Abstract
Identification and characterization of p53 target genes would lead to a better understanding of p53 functions and p53-mediated signaling pathways. Two putative p53 binding sites were identified in the promoter of a gene encoding PTGF-β, a type β transforming growth factor (TGF-β) superfamily member. Gel shift assay showed that p53 bound to both sites. Luciferase-coupled transactivation assay revealed that the gene promoter was activated in a p53 dose- as well as p53 binding site-dependent manner by wild-type p53 but not by several p53 mutants. The p53 binding and transactivation of the PTGF-β promoter was enhanced by etoposide, a p53 activator, and was largely blocked by a dominant negative p53 mutant. Furthermore, expression of endogenous PTGF-β was remarkably induced by etoposide in p53-positive, but not in p53-negative, cell lines. Finally, the conditioned medium collected from PTGF-β-overexpressing cells, but not from the control cells, suppressed tumor cell growth. Growth suppression was not, however, seen in cells that lack functional TGF-β receptors or Smad4, suggesting that PTGF-β acts through the TGF-β signaling pathway. Thus, PTGF-β, a secretory protein, is a p53 target that could mediate p53-induced growth suppression in autocrinal as well as paracrinal fashions. The finding made a vertical connection between p53 and TGF-β signaling pathways in controlling cell growth and implied a potential important role of p53 in inflammation regulation via PTGF-β.
Keywords: growth suppression, inflammation, transcription regulation
Human p53 is a 393-amino acid nuclear protein that acts biochemically as a transcription factor. The p53 molecule consists of three major domains: the N-terminal transactivation domain; a DNA-binding domain in the center portion of the molecule; and a C-terminal oligomerization domain (1). p53 specifically binds to its consensus DNA binding sequence, consisting of two repeats of 10-bp motif 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′(2), and transactivates expression of the target genes. Many biological functions of p53 are mediated through its downstream target genes. For example, p53-induced growth arrest is achieved mainly by transactivation of Waf-1/p21 (for G1 arrest) (3) or by activation of 14-3-3σ (for G2 arrest) (4). p53-induced apoptosis, on the other hand, was mediated by activation of Bax (5, 6), KILLER/DR5 (7), and the genes involved in generation of reactive oxygen species (8). p53 also regulates angiogenesis and tumor metastasis by transcriptional regulation of the genes encoding epidermal growth factor receptor (EGFR), thrombospondin, matrix metalloproteinases (MMPs), cathepsin D, Kang ai (KAI1), basic fibroblast growth factors, and multidrug resistant gene 1 (MDR1) (9, 10). The p53 mutations found in many human cancers were clustered in the specific DNA binding domain of the p53 molecule (1). This leads to an inactivation of p53 function through abolishing p53 specific DNA binding and transactivation.
The type β transforming growth factor (TGF-β) superfamily has more than 40 members (11). The members of this family are involved in regulation of many cellular functions and biological processes, including proliferation, apoptosis, extracellular matrix secretion and adhesion, terminal differentiation, and development (12). The protein PTGF-β was recently identified as a TGF-β family member that has a very high expression in placenta (13–15). Biological functions of PTGF-β appear to induce cartilage and bone formation (16). In addition, PTGF-β may suppress inflammation through inhibition of macrophage activation (17) and inhibits the proliferation of primitive hematopoietic progenitors (14).
In an attempt to achieve a better understanding of p53 signaling pathways and mechanism of action, we have used DNA chip technology to identify p53 target genes (18). Two p53 target genes, encoding antioxidant enzyme glutathione peroxidase (GPX) and calcium binding protein S100A2, were identified and characterized (19, 20). Here, we report the identification and characterization of PTGF-β, a TGF-β family member, as a p53 downstream target, determined by assays for DNA binding, transcriptional activation, and endogenous gene induction. Significantly, PTGF-β-induced tumor cell growth inhibition requires functional TGF-β receptors/Smad4 proteins, suggesting that PTGF-β acts through TGF-β signaling pathway. Thus, PTGF-β is a growth inhibitory cytokine that could mediate p53-induced growth inhibition on neighboring cells (21).
Materials and Methods
Cell Cultures.
U2-OS cells were grown in McCoy's 5A medium, supplemented with 10% fetal calf serum (FCS). Grown in 10% FCS containing DMEM are the lines of Saos-2, NCI-H460, H1299, MDA-MB468, and RKO. Mv1Lu and R1B/L17 were grown in Eagle's minimum essential medium containing 10% FCS. For drug treatment, subconfluent cells were incubated with etoposide (25 μM) (Sigma) for various periods of time up to 48 hr. The control cells were treated with DMSO.
Gel Shift Assay.
The assay was performed as described (22, 23). Two 20-bp synthetic oligonucleotides, PTGF-FBS01:5′-CATCTTGCCCAGACTTGTCT-3′ and PTGF-SBS01:5′-AGCCATGCCCGGGCAAGAAC-3′, which consist of the putative p53 binding sites found in the PTGF-β promoter, were labeled and used as the probes.
