and stained with primary and secondary antibodies (Molecular Probes A11029 and A21443) in phosphate-buffered saline, 1% bovine serum albumin. An anti-HA antibody (BioSite MMS-101R) was used to detect overexpressed proteins. Histone modifications were analyzed with antibodies described in the immunoblot protocol. A minimum of 10,000 events were analyzed using FlowJo software.
To confirm staining specificity for H3K9me2 and H3K9me3, fluorescence-activated cell sorting (FACS) was performed with and without preincubation of antibodies with H3 peptides containing methylated lysine residues, according to the procedure described by Chadwick et al. (60). Briefly, antibodies were incubated for 2 h at ambient temperature with a 50 m excess of peptides (Abcam) presenting H3K9me2 (ab1772) or H3K9me3 (ab1773), respectively.
Immunofluorescence—HeLa cells transfected with pCMV-HA-hJMJD1A or pCMV-HA-hJMJD2B using FuGENE (Roche) were immediately transferred into hypoxia and after 24 h fixed in 4% paraformaldehyde/phosphate-buffered saline. Staining was performed as described in Ref. 14 with the antibodies used for flow cytometry. Images were obtained with Cool Snap cf camera (Photometrics) and processed with Metamorph software.
Statistical Analysis—If not otherwise indicated all values are presented as mean ± S.D. Student's paired t test was applied to reveal statistical significances. p values less than 0.05 were considered significant.
RESULTS
Induction of JMJD1A and JMJD2B mRNA by Hypoxia—In an attempt to identify novel hypoxia-controlled genes in epithelial cells, we performed expression microarray analysis on human primary renal proximal epithelia cells that were treated with 1% oxygen for 24 h compared with cells cultured in normal atmospheric conditions. Among the group of 160 significantly up-regulated mRNAs in the hypoxia-treated population we found the transcripts of the JmjC proteins JMJD1A and JMJD2B (supplemental Table S1). A survey of published microarray studies revealed that besides these two (24, 61, 62) also other JmjC family members had been found up-regulated by hypoxia in a variety of normal and cancer cell types, such as JARID1 in neuroblastoma cells (61), JMJD2A in mammary epithelial cells (63), and JMJD2C in astrocytes (64).
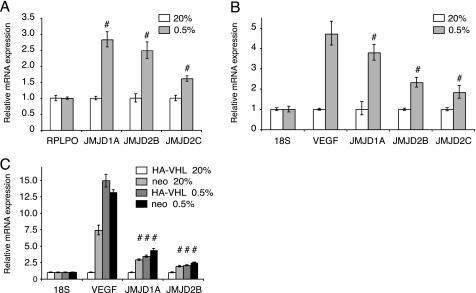
We speculated that HIF could induce the expression of some JmjC proteins in normal and transformed cells and that they might contribute to cancer progression. Therefore, we tested the expression of the mRNAs of JMJD1A, JARID1B/PLU-1, and the JMJD2 family members (JMJD2A, JMJD2B, JMJD2C, and JMJD2D) in human cancer cell lines cultured in normoxia and hypoxia in quantitative real time reverse transcription-PCR experiments. The prostate cancer line LNCaP was included due to the finding that JMJD1A can serve as a co-activator of the androgen receptor (14). As shown in Fig. 1, A and B, the mRNA levels of JMJD1A in LNCaP and HeLa cells increased significantly upon 16 h of hypoxia treatment (0.5% O2) and thereby followed the expression of vascular endothelial growth factor (VEGF), a well established HIF target gene (Fig. 1B). JMJD2B mRNA also accumulated in low oxygen, although to lower steady state levels than JMJD1A. A moderately elevated expression was observed for the JMJD2C message (Fig. 1, A and B). To confirm our results, we used immortalized human embryonic kidney (HEK293) cells, which responded to low oxygen with an up-regulation of JMJD1A, whereas we did not detect any induction of JMJD2B or JMJD2C (data not shown). Of note, a tissue-specific gene regulation in response to low oxygen has been described before (65). The levels of JMJD2A and JMJD2D mRNA either did not respond to changes in ambient oxygen or slightly decreased (data not shown). In the case of JARID1B/PLU-1, we detected a 1.5-fold induction of its mRNA only in HeLa cells (data not shown). Given these results, we decided to focus in our further studies on JMJD1A and JMD2B, because these two genes showed the most robust induction in hypoxia.
FIGURE 1.
Induction of expression of JmjC mRNAs in hypoxia or upon loss of VHL. A, quantitative real time reverse transcriptase-PCR analysis of relative transcript levels of JMJD1A, JMJD2B, and JMJD2C in LNCaP cells upon 16 h incubation in 20 or 0.5% oxygen. B, relative mRNA levels including VEGF in HeLa cells treated as in A. C, relative mRNA levels in control (neo) and VHL-reconstituted (HA-VHL) 786-0 RCC cells treated as in A. For each mRNA the values were normalized to RPLPO or 18S transcripts and levels in normoxic control samples were set to 1. Values are mean ± S.D. and error bars are representative of 3 replicates. #, p < 0.01 using a two-tailed t test.
Induction of JMJD1A and JMJD2B mRNA by Loss of VHL—Loss of functional VHL, a frequent event in RCC cells, leads to constitutive, oxygen-independent expression of HIF. The cell line 786-0 was derived from a human renal adenocarcinoma and lacks wild-type VHL. 786-0 cells constitutively express HIF-2α but no HIF-1α in normoxia (41). These cells were previously stably transfected with an empty vector (designated 786-0(neo)) or a HA-tagged wild-type VHL (designated 786-0 (HA-VHL)) that re-establishes the oxygen responsiveness of the expression of HIF-2α (58) and of the HIF target gene VEGF (66). We found increased amounts of JMJD1A and JMJD2B mRNA in normoxic 786-0(neo) cells compared with 786-0(HA-VHL) cells (Fig. 1C). Moreover, reconstitution of functional VHL reduced normoxic expression and restored inducibility of both the jumonji mRNAs and the VEGF message by hypoxia. Taken together, these observations suggest that both HIF-1α (which is predominant in HeLa and LNCaP cells) and HIF-2α (in 786-0 cells) can induce the expression of JMJD1A and JMJD2B mRNA. Also, loss of functional VHL in kidney cancer cells leads to elevated mRNA expression levels, which is a hall-mark of HIF-regulated genes.
