Abstract
Type II protein secretion plays a role in a wide variety of functions that are important for the ecology and pathogenesis of Legionella pneumophila. Perhaps most dramatic is the critical role that this secretion pathway has in L. pneumophila intracellular infection of aquatic protozoa. Recently, we showed that virulent L. pneumophila strain 130b secretes RNase activity through its type II secretion system. We now report the cloning and mutational analysis of the gene (srnA) encoding that novel type of secreted activity. The SrnA protein was defined as being a member of the T2 family of secreted RNases. Supernatants from mutants inactivated for srnA completely lacked RNase activity, indicating that SrnA is the major secreted RNase of L. pneumophila. Although srnA mutants grew normally in bacteriological media and human U937 cell macrophages, they were impaired in their ability to grow within Hartmannella vermiformis amoebae. This finding represents the second identification of a L. pneumophila type II effector being necessary for optimal intracellular infection of amoebae, with the first being the ProA zinc metalloprotease. Newly constructed srnA proA double mutants displayed an even larger infection defect that appeared to be the additive result of losing both SrnA and ProA. Overall, these data represent the first demonstration of a secreted RNase promoting an intracellular infection event, and support our long-standing hypothesis that the infection defects of L. pneumophila type II secretion mutants are due to the loss of multiple secreted effectors.
INTRODUCTION
The Gram-negative bacterium Legionella pneumophila is a ubiquitous inhabitant of natural and man-made water systems, where it survives planktonically inside protozoan hosts, and as a part of biofilms (Fields et al., 2002). It is best known as the primary agent of legionnaires' disease, which is a serious form of pneumonia (Diederen, 2008). Following its transmission by the inhalation of contaminated aerosols, L. pneumophila enters the lungs, and then invades and grows within alveolar macrophages. Type II protein secretion is involved in a broad array of processes that influence the ecology and pathogenesis of L. pneumophila (Cianciotto, 2005; De Buck et al., 2007; Shin & Roy, 2008; Steinert et al., 2007). Based upon the phenotypes of L. pneumophila type II secretion (lsp) mutants, the pathway is necessary for optimal growth in buffered yeast extract (BYE) broth or on buffered charcoal yeast extract (BCYE) agar at 12–25 °C, colony morphology at 12–37 °C, survival in tap water at 4–17 °C, growth in protozoa at 22–37 °C, infection of human macrophages at 37 °C, and persistence in the lungs of mice (DebRoy et al., 2006b; Hales & Shuman, 1999; Liles et al., 1999; Lucas et al., 2006; Polesky et al., 2001; Rossier & Cianciotto, 2001; Rossier et al., 2004, 2008; Söderberg et al., 2004, 2008). Present in many, but not all, Gram-negative bacteria (Cianciotto, 2005; Peabody et al., 2003), type II secretion is a two-step process in which nascent proteins are first translocated across the inner membrane by the Sec or Tat machinery, and then, after what is probably a short period of time, transported from the periplasm to the exterior via a dedicated outer-membrane pore (Filloux, 2004; Forest, 2008; Johnson et al., 2006). Based upon proteomic and enzymic analyses of lsp mutant supernatants, the type II system of L. pneumophila mediates the secretion of at least 25 proteins, including a metalloprotease, aminopeptidases, acid phosphatases, a chitinase, lipases, phospholipase A, phospholipase C, lysophospholipase A, cholesterol acyltransferase, a peptidyl-prolyl isomerase and an RNase (Aragon et al., 2000, 2001, 2002; Banerji et al., 2005; DebRoy et al., 2006a, b; Flieger et al., 2001, 2002; Hales & Shuman, 1999; Liles et al., 1999; Rossier & Cianciotto, 2001; Rossier et al., 2004, 2008; Söderberg & Cianciotto, 2008). In silico analysis of the L. pneumophila genome suggests the type II secretome includes at least 35 more proteins (DebRoy et al., 2006b). With the exception of the RNase, genes encoding each of the known type-II-dependent enzymic activities have been identified and mutated (Aragon et al., 2001, 2002; Banerji et al., 2005; DebRoy et al., 2006b; Flieger et al., 2002; Quinn & Tompkins, 1989; Rossier et al., 2008; Söderberg & Cianciotto, 2008; Szeto & Shuman, 1990). As a result, we have shown that the chitinase promotes persistence in the lung, and that the secreted metalloprotease promotes infection of Hartmannella vermiformis amoebae (DebRoy et al., 2006b; Rossier et al., 2008). We have now characterized the gene (srnA) encoding the L. pneumophila secreted RNase activity, and here we document that it is necessary for optimal intracellular infection of Hartmannella amoebae.
METHODS
Strains, growth media, and chemicals
L. pneumophila strain 130b (ATCC strain BAA-74, also known as AA100) served as our wild-type control (Engleberg et al., 1984; Saito et al., 1981). Mutants of 130b containing a kanamycin-resistance (KmR) cassette inserted into lspF (NU275) or proA (AA200) have been described previously (Moffat et al., 1994; Rossier et al., 2004). Legionellae were cultured in BYE broth or on BCYE agar (Rossier et al., 2004). Growth in broth was assessed by measuring the optical density of cultures at 660 nm. Whereas Escherichia coli DH5α and DH10B (Invitrogen) were hosts for most of the plasmids, C41(DE3) and C43(DE3) (Miroux & Walker, 1996) were used to maintain the plasmid containing intact srnA. E. coli strains were grown on Luria–Bertani agar (Ausubel et al., 1989). Antibiotics were used at the following concentrations (μg ml−1): ampicillin, 100; chloramphenicol (Cm), 6 for L. pneumophila, and 30 for E. coli; gentamicin, 2.5; and kanamycin, 25 for L. pneumophila, and 50 for E. coli. Chemicals were from Sigma.
RT-PCR analysis of gene expression
RT-PCR was done as described previously (Allard et al., 2006; Liles et al., 1998; Viswanathan et al., 2002). L. pneumophila RNA was isolated using RNA STAT-60 (TEL-TEST B). The primers (Integrated DNA Tech) used were: OR145srnA (5′-ATCAACCTAATCTGCGCTGG-3′) and OR147srnA (5′-TCATGGTGTTGTGAGTGGCT-3′) to amplify a 703 bp internal fragment of srnA; OR148lpg2847 (5′-AACACCTCCAGCGGAAGTTA-3′) and Rnase3′ (5′-GCCCTCATAGCGAAACCGGTTG-3′) to amplify a 289 bp internal fragment of lpg2847; and OR146srnA (5′-AAGCCAGCCACTCACAACAC-3′) and Rnase3′ to amplify a 577 bp fragment encompassing the 3′ end of srnA, an intragenic region, and the 5′ end of lpg2847. Controls in which reverse transcriptase was omitted from the PCR were done to rule out contributions of contaminating DNA in the DNase-treated RNA samples.
