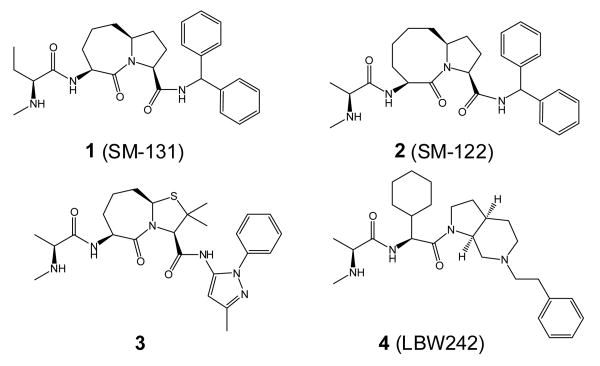
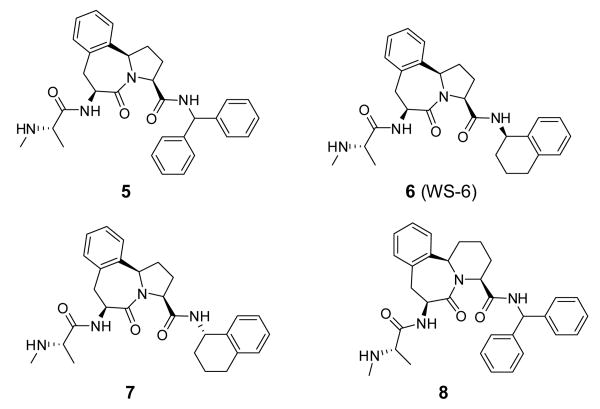
Abstract
A series of tricyclic, conformationally constrained Smac mimetics have been designed, synthesized and evaluated. The most potent compound 6 (WS-5) binds to XIAP, cIAP-1 and cIAP-2 with Ki values of 18, 1.1 and 4.2 nM, respectively. Compound 6 antagonizes XIAP in a functional assay and induces cIAP-1 degradation. Compound 6 inhibits cell growth with an IC50 value of 68 nM in the MDA-MB-231 cancer cell line and effectively induces cancer cells to undergo apoptosis.
Evasion of apoptosis, or programmed cell death, is commonly recognized as a hallmark of all cancers.1-2 Targeting critical apoptosis regulators with a goal to promote apoptosis in cancer cells is an attractive new cancer therapeutic strategy.2
Inhibitor of apoptosis proteins (IAPs) are a class of key apoptosis regulators characterized by the presence of one to three domains known as baculoviral IAP repeat (BIR) domains.3,4 Among these IAP proteins, X-linked IAP (XIAP) inhibits apoptosis by binding to and inhibition of two effectors, caspase-3/-7 and an initiator, caspase-9.4 While the third BIR domain (BIR3) of XIAP selectively targets caspase-9, the BIR2 domain, together with the linker immediately preceding it, inhibits caspase-3/-7.4,5 Cellular IAP-1 (cIAP-1) and cIAP-2 play a critical role in regulation of tumor necrosis factor (TNF) receptor-mediated apoptosis.4 Because of their central role in regulation of apoptosis, these IAP proteins are considered as promising new cancer therapeutic targets.5,6
Smac (Second Mitochondria-derived Activator of Caspases) was discovered as a potent pro-apoptotic protein and an endogenous antagonist of IAP proteins.7,8 Through direct binding, Smac antagonizes XIAP and abrogates the inhibition of caspase-3/-7 and caspase-9 by XIAP.7,9 Smac also binds to cIAP-1/29 and can reduce the levels of cIAP-1/2 in cells.10
Previous studies have firmly established that Smac interacts with XIAP and cIAP-1/2 proteins via its AVPI tetra-peptide motif.9,11,12 In the last few years, a number of laboratories, including ours, have engaged in the design of small molecules, which are called Smac mimetics, to mimic the AVPI binding motif as antagonists of IAP proteins.15-24 Two types of Smac mimetics have been reported, namely monovalent and bivalent Smac mimetics. While monovalent Smac mimetics are designed to mimic the binding of a single AVPI binding motif to IAP proteins16-20, bivalent compounds contain two AVPI binding motif mimetics tethered together through a linker.15,21-24 We have shown that bivalent Smac mimetics can achieve much higher affinities to XIAP and can be much more potent than their corresponding monovalent Smac mimetic in induction of apoptosis in tumor cells.21 However, monovalent Smac mimetics may hold certain advantages as potential drug candidates due to their small molecular weight (∼500). Furthermore, monovalent Smac mimetics provide the basic templates for the design of bivalent Smac mimetics. Herein, we wish to report the design, synthesis and evaluation of a series of conformationally constrained Smac mimetics containing a tricyclic core structure.