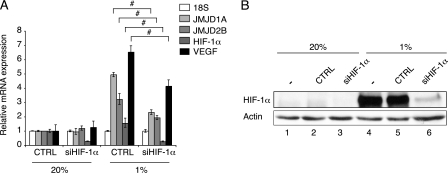
HIF Mediates the Hypoxic Induction of JMJD1A and JMJD2B mRNA—To further evaluate the effect of HIF-1α on JMJD1A and JMJD2B transcript expression, we transfected HeLa cells with a specific siRNA oligonucleotide that targeted HIF-1α. This led to a more than 70% reduction of HIF-1α mRNA (Fig. 2A) and markedly diminished, but not completely suppressed HIF-1α protein levels in hypoxic HeLa cells, as shown in the immunoblot in Fig. 2B. The suppression of HIF-1α in hypoxia caused a 53% decrease of JMJD1A mRNA and a 40% decline in JMJD2B message compared with control, but did not significantly affect their normoxic expression (Fig. 2A). The well established HIF target gene VEGF showed a similar reduced inducibility upon HIF-1α siRNA treatment. However, because gene silencing was incomplete (Fig. 2B) and hypoxic HeLa cells also express low levels of HIF-2α (compare Fig. 3A), we could not completely suppress the hypoxic induction of the JmjC protein (and VEGF) transcripts. From these experiments we conclude that the increased abundance of JMJD1A and JMJD2B transcripts in low oxygen tension is mediated by HIF.
FIGURE 2.
Suppression of HIF-1α by siRNA compromises hypoxic induction of JMJD1A and JMJD2B. A, quantitative real time reverse transcriptase-PCR analysis of relative transcript levels of HIF-1α, VEGF, JMJD1A, and JMJD2B in HeLa cells transfected with control (CTRL) or HIF-1α siRNA prior to 16 h incubation in 20 or 1% oxygen. Values were normalized to 18S transcripts and levels in normoxic samples transfected with control siRNA were set to 1. Data represent the mean ± S.D. and error bars represent n = 3. #, p < 0.01 for the indicated comparisons. B, representative immunoblot analysis for HIF-1α andβ-actin using whole cell extracts from HeLa cells that were either treated with transfection reagent only (-), transfected with control siRNA or HIF-1α siRNA prior to parallel incubation in normoxia (20% O2) or hypoxia (1% O2, 16 h).
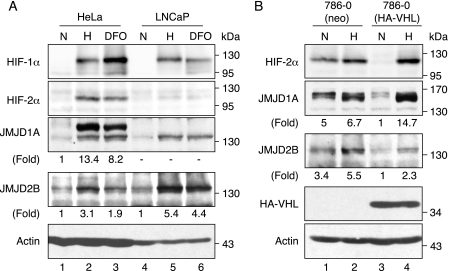
FIGURE 3.
Hypoxia and loss of VHL induce the expression of JMJD1A and JMJD2B. A, immunoblot analysis for JMJD1A, JMJD2B, HIF-1α, and HIF-2α in whole cell extracts from HeLa and LNCaP cells incubated in normoxia (N, 20% O2), hypoxia (H, 0.5% O2, 16 h) or treated with 100 μm DFO in normoxia (DFO, 16 h). Numbers below panels indicate fold induction compared with untreated cells normalized to actin loading control. B, JMJD1A and JMJD2B protein expression in 786-0 RCC cells. Control (neo) and VHL reconstituted cells (HA-VHL) were incubated in normoxia (N) or 0.5% oxygen (H) for 16 h. Fold inductions relative to normoxic cells expressing HA-VHL are indicated.
Hypoxic Induction of JMJD1A and JMJD2B Protein—To investigate if the induction of JMJD1A and JMJD2B mRNAs in hypoxic cells resulted in increased protein expression, we performed immunoblot analysis. HeLa and LNCaP cells were exposed to either 0.5% O2 or 100 μm desferrioxamine mesylate (DFO, an iron chelator and chemical hypoxia mimetic) for 16 h, and assayed for the expression of HIF-1α, HIF-2α, JMJD1A, and JMJD2B. As shown in Fig. 3A, low oxygen tension or DFO treatment led to a stabilization of HIF-1α in both cell lines. An accumulation of HIF-2α was also detected in HeLa cells, whereas LNCaP cells showed a very low induction of HIF-2α. Both hypoxia and DFO induced the expression of JMJD1A in HeLa cells. Furthermore, a second band at ∼135 kDa was induced. We speculate that this second band might represent a product of intracellular proteolysis or of alternative splicing, although the existence of a shorter splice version has only been reported for the mouse JMJD1A protein (67). Interestingly, we found that this faster migrating band is also induced in hypoxic LNCaP cells, but we could not detect the full-length 150-kDa JMJD1A protein in the prostate cancer cell lysates (Fig. 3A).
JMJD2B was also induced by both hypoxia and DFO in HeLa and LNCaP cells. Notably, it accumulated to higher steady state levels in the prostate cancer cell line (Fig. 3A, lanes 5 and 6). Taken together, the immunoblot analysis largely confirmed our previous results of the real time reverse transcription-PCR experiments.
The RCC cell line 786-0(neo) expressed HIF-2α independently of oxygenation (Fig. 3B, top panel), whereas 786-0(HA-VHL) cells showed detectable levels only after incubation in hypoxia. In general, JMJD1A levels were correlated with the expression of HIF-2α in both cell lines (Fig. 3B). In the presence of functional VHL the expression of JMJD1A was low in normoxia and induced by hypoxia. Absence of VHL resulted in constitutively high JMJD1A levels, which were only marginally influenced by oxygen availability. Interestingly, the JMJD1A protein extracted from 786-0 cells migrated as a doublet band, as previously observed for other cell types (68, 69), which might indicate a yet uncharacterized post-translational modification.
JMJD2B protein expression was also influenced by the amount of HIF-2α. However, it accumulated to relatively lower levels in hypoxia-treated 786-0(HA-VHL) compared with 786-0(neo) cells, which is opposite to the behavior of JMJD1A. Therefore, we cannot exclude a HIF-independent, but VHL-dependent component in the hypoxic stabilization of JMJD2B. Taken together, our results suggest that JMJD1A and JMJD2B are hypoxia-inducible genes, whose expression is controlled by the activity of HIF-1α and HIF-2α in a variety of cell lines.