Sequence analysis, gene cloning and mutant constructions
DNA and protein sequences were analysed using Lasergene (dnastar). The clustal method of Lasergene megalign was used for protein alignments. Protein homologues were identified in the genome database using programs based on the blast algorithm (Altschul et al., 1990). Conserved domains were identified by searching the conserved domain database (Marchler-Bauer et al., 2007). Signal sequences were identified using SignalP (Nielsen et al., 1997), and further predictions concerning the cellular location of proteins were done by using PSORTb (Gardy et al., 2005). DNA was isolated as described previously (Cianciotto et al., 1989). Primers for amplifying DNA from strain 130b were designed based on the L. pneumophila Philadelphia-1 genome database (Chien et al., 2004). RnaA-F-NdeI (5′-TCATATGGGCTTACCAAAGACAAGCC-3′) and RnaA-R-BamHI (5′-GTGAAGGATCCATCTCCTCCACTTGAAT-3′) were used to obtain a 1 kb fragment containing the srnA ORF only. The fragment was ligated into pGEM-T Easy (Promega), yielding pGsrnA. Plasmid pGsrnA was then digested with NdeI and BamHI, and the released fragment containing srnA was ligated into pET28α (Novagen) to yield pETsrnA, which was used to monitor SrnA activity in E. coli. In order to mutate srnA in strain 130b, Rnase5′ (5′-TGTACCCTTCCTTGTTGCCTTAGAGC-3′) and OR144srnA (5′-GCTCCAGACAACTGCCATAA-3′) were used to obtain a 1139 bp piece containing the first 640 bp of srnA. This fragment was then cloned into pGEM-T Easy, yielding pG5′srnA. To disrupt srnA, pG5′srnA was digested with BstBI, which cuts 110 bp after the start codon. Following Klenow treatment, the resulting fragment was ligated to a KmR gene isolated from pMB2190 upon HincII digestion (Grindley & Joyce, 1980), or to a gentamicin resistance (GmR) gene isolated from pX1918GT after HincII and PvuII digestion (Schweizer & Hoang, 1995), to give pGsrnA : : Km and pGsrnA : : Gm, respectively. These plasmids were digested with NotI, and following Klenow treatment, disrupted srnA was cloned into SmaI-digested pRE112 (Edwards et al., 1998), yielding pRsrnA : : Km and pRsrnA : : Gm. lpg2847-Rev (5′-AAGCTTATGAATTAAATGCATCAAAAATTGC-3′) and lpg2847-3′ (5′-GGGTTGTGACAGGGAATAATTTACGG-3′) amplified a 846 bp region containing the 3′ end of lpg2847. This fragment was ligated into pGEM-T Easy, yielding pG2847-3′. To disrupt lpg2847, pG2847-3′ was digested with EcoRV, resulting in a blunt cut 554 bp after the start codon, and a KmR gene was ligated in, yielding pG2847-5′ : : Km. Plasmid pG2847-3′ : : Km was then digested with NotI to release the fragment, and, after Klenow treatment, the disrupted gene was cloned into SmaI-digested pRE112, yielding pR2847-3′ : : Km. L. pneumophila was transformed with plasmids by electroporation as previously described (Cianciotto & Fields, 1992). Following electroporation of pRsrnA : : Km, pRsrnA : : Gm and pR2847-3′ : : Km into strain 130b, mutants were selected based on Cm sensitivity, sucrose resistance, and KmR (pRsrnA : : Km and pR2847-3′ : : Km) or GmR (pRsrnA : : Gm), indicative of the introduction of the mutated gene into the chromosome, and loss of pRE112 by allelic exchange. To construct a srnA lpg2847 double mutant, a GmR srnA mutant was electroporated with pR2847-3′ : : Km. Similarly, the isolation of a srnA proA double mutant was achieved by electroporation of a GmR srnA mutant with pRproA : : Km (Rossier et al., 2008). Verification of mutant genotypes was carried out by PCR analysis (data not shown).
Detection of enzymic activities
Cell-free, filter-sterilized supernatants were obtained from L. pneumophila cultures grown in BYE broth to late-exponential phase (Aragon et al., 2000). Secreted protease, phosphatase and lipolytic activities were detected as outlined previously (Aragon et al., 2000, 2001, 2002). Secreted RNase activity was initially assayed by monitoring the release of nucleotides from type III RNA, as previously described (Kar et al., 2000; Rossier et al., 2004). Briefly, a 40 μl volume of L. pneumophila culture supernatant was incubated with 400 μl assay buffer (50 mM Tris, pH 8) containing 10 mg ml−1 Baker's yeast type III RNA. At the beginning of the reaction, half of the sample was transferred to a fresh tube, and then chilled and precipitated with 2 vols ice-cold 10% TCA for 25 min. The remainder of the sample was incubated at 37 °C for 40 min, at which point the reactions were chilled and precipitated with TCA. Following centrifugation (12 min at 18 000 g) of the precipitated samples, the absorbance of the supernatants at 260 nm, reflective of released soluble nucleotides, was determined. Bovine pancreas RNase A was used as a positive control. To confirm the presence or absence of RNase activity in L. pneumophila supernatants, as well as in lysates of recombinant E. coli, we performed a negative-staining RNase zymogram (Bravo et al., 1994). Briefly, supernatants and cell lysates, which were obtained as described previously (DebRoy et al., 2006b), were electrophoresed through SDS-12 % polyacrylamide containing the yeast RNA. Following a renaturation step, RNase activity was detected by Toluidine Blue staining of the gel (Bravo et al., 1994).