In our previous study, we showed that compound 1, which contains a [7,5] bicyclic core structure, binds to the XIAP BIR3 protein with a Ki value of 61 nM.19 Our subsequent binding studies determined that 1 binds to cIAP-1 and cIAP-2 BIR3 proteins with very high affinities and has Ki values of 1.3 nM and 4.8 nM, respectively (Table 1). Furthermore, 1 potently inhibits cell growth and effectively induces apoptosis in the MDA-MB-231 breast cancer cell line but shows minimal toxicity to normal cells.19 Hence, compound 1 represents a promising lead compound for further design and optimization.
Table 1.
Binding affinities of Smac mimetics to XIAP, cIAP-1 and cIAP-2 BIR3 proteins, as determined using competitive fluorescence-polarization assays. Ki and standard deviation (SD) values were determined by 3-5 independent experiments.
| Compounds | (Ki ± SD, nM) | ||
|---|---|---|---|
| XIAP BIR3 | cIAP-1 BIR3 | cIAP-2 BIR3 | |
| 1 | 61 ± 6.0 | 1.3 ± 0.2 | 4.8 ± 1.2 |
| 5 | 30 ± 4.4 | 3.0 ± 0.5 | 5.9 ± 1.0 |
| 6 | 18 ± 10 | 1.1 ± 0.5 | 4.2 ± 1.1 |
| 7 | 1,200 ± 500 | 150 ± 35 | 370 ± 30 |
| 8 | 690 ± 200 | 13 ± 2 | 20 ± 10 |
Our predicted binding model of 1 in complex with XIAP BIR3 based upon the crystal structure of Smac in complex with XIAP BIR3 (PDB ID: 1G73)11 showed that the 7-membered ring in 1 has van der Waals contacts with Trp323 in XIAP BIR3 and may contribute to its high binding affinity (Figure 3). We have designed compound 5, which has a phenyl ring fused to the 7-membered ring, to investigate if further conformational restriction is tolerated and if this phenyl ring can further enhance the binding to XIAP.
Figure 3.
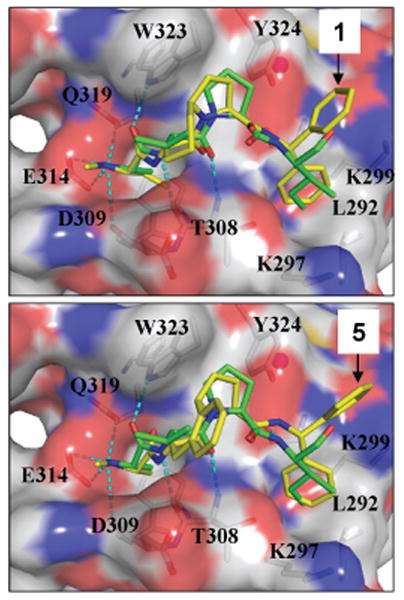
Predicted binding models of compounds 1 and 5 in complex with XIAP BIR3, in superposition on the crystal structure of Smac in complex with XIAP BIR3. Protein is displayed in surface model with key binding residues shown and labeled. Compounds 1, 5, and AVPI peptide are shown in stick. Carbon atoms of AVPI peptide are depicted in green and carbon atoms of compounds 1 and 5 are depicted in yellow. Oxygen and nitrogen atoms in these compounds are shown in red and blue, respectively.
Compound 5 was synthesized (Scheme I) and evaluated for its binding to XIAP, cIAP-1 and cIAP-2 BIR3 proteins using fluorescence-polarization based binding assays.25 Compound 5 binds to these three IAP proteins with high affinities, having Ki values of 30 nM, 3.0 nM and 5.9 nM to XIAP, cIAP-1 and cIAP-2, respectively (Table 1). Hence, the high binding affinities of compound 5 to XIAP and cIAP-1/2 clearly indicated that further conformational restriction of the [7,5] core structure in 1 by a fused phenyl ring is not detrimental for binding to these IAP proteins.