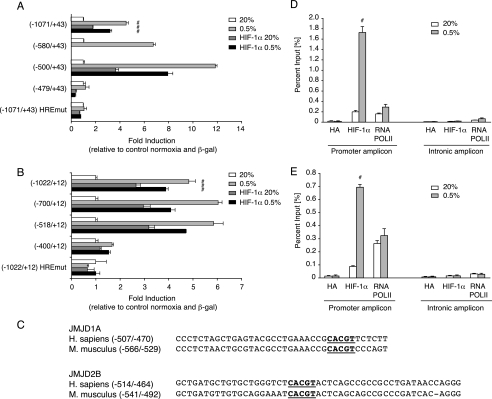
HIF Binding Sites in the Promoters of JMJD1A and JMJD2B Are Required for Hypoxic Gene Activation—Analysis of the human JMJD1A sequence revealed 2 potential hypoxia-response elements (HREs) located within 1 kilobase upstream of the transcriptional start site and one at position +30 in the transcribed sequence. Four putative HIF binding sites were found in the JMJD2B promoter sequence within 1 kilobase upstream of the transcription start. We cloned the human JMJD1A and JMJD2B promoter regions, generated luciferase reporter plasmids containing the respective fragments (designated pGL2-JMJD1A(-1071/+43) and pGL2-JMJD2B(-1022/+12)) and transfected them into HeLa cells. The JMJD1A reporter was activated 4.5-fold by hypoxia and about 1.8-fold by co-transfection of HIF-1α (Fig. 4A). The JMJD2B construct showed 5-fold activation in response to hypoxia and 2.7-fold induction after co-transfection of HIF-1α in normoxia (Fig. 4B). Both reporters responded maximally to hypoxia, a combined transfection of HIF-1α and hypoxia treatment failed to further increase their activity above the levels reached by hypoxia alone, most likely indicating a saturation of the system. A progressive deletion analysis revealed that the HREs located at position -480 bp upstream of the transcriptional start within the JMJD1A promoter (sequence ACGTG on the antisense strand), and at position -494 bp within the JMJD2B promoter (ACGTG on the antisense strand) were critical for hypoxia/HIF-1α-inducible reporter activity (Fig. 4, A and B). Mutations of these sequences in the context of the full-length constructs rendered them insensitive to hypoxia and HIF-1α (Fig. 4, A and B). Moreover, sequence analysis of the murine Jmjd1a and Jmjd2b genes revealed conservation of these HRE elements and, to different extents, of their flanking sequences (Fig. 4C).
FIGURE 4.
The JMJD1A and JMJD2B promoter sequences contain functional HIF binding sites. A, luciferase reporter assay with constructs containing the indicated sequences from the human JMJD1A gene promoter region. Positions in base pairs relative to the start site of transcription are indicated. Plasmids were co-transfected into HeLa cells along with a CMV-lacZ control plasmid and a HIF-1α expression vector where indicated. 24 h after transfection, cells were incubated in normoxia (20%) or 0.5% oxygen (0.5%) for 16 h. Indicated are fold induction values normalized to β-galactosidase activity and relative to normoxic control samples. Data are shown as the mean ± S.D. of 3 replicates. #, p < 0.01 relative to control samples. B, same experimental setup as in A but with reporter gene vectors containing the indicated JMJD2B promoter regions. C, comparison of the identified hypoxia response elements (highlighted) and flanking nucleotides in the human and mouse JMJD1A and JMJD2B gene promoter sequences. D, ChIP assay to determine binding of endogenous HIF-1α and RNA polymerase II to the JMJD1A promoter. HeLa cells were kept in normoxia (20% O2) or incubated in 0.5% O2 for 8 h. The precipitated DNA was amplified by real time quantitative PCR using specific primers for regions within the promoter (left) and the third intron of JMJD1A (right). Enrichments are presented as percentages of total input. Data are shown as the mean ± S.D. of 3 replicates. #, p < 0.01 relative to normoxic samples. E, ChIP analysis as in D on the JMJD2B locus. Amplicons are located within the promoter and within the second intron of JMJD2B, respectively.
The binding of endogenous HIF-1α protein to the HREs within the JMJD1A and JMJD2B genes in vivo was analyzed by quantitative ChIP. HIF-1α was bound to the JMJD1A and JMJD2B promoters, but not to downstream sequences in intronic regions of both genes (Fig. 4, D and E). Significantly, HIF-1α showed a strong enrichment on the HRE sequences in cells treated with 0.5% O2. In case of the JMJD1A promoter this was paralleled by an increased recruitment of RNA polymerase II (Fig. 4D), whereas a significant enrichment of RNA polymerase II on the JMJD2B promoter was not detected (Fig. 4E). Taken together, the results strongly suggest that the genes encoding both JmjC domain proteins are direct targets of HIF.
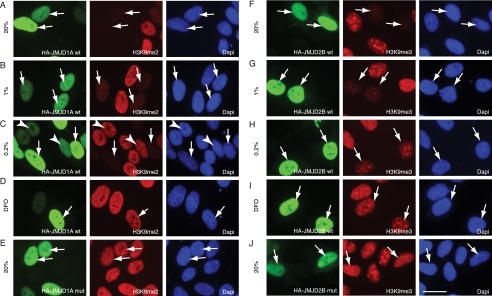
JMJD1A and JMJD2B Remain Active in Hypoxic Cells—The abundance of HIF is controlled by the HIF PHDs that are Fe(II)- and α-ketoglutarate-dependent dioxygenases. Also JmjC domain proteins hydroxylate their methylated histone substrates, which then spontaneously degrade to release formaldehyde. This reaction, analogous to HIF proline hydroxylation, requires Fe(II), α-ketoglutarate, and molecular oxygen. Moreover, the factor inhibiting HIF (FIH), an asparagine hydroxylase that specifically compromises gene activation by HIF-1α and HIF-2α in normoxia, is a JmjC domain protein. Inactivation of FIH in low oxygen is required to enable maximal HIF target gene activation (70, 71). Given the general dependence of these hydroxylation reactions on oxygen, we asked whether the hypoxia-inducible JmjC domain proteins are enzymatically active in conditions of low oxygen. To address this question, we chose a transient overexpression approach with subsequent indirect immunofluorescence staining using highly specific antibodies for H3K9me3 (the preferred JMJD2B target) and H3K9me2 (targeted by JMJD1A) (10, 14, 72). As already demonstrated for FLAG-tagged JMJD1A (14), immunofluorescence analysis revealed a nuclear localization of HA-JMJD1A. This localization was not altered when cells were incubated in 1 or 0.2% oxygen (Fig. 5A). Moreover, overexpression of the protein led to a strong decrease of detectable H3K9me2 levels in vivo. Cells that expressed a catalytically inactive protein that carries two mutations in the residues necessary for binding of Fe(II), JMJD1A(H1120G/D1122N), showed high levels of H3K9me2 despite a strong nuclear expression of the mutant protein (Fig. 5E).
FIGURE 5.
Ectopic expression of JMJD1A and JMJD2B leads to loss of H3K9me2 and H3K9me3 in normoxia and hypoxia. HeLa cells were transfected with HA-tagged JMJD1A (A-D) or HA-tagged JMJD2B (F-I) and incubated in normoxia (20% O2; A and F), 1% O2 (B and G), 0.2% O2 (C and H), or in the presence of 100 μm DFO (D and I) for 24 h. E, cells transfected with JMJD1A mutant. J, cells transfected with JMJD2B mutant. The cells were fixed, costained for the indicated histone modifications and for the expression of jumonji proteins (anti-HA), and analyzed by indirect immunofluorescence microscopy. White arrows indicate cells expressing the tested protein. Arrowheads in C indicate cells with lower expression levels of HA-JMJD1A. The cells were counterstained with 4′,6-diamidino-2-phenylindole to visualize cell nuclei. Scale bar in J corresponds to 20 μm.