Infection assays
H. vermiformis (ATCC 50237) amoebae were infected with L. pneumophila, as previously described (Cianciotto & Fields, 1992; Rossier et al., 2008). Briefly, 104 c.f.u. were used to infect 105 amoebae, and then, at various times, the numbers of bacteria per co-culture were determined by plating. U937 cells were also infected, as described previously (Cianciotto et al., 1989; Rossier & Cianciotto, 2001; Rossier et al., 2004): monolayers containing 106 macrophages were inoculated with 105 c.f.u. L. pneumophila, and, at various times, the numbers of bacteria in the monolayer were determined by plating lysates on BCYE agar, and the numbers of remaining host cells were determined by vital staining. For infection of A/J mice, 6- to 8-week-old females (Jackson Laboratory) were inoculated intratracheally with a 25 μl suspension containing 106 c.f.u. of a 1 : 1 ratio of wild-type and mutant strains of L. pneumophila (DebRoy et al., 2006b; Rossier et al., 2004). One and 3 days later, infected lungs were homogenized, and the numbers of bacteria, and the ratio of wild-type to mutant, were determined by plating. Animal experiments were approved by the Animal Care and Use Committee of Northwestern University.
RESULTS
Identification and mutation of srnA in L. pneumophila
Using 2D PAGE and proteomic analysis, we identified a protein that is present in wild-type serogroup-1 strain 130b culture supernatants, but not in type II secretion (lspF) mutant culture supernatants; the gene encoding the protein has been annotated in L. pneumophila databases as encoding an RNase (DebRoy et al., 2006b). We hypothesized that the gene, now designated srnA (secreted ribonuclease A), encodes the type-II-dependent RNase activity that we observed in earlier studies (Aragon et al., 2000; Rossier et al., 2004). In the sequenced L. pneumophila serogroup-1 strains Philadelphia-1, Paris, Lens and Corby, srnA is denoted as lpg2848, lpp2906, lpl2760 and lpc3133, respectively (Cazalet et al., 2004; Chien et al., 2004; Glockner et al., 2008). Recent proteomic analysis of culture supernatants of the Philadelphia-1 strain has further confirmed the secreted nature of SrnA (Galka et al., 2008). Compatible with its type II secretion, 38 kDa SrnA contains a Sec-dependent signal sequence (DebRoy et al., 2006b). In the L. pneumophila genomes, srnA exists in a two-gene operon, with the second gene, denoted as lpg2847 in Philadelphia-1, situated 137 bp from srnA. Using RT-PCR, we confirmed that srnA and lpg2847 are co-transcribed in strain 130b (data not shown). The Lpg2847 protein did not exhibit significant similarity to any known protein. In order to determine if SrnA is, in fact, an RNase, we cloned srnA, and used allelic exchange to construct a corresponding set of specific mutants of strain 130b. Three independent srnA mutants were obtained: GmR NU328 and NU329, and KmR NU330. Similar to other Lsp mutants that have been studied (Rossier et al., 2004), these mutants grew normally at 37 °C in BYE broth and on BCYE agar (data not shown), indicating that SrnA is not needed for extracellular growth under standard laboratory conditions. The srnA mutant supernatants had normal levels of acid phosphatase, lipase and protease (data not shown), indicating that the strains do not have general defects in type II secretion. The srnA mutants did not exhibit the altered colony morphology or reduced efficiency of plating at 25–17 °C displayed by lsp mutants (Rossier et al., 2004; Söderberg et al., 2004; Söderberg & Cianciotto, 2008).
Influence of srnA on RNase activity
In order to examine secreted RNase activity, we grew the legionellae in BYE broth to late-exponential phase, and then assayed culture supernatants for the ability to cleave purified yeast RNA. As observed previously (Rossier et al., 2004), strain 130b supernatants contained significant levels of RNase activity, whereas those of its type II secretion mutant NU275 lacked RNase activity (Fig. 1). The srnA mutants NU328, NU329, and NU330 lacked the activity (Fig. 1, and data not shown). That three independently derived mutants behaved identically indicated that this loss of secreted activity was due to the inactivation of srnA rather than spontaneous second site mutation(s). To also rule out the possibility of a polar effect associated with the insertional mutation of srnA, we made mutants with an insertion in lpg2847 (NU331 and NU332) and tested them for secreted activity. Neither of these mutants, which also grew normally in BYE broth and on BCYE agar at 37, 25 and 17 °C (data not shown), lacked any secreted RNase activity (Fig. 1, and data not shown). Double mutants inactivated for both srnA and lpg2847 (NU333 and NU334) lacked RNase activity in the same way as did the srnA mutants (Fig. 1, and data not shown). When an in-gel assay for RNase (zymogram) was performed, the supernatant of the wild-type, but not the supernatants of srnA mutants NU328 and NU329, exhibited a single protein band that had activity (Fig. 2a). The protein detected was approximately 38 kDa in size (Fig. 2a), and this indicated that SrnA, predicted to be 38 kDa, is directly responsible for the secreted RNase activity that is in L. pneumophila supernatants. That SrnA is an RNase was further confirmed when cloned srnA conferred RNase activity upon recombinant E. coli (Fig. 2b). These various data indicate that the Lsp-dependent SrnA represents the major secreted RNase of L. pneumophila. When a proA mutant of strain 130b was examined, we did not observe a loss of RNase (data not shown), indicating that SrnA, unlike some of the other L. pneumophila type II effectors (Banerji et al., 2005; Flieger et al., 2002; Rossier et al., 2008), is not cleaved by the secreted metalloprotease in order to be active.
Fig. 1.
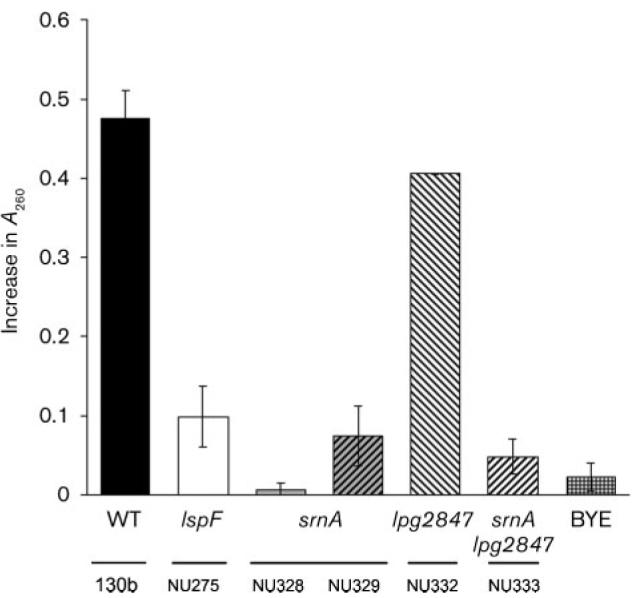
Secreted RNase activity of wild-type vs srnA mutants of L. pneumophila. Cell-free supernatants from cultures of wild-type strain 130b, lspF mutant NU275, srnA mutants NU328 and NU329, lpg2847 mutant NU332, and srnA lpg2847 double mutant NU333 were incubated with yeast RNA, and the release of nucleotides was measured by the increase in A260 over a 40 min period. A BYE medium control was also included. The values presented are the means±SD obtained from duplicate samples and are representative of at least four independent experiments. The reduced levels of activity seen for the lspF, srnA and srnA lpg2847 mutants relative to wild-type were statistically significant (P<0.05; Student's t-test).