Scheme I.
Synthesis of compounds.
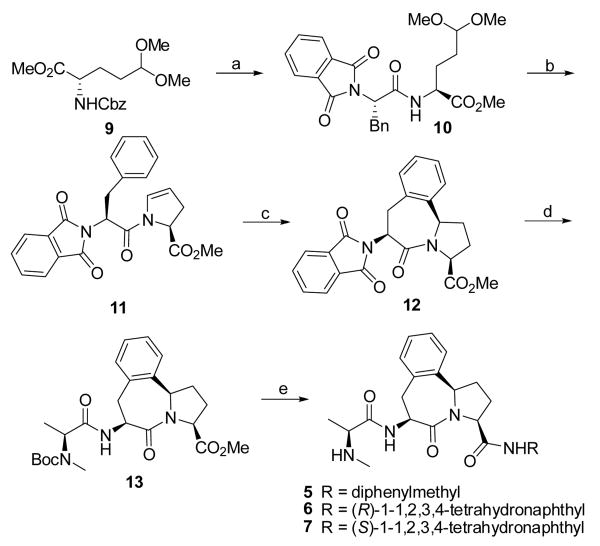
5-7. Reagents and conditions: (a) i. H2, 10% Pd-C, methanol; ii. N-phthaloyl-L-phenylpropanoic acid, EDC, HOBt, N-methylmorpholine, CH2Cl2-DMF 1:1, 0°C - rt, overnight, 92% over two steps; (b) CF3COOH, 4 Å molecular sieve, CHCl3, reflux, 88%; (c) trifluorosulfonic acid, trifluorosulfonic anhydride, CH2Cl2, 98%; (d) i. hydrazine hydrate, methanol, 3 days; ii. L-N-Boc-N-methyl-alanine, EDC, HOBt, N-methylmorpholine, CH2Cl2-DMF 1:1, 0 °C - rt, overnight, 92% over two steps; (e) i. 2 N LiOH, then 1 N HCl; ii. amine, EDC, HOBt, N-methylmorpholine, CH2Cl2-DMF 1:1, 0 °C - rt, overnight; iii. ZnBr2, CH2Cl2.
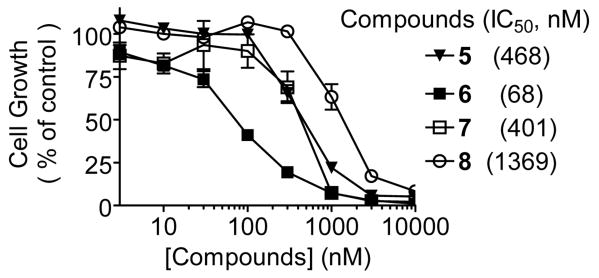
Compound 5 was evaluated for its ability to inhibit cancer cell growth in the MDA-MB-231 breast cancer cell line, a sensitive cell line used in previous studies.18-20 Indeed, compound 5 potently inhibits cell growth in this cell line with an IC50 value of 468 nM (Figure 4). Hence, compound 5 is a potent and cell-permeable Smac mimetic and a promising new lead compound for optimization and structure-activity relationship studies.
Figure 4.
Inhibition of cell growth by Smac mimetics in the MDA-MB-231 human breast cancer cell line. Cells were treated for 4 days and cell growth was determined using a WST-8 assay.
We next investigated if the diphenylmethyl group in compound 5 can be replaced by a tetrahydronaphthyl group, which was first employed in the design of potent Smac peptidomimetics.18 Modeling showed that the 1-(R)-tetrahydronaphthyl group, but not the (S)-isomer, can effectively interact with the hydrophobic pocket in XIAP BIR3 (Supporting Information). To test the modeling prediction, compounds 6 and 7 with either the (R)- or the (S)- tetrahydronaphthyl group were synthesized and evaluated.