A reduction of atmospheric oxygen down to 1% did not result in an obvious loss of demethylase activity of transfected JMJD1A (Fig. 5B). Only severely hypoxic cells (0.2% O2) that showed a weak staining for HA-JMJD1A displayed a detectable H3K9me2 signal (Fig. 5C, arrowheads). However, H3K9me2 was not increased in cells with a strong expression of HA-JMJD1A, suggesting that the protein has discernable activity even at very low oxygen levels (Fig. 5C, arrows). In contrast to hypoxia, treatment with the iron chelator DFO abrogated enzymatic activity also in cells that display strong JMJD1A expression, as documented in Fig. 5D.
HA-JMJD2B was also located in the nuclei of transfected cells and efficiently removed the H3K9me3 mark (Fig. 5F), which is in accordance with published data (10, 72). Although 1% O2 did not markedly compromise the JMJD2B demethylase activity, a further reduction of oxygen tension down to 0.2% resulted in a partial increase in H3K9me3 immunostaining even in cells that strongly expressed HA-JMJD2B (Fig. 5H). Again, treatment with DFO or two point mutations within the JmjC domain (H189G/E191Q) abolished JMJD2B demethylase activity (Fig. 5, I and J). These data demonstrate that both JmjC domain proteins retain at least a partial histone demethylase activity in conditions of strong (1% O2) and even severe hypoxia (0.2% O2). Of note, HIF-1α is strongly activated at levels above 1% O2 and induces target gene transcription in the majority of cell types (see Fig. 2B and e.g. Refs. 24 and 46). Therefore, our data suggest that HIF-mediated induction of the JmjC domain proteins can occur at oxygen tensions that are sufficient for their activity.
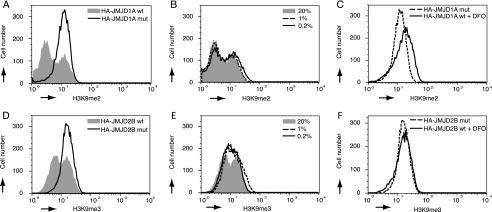
The impact of different oxygen concentrations on the activity of JMJD1A and JMJD2B was also quantified by FACS scan experiments. HeLa cells were transfected with HA-JMJD1A and HA-JMJD2B and endogenous histone methylation marks were detected by immunostaining. Control experiments (supplemental Fig. 1, A and B) showed that antibody preincubation with the matching peptides reduced staining to background levels. No effect was observed when the H3K9me3-specific antibody was incubated with the H3K9me2 peptide and vice versa (data not shown), indicative of a high specificity. Fig. 6A shows that the expression of HA-JMJD1A led to a reduced di-methylation of H3K9 in vivo compared with cells expressing the inactive mutant HA-JMJD1A(H1120G/D1122N). However, not all of the cells that expressed HA-JMJD1A displayed a reduced staining for H3K9me2, these cells appear as the smaller right peak (Fig. 6A). The persistence of H3K9me2 in these cells did not correlate with expression levels of JMJD1A and is being investigated.3 Hypoxia had a modest effect on the activity of JMJD1A, which is apparent in a slight decrease in the number of cells with a low H3K9me2 signal especially at 0.2% O2 as shown in Fig. 6B. However, a pronounced increase of intermediate signal intensities for H3K9me2 was not seen, implying that JMJD1A-mediated demethylation occurs, if once initiated, at the majority of the accessible residues. Treatment with DFO not only abrogated demethylase activity of HA-JMJD1A but increased the maximum H3K9me2 signal beyond the one detected in cells that express HA-JMJD1A(H1120G/D1122N), as shown in Fig. 6C. A possible explanation for this observation is that the depletion of intracellular iron pools results in an inhibition of endogenous H3K9me2-specific demethylases.
FIGURE 6.
Flow cytometry analysis of the effect of different ambient oxygen levels on the activity of JMJD1A and JMJD2B. HeLa cells were transfected with HA-tagged JMJD1A, mutant JMJD1A, JMJD2B, or mutant JMJD2B and incubated in normoxia (20% O2), 1% O2, 0.2% O2, or in the presence of 100 μm DFO in normoxia for 24 h. The cells were fixed, costained for H3K9me2 (A-C) or H3K9me3 (D-F) and for the expression of jumonji proteins (anti-HA) and analyzed by flow cytometry. Depicted are results for transfected (HA positive) cells only. A, effect of overexpression of JMJD1A compared with mutant JMJD1A on cellular H3K9me2 levels. B, impact of oxygen tension on JMJD1A demethylase activity. C, comparison of mutant JMJD1A and DFO treatment. D, H3K9me3 levels in JMJD2B and mutant JMJD2B overexpressing cells. E, impact of oxygen availability on JMJD2B demethylase activity. F, effect of DFO compared with mutant JMJD2B on H3K9me3.
The FACS assay also confirmed our previous observation that overexpression of HA-JMJD2B induced a decrease in the number of cells with high levels of H3K9me3 (Fig. 6D), an effect that was clearly correlated with expression levels of HA-JMJD2B (data not shown). A reduction in the oxygen tension from 20% to either 1 or 0.2% O2 resulted in a partial increase of trimethylated H3K9 in transfected cells, as shown in Fig. 6E. In contrast to what was observed for JMJD1A and H3K9me2, the number of cells expressing JMJD2B that showed intermediate levels of histone H3K9 trimethylation increased substantially in hypoxia, leading to a bell-shaped curve in the histogram instead of the two maxima observed in normoxia (Fig. 6E). The strongest inhibition was achieved by DFO treatment, which resulted in a H3K9 trimethylation status similar to cells expressing the catalytically inactive JMJD2B mutant, but did not lead to a further gain of H3K9me3 signal (Fig. 6F). Taken together, the results of our FACS analysis indicate that both JMJD1A and JMJD2B are catalytically active in hypoxic conditions and remove methyl groups from endogenous histone H3K9 residues, although very low oxygen levels (0.2% O2) compromise this activity to varying degrees.