Fig. 2.
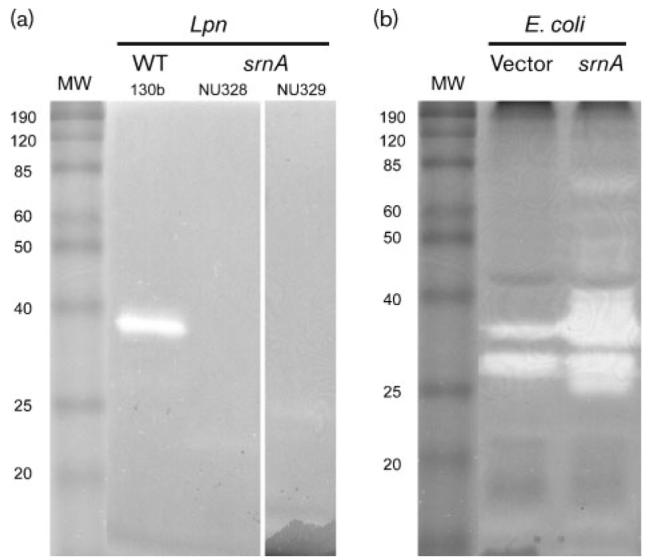
SrnA in L. pneumophila (Lpn) supernatants and recombinant E. coli lysates. (a) Neat supernatants obtained from wild-type (WT) 130b, and srnA mutants NU328 and NU239, were subjected to SDS-PAGE in the presence of RNA substrate, and then RNase was detected as a clear band following toluidine blue staining of the gel. MW, molecular mass markers (kDa) (BenchMark Protein Ladder; Invitrogen). The results depicted were obtained on two separate occasions using neat supernatants, and a further two times using 40× concentrated supernatants. (b) Lysates obtained from E. coli C41 (DE3) containing pET28α (vector) or pETsrnA that had srnA cloned into pET28α (srnA) were subjected to SDS-PAGE, and then RNase staining was done as described above. The two bands in the vector control lane probably represent endogenous E. coli RNase(s), whereas the band of approximately 25 kDa that is present in the sample from the srnA clone is probably a breakdown product of 38 kDa SrnA; results similar to each other were obtained on two separate occasions.
Importance of srnA for infection of mammalian hosts
Lsp mutants of strain 130b are impaired for infection of human macrophages and murine lungs (Rossier et al., 2004). Thus, to begin to judge the role of the Lsp-dependent RNase in L. pneumophila infection, we compared wild-type and srnA mutants for their ability to infect U937 cells. The srnA mutants showed no defect in the ability to grow in the macrophage cell line (Fig. 3a). Compatible with this result, infection with mutant bacteria ultimately produced death of the macrophage monolayer in a way that was similar to that resulting from wild-type infection (data not shown). When inoculated into the lungs of A/J mice, the srnA mutant grew and exhibited a recoverability from the lungs that was similar to wild-type 130b (Fig. 3b). These data indicate that SrnA is not required for optimal infection of mammalian host cells or tissue.
Fig. 3.
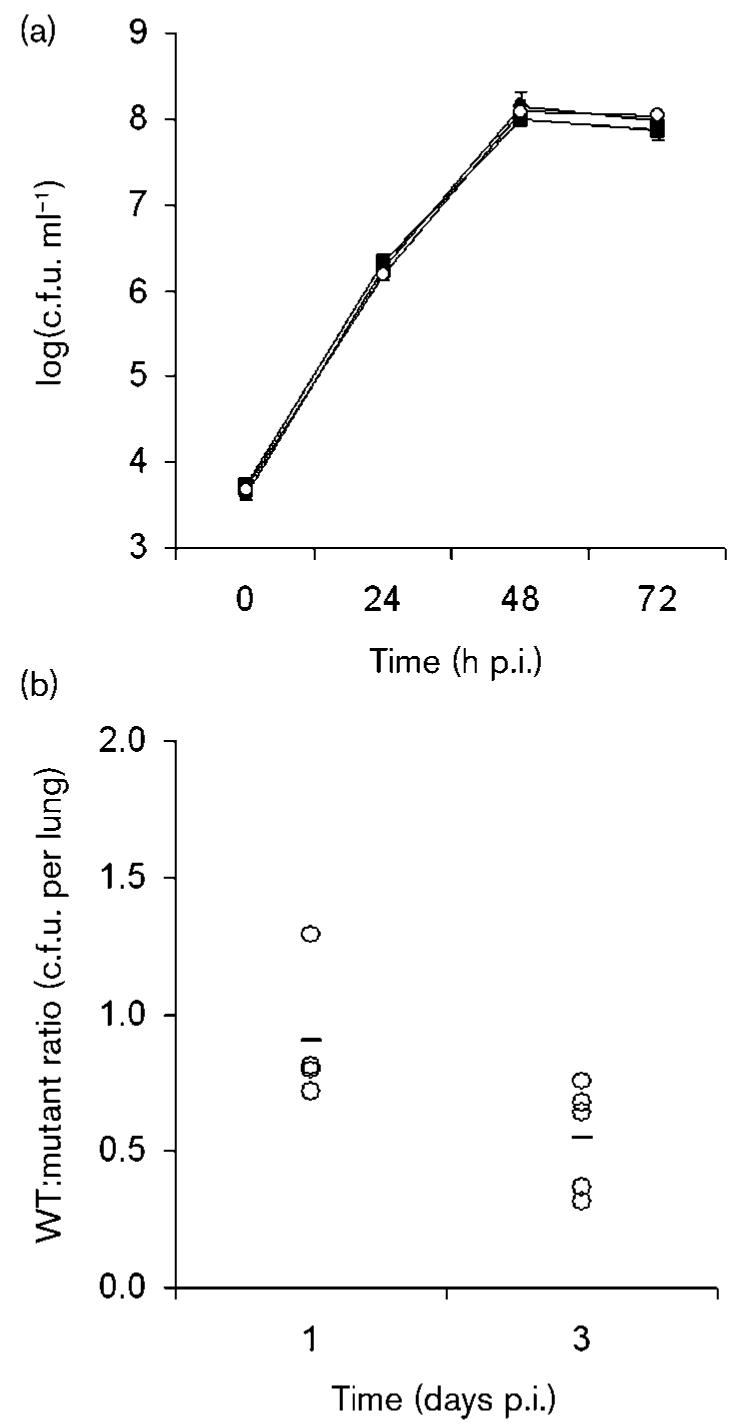
Infection of macrophages and mouse lungs by srnA mutants of L. pneumophila. (a) U937 cells were infected with wild-type strain 130b (■), srnA mutant NU328 (○), and srnA mutant NU329 (●). At various times post-inoculation, the numbers of bacteria per well were determined. The values presented are the means (±sd) obtained from four infected wells, and are representative of at least two independent experiments. (b) A/J mice were inoculated intratracheally with equal numbers of wild-type 130b and srnA mutant NU328, and then the ratio of wild-type to mutant was determined at days 1 and 3 after inoculation. Data are representative of actual values obtained per mouse (n=5), and the horizontal bar indicates the mean value.