Compounds 6 and 7 bind to XIAP BIR3 with Ki values of 18 nM and 1200 nM, respectively. Thus, 6 is 67 times more potent than 7 in binding to XIAP. Furthermore, 6 binds to cIAP-1 and cIAP-2 proteins with Ki values of 1.1 nM and 4.2 nM, respectively, and is >80 times more potent than 7. Consistent with its high binding affinity to these IAP proteins, 6 achieves an IC50 value of 68 nM in inhibition of cell growth in the MDA-MB-231 cell line, while 7 has an IC50 value of 401 nM (Figure 4).
We next designed and synthesized compound 8, in which the 5-membered proline ring in 5 is replaced by a 6-membered ring, to investigate the importance of the ring size. Modeling showed that replacement of the 5-membered proline ring in 5 by a 6-membered ring in 8 significantly weakens the contacts of compound 6 with Trp323, although all the hydrogen bonds are maintained (Figure S2, Supporting Information). Consistent with modeling prediction, compound 8 binds to XIAP, cIAP-1 and cIAP-2 proteins with Ki values of 692, 13, and 20 nM, respectively, substantially less potent than 5. These data showed that the 5-membered ring in 5 is critical in maintaining the optimal conformation for interactions with these IAP proteins. Compound 8 is 20-times less potent than 5 in cell growth inhibition in the MDA-MB-231 cancer cell line (Figure 4).
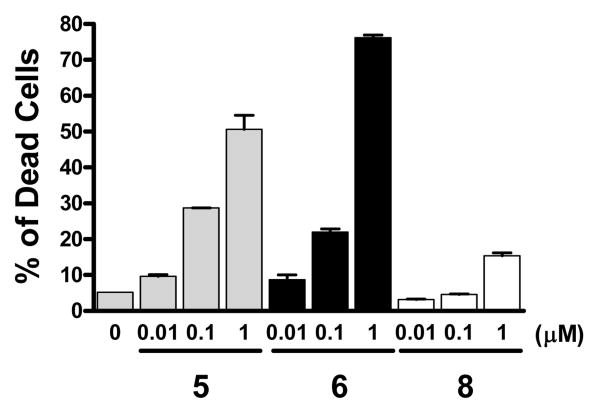
We next investigated if compounds 5, 6 and 8 can effectively induce cell death in the MDA-MB-231 cell line. Our data showed that while all these three Smac mimetics are capable of inducing cell death in a dose-dependent manner, compound 6 is most effective, and 8 is least effective (Figure 5). Treatment of the MDA-MB-231 cancer cells with 1 μM of compound 6 for 48 hours induced 75% of cells to undergo cell death but the treatment by compound 8 caused less than 20% of the cells to die. Thus, our data showed that compound 6 is a potent and effective inducer of cell death in the MDA-MB-231 cancer cell line.
Figure 5.
Induction of cell death by compounds 5, 6 and 8 in the MDA-MB-231 breast cancer cell line. Cells were treated with different concentrations of the compounds for 48 hours. Cell viability was determined using a trypan blue exclusion assay.
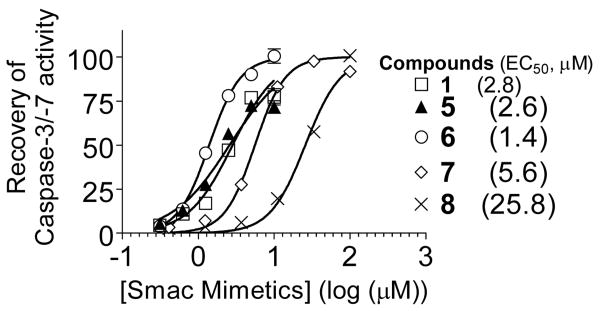
These new Smac mimetics were evaluated as antagonists of XIAP in a cell-free functional assay. While the XIAP BIR3 protein effectively inhibits the activity of caspase-9 and caspase-3/-7, these Smac mimetics dose-dependently antagonize the inhibition of XIAP to caspase activity (Figure 6). Consistent with their binding affinity data, 5 and 6 are the most potent Smac mimetics in relieving the inhibition of XIAP in this functional assay, while 8 is the least potent.
Figure 6.
Functional antagonism of Smac mimetics against XIAP BIR3 in a cell-free functional assay. Addition of dATP and cyctochrome c into the MDA-MB-231 cell lysates induced activation of caspase-3/-7 and XIAP BIR3 at 500 nM completely inhibited the caspase-3/-7 activity. Compounds 1, 5, 6, 7 and 8 dose-dependently recovered the caspase activity.