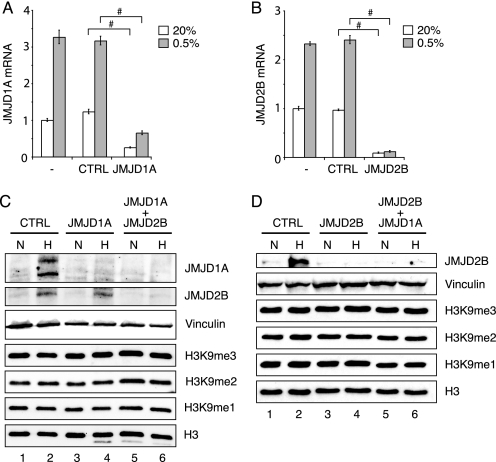
JMJD1A and JMJD2B Do Not Affect Global H3K9 Methylation Levels in Hypoxic Cells—To test whether the induction of JMJD1A and JMJD2B by HIF has an effect on global histone methylation levels, HeLa cells were transfected with siRNAs that target JMJD1A and JMJD2B specifically and the histone methylation status analyzed by immunoblotting. As shown in Fig. 7, A and B, siRNA efficiently suppressed both jumonji mRNAs and moreover impaired their hypoxic induction. Accordingly, protein levels of JMJD1A and JMJD2B were strongly reduced in cells treated with the specific siRNAs (Fig. 7, C and D).
FIGURE 7.
siRNA-mediated down-regulation of JMJD1A and JMJD2B does not change global histone H3K9 methylation levels. A, transcript levels of JMJD1A in HeLa cells after treatment with Oligofectamine alone (-), control siRNA (CTRL), or JMJD1A-directed siRNA. 24 h after transfection, cells were incubated in normoxia (N, 20% O2) or hypoxia (H, 0.5% O2) for 16 h. Values were normalized to 18S transcripts and levels in untransfected normoxic samples were set to 1. Error bars represent mean ± S.D. of 3 replicates; # p < 0.01. B, transcript levels of JMJD2B after treatment with JMJD2B-directed siRNA. C, immunoblot analysis for JMJD1A, JMJD2B, vinculin (loading control), histone H3K9me3, H3K9me2, H3K9me1, and total histone H3 in extracts from HeLa cells after treatment with control siRNA, JMJD1A-directed siRNA, or a mixture of JMJD2B- and JMJD1A-directed oligonucleotides. Hypoxia treatment was performed as in A. D, immunoblot analysis for JMJD2B, vinculin, histone H3K9me3, H3K9me2, H3K9me1, and total histone H3 in extracts from cells treated with JMJD2B-directed siRNA or a combination of JMJD2B- and JMJD1A-siRNA in normoxia and hypoxia.
In contrast to what has been reported for A549 lung carcinoma and HEK293 cells (73), we could not detect a consistent alteration in total H3K9 di- or trimethylation levels in hypoxic HeLa cells. In addition, suppression of either JMJD1A or JMJD2B or of both proteins simultaneously had no detectable effect on bulk histone H3K9 methylation levels (Fig. 7, C and D). These results are not unexpected, because the deletion of JMJD1A in mice did not result in a global effect on H3K9 methylation (68). Also, it has been reported that RNA interference against JMJD2A and JMJD2C, two close relatives of JMJD2B, did not impact global levels of H3K9me3 (10, 11). However, knockdown of JMJD2A resulted in a significant increase in local H3 methylation at the target gene ASCL2 (11), whereas siRNA against JMJD2C impaired cell proliferation (10). From these and our results we conclude that we need to identify specific target genes of JMJD1A and JMJD2B in hypoxic cells to study the consequences of their induction on histone methylation and gene regulation, respectively.
DISCUSSION
The hypoxia-inducible factor HIF is the major regulator of cellular adaptation to low oxygen levels. HIF impacts transcription of several hundred genes to adjust cellular metabolism and signaling to cope with oxygen limitation and acidosis (74), and the list of studies that define direct HIF target genes is constantly growing (31). In addition, secondary transcriptional programs are initiated by HIF via activation of other DNA-binding transcription factors (31).
Here we demonstrate HIF-mediated induction of a novel class of chromatin regulators, the JmjC domain histone demethylases. While this manuscript was under revision, two other groups reported similar results (69, 75). Because tri- and dimethylation of H3K9 contribute to the establishment of a repressive chromatin structure, the induction of H3K9-specific demethylases by HIF may directly and indirectly impact the hypoxic transcriptome. The latter is supported by findings that JMJD1A interacts with the androgen receptor and facilitates transcriptional activation in response to androgens (14). Moreover, it contributes to the expression of several pluripotency marks in F9 cells (14) and in murine ES cells (76), as well as to the expression of factors required for spermatogenesis (68) and smooth muscle cell-specific transcripts (77). Until now, no interaction partner that can recruit JMJD2B to specific DNA sequences has been identified. However, the closely related JMJD2C also associates with the androgen receptor and facilitates gene activation (20).
Current knowledge on the role of histone methylation in HIF-controlled gene expression is limited. It remains to be investigated if HIF itself can recruit histone demethylases to promoter sequences. In addition, the dynamics of subsequent changes in the histone modification patterns upon binding of HIF need to be carefully characterized. Previously, it has been reported that hypoxia increases the dimethylated state of H3K9 by stabilization of the histone methyltransferase G9a (73), however, in our cell system we could not detect significant changes in global H3K9 methylation levels in hypoxia.
A recent publication (78) described a global increase in a broad range of histone methylation marks indicative of both transcriptional activation and repression in 0.2% oxygen. Furthermore, the authors observed a comparatively low histone H3 occupancy of hypoxia-activated promoters accompanied by a down-regulation of H3K9/27me2 and an increase in H3K9 acetylation, all compatible with a less compact chromatin environment permissive for binding of transcriptional activators and increased transcription. Surprisingly, H3K27me3, a histone mark that is highly correlated with genomic silencing, decreased not only at hypoxia-activated but also at hypoxia-repressed promoter sequences. Also H3K4me3, implicated in activation of transcription, accumulated to equal levels at both activated and repressed promoters after 48 h of hypoxia treatment. Despite these divergent results, it is tempting to speculate that the HIF-mediated induction of jumonji histone demethylases targeting H3K9 methylation may contribute to the expression of HIF target genes in the background of a generally more repressive chromatin environment in hypoxic cells.
It has been proposed that hypoxia can compromise the enzymatic activity of JmjC domain proteins (73, 78), implicating that their transcriptional up-regulation would serve to maintain sufficient demethylase activity in low oxygen. This raises the question as to why JMJD1A and JMJD2B are induced preferentially in a variety of cellular systems (69, 75). In addition, our data indicate a different sensitivity of individual JmjC demethylases toward decreased oxygen availability. However, both enzymes were active in 1% O2, a condition that occurs in hypoxic tumor tissue (79, 80) and activates HIF target genes. Further studies are needed to determine oxygen requirements for additional JmjC family members. The capacity of jumonji proteins to remain active in low oxygen has most notably been demonstrated for FIH: endogenous FIH limits transcriptional effects of HIF at 1% atmospheric O2, and moderately overexpressed FIH exerts this effect down to levels of 0.2% O2 (81). Upon adaptation to hypoxia, intracellular redistribution of the available oxygen might also re-activate oxygen-dependent enzymes (82).