Importance of srnA for intracellular infection of protozoan hosts
In the past, we and others have shown that type II secretion mutants of L. pneumophila are severely impaired for infection of protozoa (Hales & Shuman, 1999; Liles et al., 1999; Polesky et al., 2001; Rossier et al., 2004, 2008). Therefore, we next compared strain 130b and its srnA mutants for their ability to infect H. vermiformis amoebae. In nine out of nine tests, RNase mutants were significantly impaired for infection of hartmannellae, as evidenced by reduced recovery of the mutants at 48 and 72 h post-inoculation (Fig. 4). The magnitude of the defect ranged from fivefold, as exhibited by the NU328 mutant at 48 h in the experiment depicted in Fig. 4(a), to 17-fold, as shown by the NU329 mutant at 48 h in the experiment depicted in Fig. 4(b). Because both of the independently derived srnA mutants were impaired, this defect was due to the srnA mutation and not a second-site mutation. The NU331 and NU332 mutants inactivated for the lpg2847 gene downstream of srnA did not show any growth defect in the H. vermiformis culture (Fig. 4a, and data not shown), and this was a further indication that the defect was due specifically to the loss of srnA. The srnA lpg2847 double mutants displayed a reduced infectivity that was similar to the srnA mutants (Fig. 4a, and data not shown). That srnA mutants displayed reduced recovery only after extended incubation indicates that SrnA does not have a role in the early stages of intracellular infection, but that it is more likely to be involved in later replicative phases. Overall, these results indicate that SrnA is necessary for optimal intracellular infection of H. vermiformis.
Fig. 4.
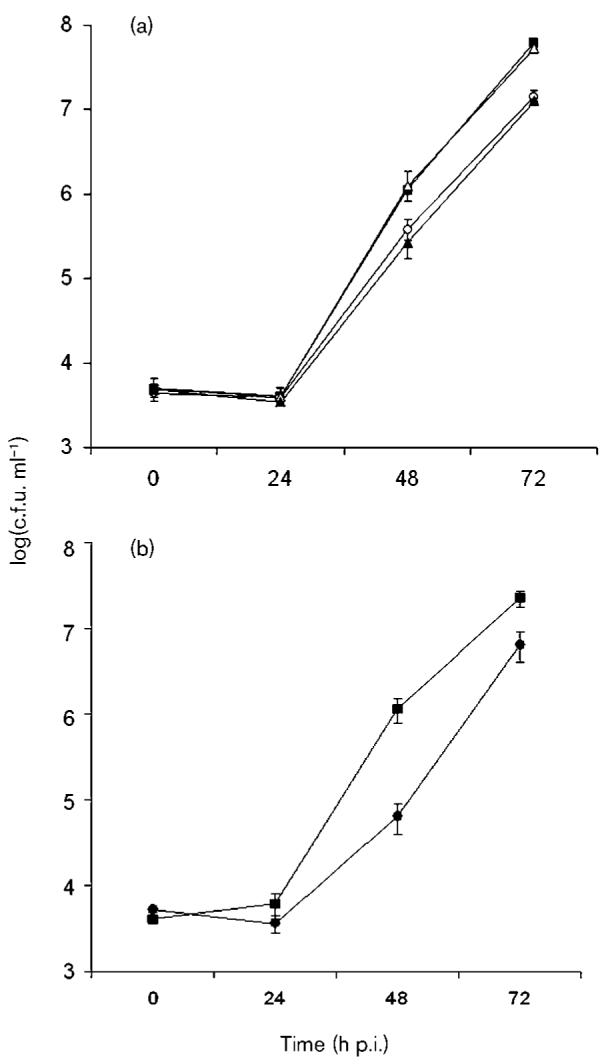
Infection of amoebae by srnA mutants of L. pneumophila. (a) H. vermiformis was infected with wild-type strain 130b (■), srnA mutant NU328 (○), lpg2847 mutant NU331 (△), and srnA lpg2847 double mutant NU333 (▲). At various times post-inoculation, the numbers of bacteria per well were determined. The values presented are the means (±sd) obtained from four wells. (b) H. vermiformis was infected with wild-type strain 130b (■) and srnA mutant NU329 (●), and then examined as described above for (a). At 48 and 72 h post-inoculation, the numbers of srnA and srnA lpg2847 mutants recovered from the wells were significantly less than numbers of the wild-type (P<0.05; Student's t test). The values shown in (a) are representative of six (for NU328), and two (for NU331 and NU333), independent experiments, and the values shown in (b) represent three independent experiments (for NU329).
Recently, we determined that a L. pneumophila 130b mutant specifically lacking the Lsp-dependent ProA/Msp metalloprotease is impaired for infection of H. vermiformis (Rossier et al., 2008). Thus, we constructed a new double mutant of strain 130b that was lacking both srnA and proA (NU335), and tested it for the ability to infect amoebae. Interestingly, the double mutant had a defect that was significantly greater than that exhibited by the single srnA and proA mutants (Fig. 5). Indeed, the defect for NU335 appeared to be the additive result of losing both SrnA and ProA, indicating that secreted RNase and protease activities contribute separately toward explaining the role of type II secretion in L. pneumophila infection of H. vermiformis. Since the srnA proA mutant was not as impaired as a lspF mutant (Fig. 5), there are likely to be important Lsp-dependent effectors in addition to SrnA and ProA.