Our binding data showed that these Smac mimetics bind to cIAP-1 with high affinities. Several recent studies have demonstrated shown that Smac mimetics induce rapid cIAP-1 degradation in cells. Compounds 5, 6 and 8 were thus evaluated for their ability to induce cIAP-1 degradation in the MDA-MB-231 cancer cell line. Western blotting showed that all these three compounds can induce cIAP-1 degradation but compound 6 is the most potent one (Figure 7). Compound 6 effectively induces cIAP-1 degradation at concentrations as low as 100 nM and is more potent than 5 and 8, consistent with their binding affinities to cIAP-1. Compound 6 also potently and dose-dependently induces processing of caspase-8 and cleavage of poly(ADP-ribose) polymerase (PARP), two biochemical markers of apoptosis, at concentrations as low as 100 nM within 24 h.
Figure 7.
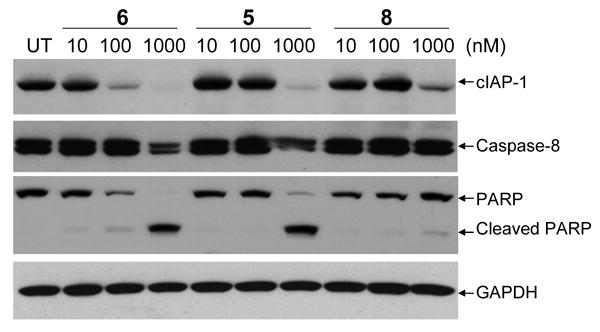
Western blot analysis of the levels of cIAP-1, caspase-8, pro- and cleaved PARP. MDA-MB-231 breast cancer cells were treated with different concentrations of Smac mimetics for 24 hours and proteins were probed with specific antibodies. GAPDH was used as the loading control.
The synthesis of compounds 5-7 is shown in Scheme I, whereas the synthesis of compound 8 is provided in Supporting Information. The synthesis of the key intermediate 13 was accomplished using a procedure similar to that published previously26 with some modifications. Briefly, removal of the Cbz protecting group in 9, followed by condensation of the resulted amine with N-phthaloyl-L-phenylpropanoic acid, yielded amide 10. Cyclization of 10 under the catalytic conditions using trifluoroacetic acid furnished 11. Cyclization of the enamide moiety with the phenyl ring in 11 in the existence of trifluorosulfonic acid and trifluorosulfonic anhydride provided 12. Conversion of the phthalimide moiety in 12 to amine, followed by condensation of this amine with L-N-Boc-N-methyl alanine, generated an amide 13. Hydrolysis of the methyl ester group in 13 furnished an acid. Condensation of this acid with corresponding amines afforded three amides, which were deprotected with ZnBr2 to give designed compounds 5-7.
In summary, we have designed and synthesized a series of novel Smac mimetics containing a tricyclic core structure. The most potent compound 6 (WS-5) binds to XIAP, cIAP-1 and cIAP-2 with low nanomoalr affinities. Consistent with its molecular mechanism of action, 6 effectively antagonizes XIAP in a cell-free functional assay and efficiently induces the degradation of cIAP-1 in cancer cells at concentrations as low as 100 nM. Compound 6 achieves an IC50 value of 68 nM in the MDA-MB-231 cell line in a cell growth assay and effectively induces cell death at 100 nM. Taken together, these data showed that 6 is a promising Smac mimetic for further evaluations and optimization for the development of a novel class of anticancer drugs. Further in vitro and in vivo studies of 6 and its analogues are being performed and the results will be reported in due course.
Supplementary Material
Figure 1.
Examples of reported Smac mimetics.
Figure 2.
Chemical structures of new Smac mimetics.
Acknowledgments
We are grateful for financial support from the National Natural Science Foundation of China (20672129, 20621062, 20772143) and the Chinese Academy of Sciences (“Knowledge Innovation Project”, KJCX2.YW.H08 and KGCX2-SW-209), the National Cancer Institute, National Institutes of Health, USA (R01CA109025), the Breast Cancer Research Foundation, the Susan G. Komen Foundation, the Prostate Cancer Foundation, and the University of Michigan Cancer Center Core grant (P30CA046592). cIAP-1 antibody is a kind gift from Dr. John Silke of La Trobe University, Victoria, Australia.