Our data indicate that loss of VHL in RCC cells can result in constitutively elevated levels of JMJD1A and JMJD2B and define them as target genes also for HIF-2α. The latter is in agreement with microarray data on a HIF-2α expressing neuroblastoma cell that indicates an induction of JMJD1A mRNA in response to even mild hypoxia (5% O2) (61).
In VHL-deficient tumors, oxygen levels should not be limiting for the activity of JmjC proteins and in these tumors one might expect the strongest effect on histone methylation. Because loss of histone H3K9 methylation promotes chromosomal instabilities (22), the induction of H3K9me3/me2 specific demethylases might also contribute to increased mutation rates in tumors with deregulated HIF expression.
It has been proposed that HIF influences cellular differentiation during embryogenesis and promotes the adoption of stem cell properties in tumor cells, in part by a cross-talk to Notch signaling and by HIF-2α-driven activation of Oct4 (83). Because JMJD1A regulates the expression of Oct4 in ES cells (76), our findings may be supportive of the proposed cross-talk between hypoxia signaling and stem cell function.
Supplementary Material
Acknowledgments
We thank Signe Breum for excellent technical assistance, Jesper Christensen and Marianne Pedersen for helpful discussions and JMJD2B cDNA, Wilhelm Krek for providing unpublished data, and Kristian Helin, James Whiteford, and John Couchman for critical comments on the manuscript.
This work was supported by the Biotech Research and Innovation Centre, the Danish Cancer Society, the Danish Cancer Research Foundation, Lykfeldts Legat, and the Novo Nordisk Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S2 and S2.
Footnotes
The abbreviations used are: JmjC, jumonji C domain; JMJD1A, jumonji domain containing 1A; JMJD2B, jumonji domain containing 2B; JMJD2C/GASC1, jumonji domain containing 2C/gene amplified in squamous cell carcinoma 1; ChIP, chromatin immunoprecipitation; FIH, factor inhibiting HIF; HIF, hypoxia-inducible factor; HRE, HIF-responsive element; DFO, desferrioxamine; H3K9, histone H3 lysine 9; H3K27, histone H3 lysine 27; H3K9me3, trimethylated histone H3 lysine 9; H3K9me2, dimethylated histone H3 lysine 9; Oct4, octamer-binding protein 4; RCC, renal clear cell carcinoma; 18S, 18 S ribosomal RNA; siRNA, small interfering RNA; VEGF, vascular endothelial growth factor; VHL, von Hippel Lindau tumor suppressor; FACS, fluorescence-activated cell sorting; HA, hemagglutinin; HEK, human embryonic kidney.
S. Beyer and P. Staller, unpublished data.
References
- 1.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., and Kouzarides, T. (2001) Nature 410 120-124 [DOI] [PubMed] [Google Scholar]
- 2.Kim, J., Daniel, J., Espejo, A., Lake, A., Krishna, M., Xia, L., Zhang, Y., and Bedford, M. T. (2006) EMBO Rep. 7 397-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001) Nature 410 116-120 [DOI] [PubMed] [Google Scholar]
- 4.Taverna, S. D., Ilin, S., Rogers, R. S., Tanny, J. C., Lavender, H., Li, H., Baker, L., Boyle, J., Blair, L. P., Chait, B. T., Patel, D. J., Aitchison, J. D., Tackett, A. J., and Allis, C. D. (2006) Mol. Cell 24 785-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wysocka, J., Swigut, T., Xiao, H., Milne, T. A., Kwon, S. Y., Landry, J., Kauer, M., Tackett, A. J., Chait, B. T., Badenhorst, P., Wu, C., and Allis, C. D. (2006) Nature 442 86-90 [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. L. (2007) Nature 447 407-412 [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides, T. (2007) Cell 128 693-705 [DOI] [PubMed] [Google Scholar]
- 8.Klose, R. J., and Zhang, Y. (2007) Nat. Rev. Mol. Cell. Biol. 8 307-318 [DOI] [PubMed] [Google Scholar]
- 9.Cloos, P. A., Christensen, J., Agger, K., and Helin, K. (2008) Genes Dev. 22 1115-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloos, P. A., Christensen, J., Agger, K., Maiolica, A., Rappsilber, J., Antal, T., Hansen, K. H., and Helin, K. (2006) Nature 442 307-311 [DOI] [PubMed] [Google Scholar]
- 11.Klose, R. J., Yamane, K., Bae, Y., Zhang, D., Erdjument-Bromage, H., Tempst, P., Wong, J., and Zhang, Y. (2006) Nature 442 312-316 [DOI] [PubMed] [Google Scholar]
- 12.Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., and Zhang, Y. (2006) Nature 439 811-816 [DOI] [PubMed] [Google Scholar]
- 13.Whetstine, J. R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z., Spooner, E., Li, E., Zhang, G., Colaiacovo, M., and Shi, Y. (2006) Cell 125 467-481 [DOI] [PubMed] [Google Scholar]
- 14.Yamane, K., Toumazou, C., Tsukada, Y., Erdjument-Bromage, H., Tempst, P., Wong, J., and Zhang, Y. (2006) Cell 125 483-495 [DOI] [PubMed] [Google Scholar]
- 15.Chang, B., Chen, Y., Zhao, Y., and Bruick, R. K. (2007) Science 318 444-447 [DOI] [PubMed] [Google Scholar]
- 16.Lu, P. J., Sundquist, K., Baeckstrom, D., Poulsom, R., Hanby, A., Meier-Ewert, S., Jones, T., Mitchell, M., Pitha-Rowe, P., Freemont, P., and Taylor-Papadimitriou, J. (1999) J. Biol. Chem. 274 15633-15645 [DOI] [PubMed] [Google Scholar]
- 17.Xiang, Y., Zhu, Z., Han, G., Ye, X., Xu, B., Peng, Z., Ma, Y., Yu, Y., Lin, H., Chen, A. P., and Chen, C. D. (2007) Proc. Natl. Acad. Sci. U. S. A 104 19226-19231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamane, K., Tateishi, K., Klose, R. J., Fang, J., Fabrizio, L. A., Erdjument-Bromage, H., Taylor-Papadimitriou, J., Tempst, P., and Zhang, Y. (2007) Mol. Cell 25 801-812 [DOI] [PubMed] [Google Scholar]