Fig. 5.
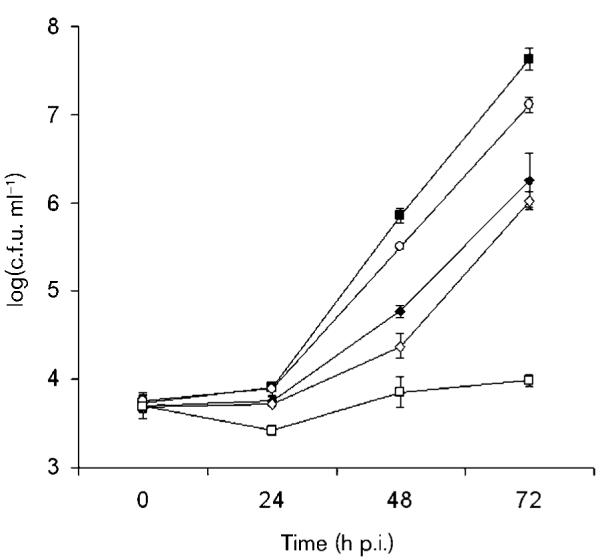
Infection of amoebae by an srnA proA double mutant of L. pneumophila. H. vermiformis was infected with wild-type strain 130b (■), srnA mutant NU328 (○), proA mutant AA200 (◆), srnA proA double mutant NU335 (◇), and lspF mutant NU275 (□). At various times, the numbers of bacteria per well were determined. The values are the means (±SD) obtained from four wells, and are representative of two independent experiments. The numbers of each of the mutants recovered from the wells were significantly less than the numbers of wild-type, at 48 and 72 h post-inoculation (P<0.05; Student's t test). The numbers of the srnA proA double mutant were also significantly less than those of the proA mutant at 48 h, and the srnA mutant at 48 and 72 h, while being significantly more than those of the lspF mutant at 48 and 72 h (P<0.05). In a separate experiment, the numbers of the srnA proA double mutant recovered were significantly less than those of the proA mutant at 72 h.
DISCUSSION
Here, we report the characterization of SrnA as a type-II-secreted RNase of L. pneumophila that promotes bacterial infection of a protozoan host. Current BLASTP results indicate that SrnA belongs to the T2 family of RNases (Deshpande & Shankar, 2002), based on a conserved domain (cd00374) located between amino acid residues 118 and 294. In the prokaryotic subfamily of T2 RNases (cd01062), SrnA has as its closest homologues proteins encoded in the genomes of Photobacterium, Shewanella, Methylococcus, Vibrio and Azotobacter species, with the corresponding E values ranging from 6×10−52 (for Photobacterium) to 1×10−43 (for Azotobacter). There are three groups of secreted RNases, i.e. RNase A, and T1 and T2 families (Aravind & Koonin, 2001). These enzymes share the ability to hydrolyse RNA to 3′ mononucleotides via 2′,3′ cyclic nucleotides (Deshpande & Shankar, 2002). The T2 RNases cleave RNA endonucleolytically, and are base non-specific (Deshpande & Shankar, 2002; Irie & Ohgi, 2001). Whereas RNase A proteins are restricted to vertebrates, and T1 RNases are restricted to fungi and Gram-positive bacteria (Dyer & Rosenberg, 2006; Sevcik et al., 2002; Yoshida, 2001), the T2 RNases are widespread across the biological kingdoms, occurring in viruses, bacteria, protozoa, fungi, plants and animals (Deshpande & Shankar, 2002; Irie & Ohgi, 2001). T2 RNases have been most extensively studied in eukaryotes, where they have been implicated in nutritional scavenging of nucleotides and/or phosphate in protozoa, fungi and plants, senescence, self-incompatibility and defence against pathogens in plants, and regulation of membrane permeability in yeast (Deshpande & Shankar, 2002; Irie & Ohgi, 2001; MacIntosh et al., 2001; McGugan et al., 2007). Additionally, some fungal and animal virus T2 RNases have been shown to possess cytotoxic activity, which has potential for anti-carcinogenic and anti-angiogenic therapy (Hulst et al., 1994; Schneider et al., 1993; Schwartz et al., 2007). In contrast to this wealth of information in eukaryotes, there are very little data on the role of T2 RNases in bacteria, even though there are clearly many T2 RNase genes in the prokaryotic genome database. In E. coli, the T2 RNase known as RNase I has been studied biochemically, and it has been defined as being localized to the periplasmic space (Messens et al., 2007; Padmanabhan et al., 2001). A similar situation exists in Aeromonas hydrophila and Shigella sp. (Favre et al., 1993). There are other secreted RNase activities in bacteria, but they represent nucleases that act on both RNA and DNA, e.g. the S1-P1, Staphylococcus nuclease, and colicin families (Desai & Shankar, 2003; Hsia et al., 2005). Secreted RNase activities have been found in some species of Klebsiella, Salmonella, Stenotrophomonas and Vibrio, but it is not clear what type of nucleases they are, or how they are secreted (Arella & Sylvestre, 1979; Favre et al., 1993). Thus, our findings involving SrnA are thought to be the first demonstration of a bacterial T2 RNase clearly localized to the extracellular milieu, and the first documentation of a role for any type of secreted RNase in an intracellular infection process.
The inability of srnA mutants to flourish optimally within H. vermiformis cultures indicates that SrnA has a significant role in L. pneumophila environmental persistence and, by extension, transmission to mammalian hosts. Several scenarios can be imagined for how SrnA promotes growth within H. vermiformis. First, SrnA might promote nutrient acquisition by degrading host cell RNA in order to obtain (portions of) nucleotides and/or phosphate. Second, SrnA might act to alter host cell function by degrading host cell RNA. Third, SrnA might possess a second enzymic activity that is relevant for intracellular infection; compatible with such a scenario, the T2 RNase of Aspergillus niger has an actin-binding activity that is separate from its nuclease activity (Schwartz et al., 2007). Finally, although we favour the hypothesis that there is a direct action of SrnA on a host cell target, it is also conceivable that the protein acts upon the bacterium itself, and in its absence intracellular legionellae are less able to persist. The location of SrnA in the infected host would influence protein function, and this, in turn, would be dictated by molecular trafficking across the phagosome membrane. Whether SrnA exits the phagosome into the cytoplasm and/or whether host cytoplasmic contents move into the phagosome over time is a key question. Relevant to that question, there are data showing type-II-secreted ProA present within the cytoplasm of infected host cells (Rechnitzer et al., 1992).