Footnotes
Abbreviations: IAP, inhibitor of apoptosis protein; XIAP, X-linked IAP; cIAP-1/-2, cellular IAP 1/2; Smac, second mitochondria-derived activator of caspases; BIR, baculoviral IAP repeats (BIR) domain; BIR2, the second BIR domain; BIR3, the third BIR domain; TNF, tumor necrosis factor; FP, fluorescence polarization.
Supporting Information Available: An experimental section including the information on the synthesis of compound 8, chemical data for compounds 5-8, details of molecular modeling, fluorescence polarization-based binding assays to XIAP, cIAP-1 and cIAP-2, the cell-free caspase functional assay, the cell growth assay, cell viability assay and Western blot analysis is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–95. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 2.Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1:111–121. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 3.Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;1:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 4.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 5.Holcik M, Gibson H, Korneluk RG. XIAP: Apoptotic brake and promising therapeutic target. Apoptosis. 2001;6:253–261. doi: 10.1023/a:1011379307472. [DOI] [PubMed] [Google Scholar]
- 6.Fulda S. Inhibitor of apoptosis proteins as targets for anticancer therapy. Expert Rev Anticancer Ther. 2007;7:1255–64. doi: 10.1586/14737140.7.9.1255. [DOI] [PubMed] [Google Scholar]
- 7.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 9.Shiozaki EN, Shi Y. Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem Sci. 2004;29:486–94. doi: 10.1016/j.tibs.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Yang QH, Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J Biol Chem. 2004;279:16963–70. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- 11.Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Sun C, Olejniczak ET, Meadows R, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 14.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 2004;305:1471–4. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 16.Sun H, Nikolovska-Coleska Z, Yang CY, Xu L, Liu M, Tomita Y, Pan H, Yoshioka Y, Krajewski K, Roller PP, Wang S. Structure-Based Design of Potent, Conformationally Constrained Smac Mimetics. J Am Chem Soc. 2004;126:16686–16687. doi: 10.1021/ja047438+. [DOI] [PubMed] [Google Scholar]
- 17.Sun H, Nikolovska-Coleska Z, Yang CY, Xu L, Tomita Y, Krajewski K, Roller PP, Wang S. Structure-based design, synthesis, and evaluation of conformationally constrained mimetics of the second mitochondria-derived activator of caspase that target the X-linked inhibitor of apoptosis protein/caspase-9 interaction site. J Med Chem. 2004;47:4147–50. doi: 10.1021/jm0499108. [DOI] [PubMed] [Google Scholar]
- 18.Oost TK, Sun C, Armstrong RC, Al-Assaad AS, Betz SF, Deckwerth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg SH, Shoemaker AR, Tomaselli KJ, Zou H, Fesik SW. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J Med Chem. 2004;47:4417–26. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Nikolovska-Coleska Z, Lu J, Qiu S, Yang CY, Gao W, Meagher J, Stuckey J, Wang S. Design, synthesis, and evaluation of a potent, cell-permeable, conformationally constrained second mitochondria derived activator of caspase (Smac) mimetic. J Med Chem. 2006;49:7916–20. doi: 10.1021/jm061108d. [DOI] [PubMed] [Google Scholar]
- 20.Zobel K, Wang L, Varfolomeev E, Franklin MC, Elliott LO, Wallweber HJ, Okawa DC, Flygare JA, Vucic D, Fairbrother WJ, Deshayes K. Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem Biol. 2006;1:525–33. doi: 10.1021/cb600276q. [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang S. Design, synthesis, and characterization of a potent, nonpeptide, cell-permeable, bivalent Smac mimetic that concurrently targets both the BIR2 and BIR3 domains in XIAP. J Am Chem Soc. 2007;129:15279–94. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 23.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 24.Petersen SL, et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–56. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wangs S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–73. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 26.Flynn GA, Giroux EL, Dage RC. An acyl-iminium ion cyclization route to a novel conformationally restricted dipeptide mimic: applications to angiotensin-converting enzyme inhibition. J Am Chem Soc. 1987;109:7914–7915. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.