- 19.Pfau, R., Tzatsos, A., Kampranis, S. C., Serebrennikova, O. B., Bear, S. E., and Tsichlis, P. N. (2008) Proc. Natl. Acad. Sci. U. S. A 105 1907-1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wissmann, M., Yin, N., Muller, J. M., Greschik, H., Fodor, B. D., Jenuwein, T., Vogler, C., Schneider, R., Gunther, T., Buettner, R., Metzger, E., and Schule, R. (2007) Nat. Cell Biol. 9 347-353 [DOI] [PubMed] [Google Scholar]
- 21.Yang, Z. Q., Imoto, I., Fukuda, Y., Pimkhaokham, A., Shimada, Y., Imamura, M., Sugano, S., Nakamura, Y., and Inazawa, J. (2000) Cancer Res. 60 4735-4739 [PubMed] [Google Scholar]
- 22.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., Opravil, S., Doyle, M., Sibilia, M., and Jenuwein, T. (2001) Cell 107 323-337 [DOI] [PubMed] [Google Scholar]
- 23.Semenza, G. L. (2006) Exp. Physiol. 91 803-806 [DOI] [PubMed] [Google Scholar]
- 24.Elvidge, G. P., Glenny, L., Appelhoff, R. J., Ratcliffe, P. J., Ragoussis, J., and Gleadle, J. M. (2006) J. Biol. Chem. 281 15215-15226 [DOI] [PubMed] [Google Scholar]
- 25.Forsythe, J. A., Jiang, B. H., Iyer, N. V., Agani, F., Leung, S. W., Koos, R. D., and Semenza, G. L. (1996) Mol. Cell. Biol. 16 4604-4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer, N. V., Kotch, L. E., Agani, F., Leung, S. W., Laughner, E., Wenger, R. H., Gassmann, M., Gearhart, J. D., Lawler, A. M., Yu, A. Y., and Semenza, G. L. (1998) Genes Dev. 12 149-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seagroves, T. N., Ryan, H. E., Lu, H., Wouters, B. G., Knapp, M., Thibault, P., Laderoute, K., and Johnson, R. S. (2001) Mol. Cell. Biol. 21 3436-3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2006) Cell Metab. 3 177-185 [DOI] [PubMed] [Google Scholar]
- 29.Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L., and Denko, N. C. (2006) Cell Metab. 3 187-197 [DOI] [PubMed] [Google Scholar]
- 30.Fukuda, R., Zhang, H., Kim, J. W., Shimoda, L., Dang, C. V., and Semenza, G. L. (2007) Cell 129 111-122 [DOI] [PubMed] [Google Scholar]
- 31.Semenza, G. L. (2007) Biochem. J. 405 1-9 [DOI] [PubMed] [Google Scholar]
- 32.Wang, G. L., Jiang, B. H., Rue, E. A., and Semenza, G. L. (1995) Proc. Natl. Acad. Sci. U. S. A 92 5510-5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian, H., McKnight, S. L., and Russell, D. W. (1997) Genes Dev. 11 72-82 [DOI] [PubMed] [Google Scholar]
- 34.Gu, Y. Z., Moran, S. M., Hogenesch, J. B., Wartman, L., and Bradfield, C. A. (1998) Gene Expr. 7 205-213 [PMC free article] [PubMed] [Google Scholar]
- 35.Bruick, R. K., and McKnight, S. L. (2001) Science 294 1337-1340 [DOI] [PubMed] [Google Scholar]
- 36.Epstein, A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., Tian, Y. M., Masson, N., Hamilton, D. L., Jaakkola, P., Barstead, R., Hodgkin, J., Maxwell, P. H., Pugh, C. W., Schofield, C. J., and Ratcliffe, P. J. (2001) Cell 107 43-54 [DOI] [PubMed] [Google Scholar]
- 37.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S., and Kaelin, W. G., Jr. (2001) Science 292 464-468 [DOI] [PubMed] [Google Scholar]
- 38.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001) Science 292 468-472 [DOI] [PubMed] [Google Scholar]
- 39.Kallio, P. J., Wilson, W. J., O'Brien, S., Makino, Y., and Poellinger, L. (1999) J. Biol. Chem. 274 6519-6525 [DOI] [PubMed] [Google Scholar]
- 40.Kamura, T., Sato, S., Iwai, K., Czyzyk-Krzeska, M., Conaway, R. C., and Conaway, J. W. (2000) Proc. Natl. Acad. Sci. U. S. A 97 10430-10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maxwell, P. H., Wiesener, M. S., Chang, G. W., Clifford, S. C., Vaux, E. C., Cockman, M. E., Wykoff, C. C., Pugh, C. W., Maher, E. R., and Ratcliffe, P. J. (1999) Nature 399 271-275 [DOI] [PubMed] [Google Scholar]
- 42.Salceda, S., and Caro, J. (1997) J. Biol. Chem. 272 22642-22647 [DOI] [PubMed] [Google Scholar]
- 43.Kaelin, W. G. (2005) Cold Spring Harbor Symp. Quant. Biol. 70 159-166 [DOI] [PubMed] [Google Scholar]
- 44.Maxwell, P. H., Dachs, G. U., Gleadle, J. M., Nicholls, L. G., Harris, A. L., Stratford, I. J., Hankinson, O., Pugh, C. W., and Ratcliffe, P. J. (1997) Proc. Natl. Acad. Sci. U. S. A 94 8104-8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pouyssegur, J., Dayan, F., and Mazure, N. M. (2006) Nature 441 437-443 [DOI] [PubMed] [Google Scholar]
- 46.Staller, P., Sulitkova, J., Lisztwan, J., Moch, H., Oakeley, E. J., and Krek, W. (2003) Nature 425 307-311 [DOI] [PubMed] [Google Scholar]
- 47.Pennacchietti, S., Michieli, P., Galluzzo, M., Mazzone, M., Giordano, S., and Comoglio, P. M. (2003) Cancer Cell 3 347-361 [DOI] [PubMed] [Google Scholar]
- 48.Erler, J. T., Bennewith, K. L., Nicolau, M., Dornhofer, N., Kong, C., Le, Q. T., Chi, J. T., Jeffrey, S. S., and Giaccia, A. J. (2006) Nature 440 1222-1226 [DOI] [PubMed] [Google Scholar]
- 49.Jogi, A., Ora, I., Nilsson, H., Lindeheim, A., Makino, Y., Poellinger, L., Axelson, H., and Pahlman, S. (2002) Proc. Natl. Acad. Sci. U. S. A 99 7021-7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jogi, A., Vallon-Christersson, J., Holmquist, L., Axelson, H., Borg, A., and Pahlman, S. (2004) Exp. Cell Res. 295 469-487 [DOI] [PubMed] [Google Scholar]