Although srnA mutants did not show a defect when grown in media, U937 cells or the A/J mouse lung, we do not conclude that SrnA and secreted RNase activity are only relevant for amoebal interactions. For example, SrnA might have an extracellular role in the environment when the legionellae experience extreme nutrient limitation and/or are faced with heightened competition or predatory microorganisms. Also, as suggested by our in silico analysis (DebRoy et al., 2006b), it is possible that L. pneumophila produces other nucleases that could serve in the absence of SrnA.
With the results presented here, we now have our second demonstration of a type II secreted protein promoting intracellular infection of amoebae. The significance of both SrnA and ProA confirms our long-standing hypothesis that the intracellular growth defects of lsp mutants are due to the loss of secreted effectors versus being solely due to potential cell envelope aberrations. That the srnA and proA mutants exhibited relatively modest defects, and that the srnA proA double mutant was not as impaired as an lsp mutant, imply that the L. pneumophila type II secretion system promotes intracellular infection through the combined action of many secreted effectors. A similar situation occurs with the well-studied type IVB secretion system of L. pneumophila, i.e. in many cases, the loss of an individual Dot/Icm effector has little, if any, effect on the numbers of legionellae produced in the course of an intracellular infection assay (Laguna et al., 2006; Liu & Luo, 2007; Ninio & Roy, 2007). Finally, at present, L. pneumophila is the only bacterium known to secrete a nuclease through a type II secretion system (Cianciotto, 2005). However, given the new-found significance of SrnA, it would be worthwhile to examine other bacteria, especially other environmental pathogens and intracellular parasites, for the production of secreted T2 RNases.
ACKNOWLEDGEMENTS
We thank past and present members of the Cianciotto laboratory for their assistance and helpful comments. Sruti DebRoy (University of Iowa) is also acknowledged for providing E. coli strains C41(DE3) and C43(DE3). This work was supported by NIH grant AI43987 awarded to N. P. C.
REFERENCES
- Allard KA, Viswanathan VK, Cianciotto NP. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J Bacteriol. 2006;188:1351–1363. doi: 10.1128/JB.188.4.1351-1363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aragon V, Kurtz S, Flieger A, Neumeister B, Cianciotto NP. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect Immun. 2000;68:1855–1863. doi: 10.1128/iai.68.4.1855-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon V, Kurtz S, Cianciotto NP. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect Immun. 2001;69:177–185. doi: 10.1128/IAI.69.1.177-185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon V, Rossier O, Cianciotto NP. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology. 2002;148:2223–2231. doi: 10.1099/00221287-148-7-2223. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. A natural classification of ribonucleases. Methods Enzymol. 2001;341:3–28. doi: 10.1016/s0076-6879(01)41142-6. [DOI] [PubMed] [Google Scholar]
- Arella M, Sylvestre M. Production of an extracellular ribonuclease by Pseudomonas maltophilia. Can J Microbiol. 1979;25:321–328. doi: 10.1139/m79-051. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Wiley; New York: 1989. [Google Scholar]
- Banerji S, Bewersdorff M, Hermes B, Cianciotto NP, Flieger A. Characterization of the major secreted zinc metalloprotease-dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophila. Infect Immun. 2005;73:2899–2909. doi: 10.1128/IAI.73.5.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Fernandez E, Ribo M, de Llorens R, Cuchillo CM. A versatile negative-staining ribonuclease zymogram. Anal Biochem. 1994;219:82–86. doi: 10.1006/abio.1994.1234. [DOI] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. other authors. [DOI] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. other authors. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci U S A. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto NP, Eisenstein BI, Mody CH, Toews GB, Engleberg NC. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Aragon V, Kurtz S, Cianciotto NP. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect Immun. 2006a;74:5152–5160. doi: 10.1128/IAI.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A. 2006b;103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck E, Anne J, Lammertyn E. The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology. 2007;153:3948–3953. doi: 10.1099/mic.0.2007/012039-0. [DOI] [PubMed] [Google Scholar]
- Desai NA, Shankar V. Single-strand-specific nucleases. FEMS Microbiol Rev. 2003;26:457–491. doi: 10.1111/j.1574-6976.2003.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Shankar V. Ribonucleases from T2 family. Crit Rev Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- Diederen BM. Legionella spp. and Legionnaires' disease. J Infect. 2008;56:1–12. doi: 10.1016/j.jinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Dyer KD, Rosenberg HF. The RNase A superfamily: generation of diversity and innate host defense. Mol Divers. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. [Google Scholar]
- Engleberg NC, Drutz DJ, Eisenstein BI. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect Immun. 1984;44:222–227. doi: 10.1128/iai.44.2.222-227.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre D, Ngai PK, Timmis KN. Relatedness of a periplasmic, broad-specificity RNase from Aeromonas hydrophila to RNase I of Escherichia coli and to a family of eukaryotic RNases. J Bacteriol. 1993;175:3710–3722. doi: 10.1128/jb.175.12.3710-3722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Flieger A, Gong S, Faigle M, Stevanovic S, Cianciotto NP, Neumeister B. Novel lysophospholipase A secreted by Legionella pneumophila. J Bacteriol. 2001;183:2121–2124. doi: 10.1128/JB.183.6.2121-2124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flieger A, Neumeister B, Cianciotto NP. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect Immun. 2002;70:6094–6106. doi: 10.1128/IAI.70.11.6094-6106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest KT. The type II secretion arrowhead: the structure of GspI-GspJ-GspK. Nat Struct Mol Biol. 2008;15:428–430. doi: 10.1038/nsmb0508-428. [DOI] [PubMed] [Google Scholar]
- Galka F, Wai SN, Kusch H, Engelmann S, Hecker M, Schmeck B, Hippenstiel S, Uhlin BE, Steinert M. Proteomic characterisation of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun. 2008;76:1825–1836. doi: 10.1128/IAI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Glockner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, Steinert M, Hacker J, Heuner K. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol. 2008;298:411–428. doi: 10.1016/j.ijmm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Grindley ND, Joyce CM. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980;77:7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales LM, Shuman HA. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect Immun. 1999;67:3662–3666. doi: 10.1128/iai.67.7.3662-3666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Li CL, Yuan HS. Structural and functional insight into sugar-nonspecific nucleases in host defense. Curr Opin Struct Biol. 2005;15:126–134. doi: 10.1016/j.sbi.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Hulst MM, Himes G, Newbigin E, Moormann RJ. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology. 1994;200:558–565. doi: 10.1006/viro.1994.1218. [DOI] [PubMed] [Google Scholar]
- Irie M, Ohgi K. Ribonuclease T2. Methods Enzymol. 2001;341:42–55. doi: 10.1016/s0076-6879(01)41144-x. [DOI] [PubMed] [Google Scholar]
- Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- Kar S, Soong L, Colmenares M, Goldsmith-Pestana K, McMahon-Pratt D. The immunologically protective P-4 antigen of Leishmania amastigotes. A developmentally regulated single strand-specific nuclease associated with the endoplasmic reticulum. J Biol Chem. 2000;275:37789–37797. doi: 10.1074/jbc.M002149200. [DOI] [PubMed] [Google Scholar]
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles MR, Viswanathan VK, Cianciotto NP. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles MR, Edelstein PH, Cianciotto NP. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CE, Brown E, Fields BS. Type IV pili and type II secretion play a limited role in Legionella pneumophila biofilm colonization and retention. Microbiology. 2006;152:3569–3573. doi: 10.1099/mic.0.2006/000497-0. [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Bariola PA, Newbigin E, Green PJ. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes? Proc Natl Acad Sci U S A. 2001;98:1018–1023. doi: 10.1073/pnas.98.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGugan GC, Jr, Joshi MB, Dwyer DM. Identification and biochemical characterization of unique secretory nucleases of the human enteric pathogen, Entamoeba histolytica. J Biol Chem. 2007;282:31789–31802. doi: 10.1074/jbc.M705975200. [DOI] [PubMed] [Google Scholar]
- Messens J, Collet JF, Van Belle K, Brosens E, Loris R, Wyns L. The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem. 2007;282:31302–31307. doi: 10.1074/jbc.M705236200. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Moffat JF, Edelstein PH, Regula DP, Jr, Cirillo JD, Tompkins LS. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Zhou K, Chu CY, Lim RW, Lim LW. Overexpression, biophysical characterization, and crystallization of ribonuclease I from Escherichia coli, a broad-specificity enzyme in the RNase T2 family. Arch Biochem Biophys. 2001;390:42–50. doi: 10.1006/abbi.2001.2359. [DOI] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Polesky AH, Ross JT, Falkow S, Tompkins LS. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect Immun. 2001;69:977–987. doi: 10.1128/IAI.69.2.977-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn FD, Tompkins LS. Analysis of a cloned sequence of Legionella pneumophila encoding a 38 kD metalloprotease possessing haemolytic and cytotoxic activities. Mol Microbiol. 1989;3:797–805. doi: 10.1111/j.1365-2958.1989.tb00228.x. [DOI] [PubMed] [Google Scholar]
- Rechnitzer C, Williams A, Wright JB, Dowsett AB, Milman N, Fitzgeorge RB. Demonstration of the intracellular production of tissue-destructive protease by Legionella pneumophila multiplying within guinea-pig and human alveolar macrophages. J Gen Microbiol. 1992;138:1671–1677. doi: 10.1099/00221287-138-8-1671. [DOI] [PubMed] [Google Scholar]
- Rossier O, Cianciotto NP. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect Immun. 2001;69:2092–2098. doi: 10.1128/IAI.69.4.2092-2098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O, Starkenburg S, Cianciotto NP. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect Immun. 2004;72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O, Dao J, Cianciotto NP. The type II secretion system of Legionella pneumophila elaborates two aminopeptidases as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl Environ Microbiol. 2008;74:753–761. doi: 10.1128/AEM.01944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Rolfe RD, Edelstein PH, Finegold SM. Comparison of liquid growth media for Legionella pneumophila. J Clin Microbiol. 1981;14:623–627. doi: 10.1128/jcm.14.6.623-627.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel HJ. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Shoseyov O, Melnikova VO, McCarty M, Leslie M, Roiz L, Smirnoff P, Hu GF, Lev D, Bar-Eli M. ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis, and metastasis. Cancer Res. 2007;67:5258–5266. doi: 10.1158/0008-5472.CAN-07-0129. [DOI] [PubMed] [Google Scholar]
- Schweizer HP, Hoang TT. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- Sevcik J, Urbanikova L, Leland PA, Raines RT. X-ray structure of two crystalline forms of a streptomycete ribonuclease with cytotoxic activity. J Biol Chem. 2002;277:47325–47330. doi: 10.1074/jbc.M208425200. [DOI] [PubMed] [Google Scholar]
- Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Söderberg MA, Cianciotto NP. A Legionella pneumophila peptidyl-prolyl cis-trans isomerase present in culture supernatants is necessary for optimal growth at low temperatures. Appl Environ Microbiol. 2008;74:1634–1638. doi: 10.1128/AEM.02512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg MA, Rossier O, Cianciotto NP. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J Bacteriol. 2004;186:3712–3720. doi: 10.1128/JB.186.12.3712-3720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg MA, Dao J, Starkenburg S, Cianciotto NP. The importance of type II secretion for Legionella pneumophila survival in tap water and amoebae at low temperature. Appl Environ Microbiol. 2008;74:5583–5588. doi: 10.1128/AEM.00067-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Heuner K, Buchrieser C, Albert-Weissenberger C, Glockner G. Legionella pathogenicity: genome structure, regulatory networks and the host cell response. Int J Med Microbiol. 2007;297:577–587. doi: 10.1016/j.ijmm.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Szeto L, Shuman HA. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan VK, Kurtz S, Pedersen LL, Abu-Kwaik Y, Krcmarik K, Mody S, Cianciotto NP. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect Immun. 2002;70:1842–1852. doi: 10.1128/IAI.70.4.1842-1852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. The ribonuclease T1 family. Methods Enzymol. 2001;341:28–41. doi: 10.1016/s0076-6879(01)41143-8. [DOI] [PubMed] [Google Scholar]