- 51.Martens, L. K., Kirschner, K. M., Warnecke, C., and Scholz, H. (2007) J. Biol. Chem. 282 14379-14388 [DOI] [PubMed] [Google Scholar]
- 52.Murata, H., Tajima, N., Nagashima, Y., Yao, M., Baba, M., Goto, M., Kawamoto, S., Yamamoto, I., Okuda, K., and Kanno, H. (2002) Cancer Res. 62 7004-7011 [PubMed] [Google Scholar]
- 53.Nilsson, H., Jogi, A., Beckman, S., Harris, A. L., Poellinger, L., and Pahlman, S. (2005) Exp. Cell Res. 303 447-456 [DOI] [PubMed] [Google Scholar]
- 54.Tacconelli, A., Farina, A. R., Cappabianca, L., Desantis, G., Tessitore, A., Vetuschi, A., Sferra, R., Rucci, N., Argenti, B., Screpanti, I., Gulino, A., and Mackay, A. R. (2004) Cancer Cell 6 347-360 [DOI] [PubMed] [Google Scholar]
- 55.Birner, P., Schindl, M., Obermair, A., Plank, C., Breitenecker, G., and Oberhuber, G. (2000) Cancer Res. 60 4693-4696 [PubMed] [Google Scholar]
- 56.Giatromanolaki, A., Koukourakis, M. I., Sivridis, E., Pastorek, J., Wykoff, C. C., Gatter, K. C., and Harris, A. L. (2001) Cancer Res. 61 7992-7998 [PubMed] [Google Scholar]
- 57.Kurokawa, T., Miyamoto, M., Kato, K., Cho, Y., Kawarada, Y., Hida, Y., Shinohara, T., Itoh, T., Okushiba, S., Kondo, S., and Katoh, H. (2003) Br. J. Cancer 89 1042-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lisztwan, J., Imbert, G., Wirbelauer, C., Gstaiger, M., and Krek, W. (1999) Genes Dev. 13 1822-1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bracken, A. P., Pasini, D., Capra, M., Prosperini, E., Colli, E., and Helin, K. (2003) EMBO J. 22 5323-5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chadwick, B. P., and Willard, H. F. (2004) Proc. Natl. Acad. Sci. U. S. A 101 17450-17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmquist-Mengelbier, L., Fredlund, E., Lofstedt, T., Noguera, R., Navarro, S., Nilsson, H., Pietras, A., Vallon-Christersson, J., Borg, A., Gradin, K., Poellinger, L., and Pahlman, S. (2006) Cancer Cell 10 413-423 [DOI] [PubMed] [Google Scholar]
- 62.Vengellur, A., Phillips, J. M., Hogenesch, J. B., and LaPres, J. J. (2005) Physiol. Genomics 22 308-318 [DOI] [PubMed] [Google Scholar]
- 63.Seigneuric, R., Starmans, M. H., Fung, G., Krishnapuram, B., Nuyten, D. S., van Erk, A., Magagnin, M. G., Rouschop, K. M., Krishnan, S., Rao, R. B., Evelo, C. T., Begg, A. C., Wouters, B. G., and Lambin, P. (2007) Radiother. Oncol. 83 374-382 [DOI] [PubMed] [Google Scholar]
- 64.Mense, S. M., Sengupta, A., Zhou, M., Lan, C., Bentsman, G., Volsky, D. J., and Zhang, L. (2006) Physiol. Genomics 25 435-449 [DOI] [PubMed] [Google Scholar]
- 65.Maina, E. N., Morris, M. R., Zatyka, M., Raval, R. R., Banks, R. E., Richards, F. M., Johnson, C. M., and Maher, E. R. (2005) Oncogene 24 4549-4558 [DOI] [PubMed] [Google Scholar]
- 66.Stratmann, R., Krieg, M., Haas, R., and Plate, K. H. (1997) J. Neuropathol. Exp. Neurol. 56 1242-1252 [DOI] [PubMed] [Google Scholar]
- 67.Knebel, J., De Haro, L., and Janknecht, R. (2006) J. Cell. Biochem. 99 319-329 [DOI] [PubMed] [Google Scholar]
- 68.Okada, Y., Scott, G., Ray, M. K., Mishina, Y., and Zhang, Y. (2007) Nature 450 119-123 [DOI] [PubMed] [Google Scholar]
- 69.Pollard, P. J., Loenarz, C., Mole, D., McDonough, M. A., Gleadle, J. M., Schofield, C., and Ratcliffe, P. J. (2008) Biochem. J., in press [DOI] [PubMed]
- 70.Lando, D., Peet, D. J., Gorman, J. J., Whelan, D. A., Whitelaw, M. L., and Bruick, R. K. (2002) Genes Dev. 16 1466-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahon, P. C., Hirota, K., and Semenza, G. L. (2001) Genes Dev. 15 2675-2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fodor, B. D., Kubicek, S., Yonezawa, M., O'Sullivan, R. J., Sengupta, R., Perez-Burgos, L., Opravil, S., Mechtler, K., Schotta, G., and Jenuwein, T. (2006) Genes Dev. 20 1557-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen, H., Yan, Y., Davidson, T. L., Shinkai, Y., and Costa, M. (2006) Cancer Res. 66 9009-9016 [DOI] [PubMed] [Google Scholar]
- 74.Brahimi-Horn, M. C., and Pouyssegur, J. (2007) Essays Biochem. 43 165-178 [DOI] [PubMed] [Google Scholar]
- 75.Wellmann, S., Bettkober, M., Zelmer, A., Seeger, K., Faigle, M., Eltzschig, H. K., and Buhrer, C. (2008) Biochem. Biophys. Res. Commun. 372 892-897 [DOI] [PubMed] [Google Scholar]
- 76.Loh, Y. H., Zhang, W., Chen, X., George, J., and Ng, H. H. (2007) Genes Dev. 21 2545-2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lockman, K., Taylor, J. M., and Mack, C. P. (2007) Circ. Res. 101 e115-e123 [DOI] [PubMed] [Google Scholar]
- 78.Johnson, A. B., Denko, N., and Barton, M. C. (2008) Mutat. Res. 640 174-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaupel, P., Schlenger, K., Knoop, C., and Hockel, M. (1991) Cancer Res. 51 3316-3322 [PubMed] [Google Scholar]
- 80.Adam, M. F., Gabalski, E. C., Bloch, D. A., Oehlert, J. W., Brown, J. M., Elsaid, A. A., Pinto, H. A., and Terris, D. J. (1999) Head Neck 21 146-153 [DOI] [PubMed] [Google Scholar]
- 81.Stolze, I. P., Tian, Y. M., Appelhoff, R. J., Turley, H., Wykoff, C. C., Gleadle, J. M., and Ratcliffe, P. J. (2004) J. Biol. Chem. 279 42719-42725 [DOI] [PubMed] [Google Scholar]
- 82.Hagen, T., Taylor, C. T., Lam, F., and Moncada, S. (2003) Science 302 1975-1978 [DOI] [PubMed] [Google Scholar]
- 83.Covello, K. L., Kehler, J., Yu, H., Gordan, J. D., Arsham, A. M., Hu, C. J., Labosky, P. A., Simon, M. C., and Keith, B. (2006) Genes Dev. 20 557-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.