Luciferase Reporter Constructions.
The luciferase reporter constructs driven by the PTGF-β promoter (see Fig. 2A for a diagram) were made based on a published sequence (15) as follows: (i) PTGF-β W/p53BS: a 923-bp DNA fragment of the PTGF-β promoter containing two p53 binding sites, generated by PCR amplification of human placenta DNA (Oncor) with primers PTGF-FBS01 and PTGF04 (5′-CATCTGAGAGCCATTCA CCG-3′); (ii) PTGF-β W/Fp53BS: an 877-bp DNA fragment with the first p53 binding site, generated with primers PTGF-FBS01 and PTGF14 (5′-GTGCAGGTTGCGGCTCTGA-3′); (iii) PTFG-β W/Sp53BS: a 903-bp DNA fragment with the second p53 binding site, generated with primers PTGF03 (5′-CGAACTCCTGGGCTCAAACA-3′) and PTGF04; (iv) PTGF-β W/O p53BS: an 857-bp DNA fragment with neither p53 binding site, generated by primers PTGF03 and PTGF14. The PCR fragments were subcloned into a TA cloning vector (Invitrogen), were sequenced, and then were resubcloned into pGL-Basic-3 luciferase reporter (Promega).
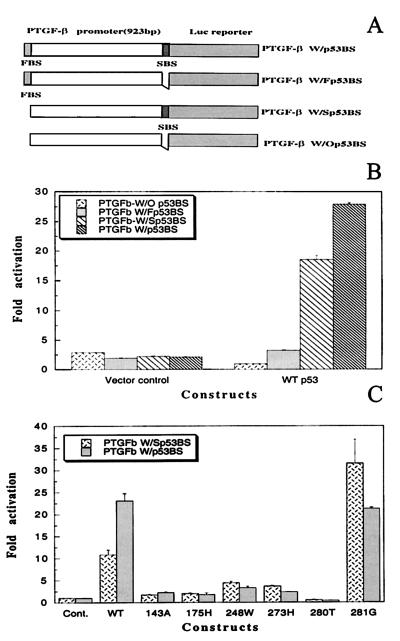
Figure 2.
p53 binding site-dependent activation of the PTGF-β promoter by wild-type p53, but not by most p53 mutants. (A) Diagrammatic presentation of four luciferase reporter constructs driven by the PTGF-β promoter, containing, respectively, two (W/p53BS), the first one (W/Fp53BS), the second one (W/Fp53BS), and none of p53 binding sites (W/Op53BS). (B) Activation of luciferase activity in p53-binding site dependent manner. (C) Lack of transactivation of PTGF-β promoter by most p53 mutants. Four constructs were transiently co-transfected with or without p53 expressing plasmids, respectively, along with a β-galactosidase-expressing vector into human Saos-2 cells, followed by luciferase assay. The results are presented as fold activation ± standard error derived from three independent transfections, each run in duplicate, after normalization with β-galactosidase activity for transfection efficiency.
DNA Transfection and Luciferase Assay.
Dispersed cells were seeded into 24-well plates at a cell concentration of 105 per well (for Saos-2) or 2 × 105 per well (for U2-OS) 16–24 hr before transfection. The calcium phosphate method was used to transiently transfect Saos-2 cells as described (24) whereas the Lipofectamine method (BRL) was used for U2-OS transfection. The luciferase reporters, along with the control plasmids, were co-transfected with a β-galactosidase construct in the presence or absence of constructs expressing wild-type or mutant p53 proteins (19, 24). Thirty-eight hours after transfection, cells were lysed and assayed for luciferase/β-galactosidase activities (24).
Northern Blot Analysis.
Subconfluent cells were treated with etoposide (25 μM) for various periods of time up to 48 hr. Total RNA was isolated by using RNAzol solution (Tel-Test, Friendswood, TX), and 15–20 μg of total RNA was subjected to Northern blot analysis (25).
Construction of PTGF-β-Expressing Plasmid, Establishment of Stable Transfectants, Collection of Conditioned Medium, and Western Blot Analysis.
An 886-bp cDNA fragment, flanking the entire open reading frame (ORF) of PTGF-β, was generated by reverse transcription–PCR amplification (26) using human placenta mRNA (Ambion) as template. The primers used are PTGF-ORF01:5′-GGAATTCGCCACCATGCTCCTGGTGTTGCTGG-3; and PTGF-ORF02:5′-GGAATTCTCACTTGTCATCGTCGTCCTTGTAGTCTATGCAGTGGCAGTCTTTGG-3′, containing a Flag-tag sequence at its 3′ end. The PCR fragment was subcloned into pcDNA3 (Invitrogen) and was sequenced to confirm the orientation and freedom of reverse transcription–PCR-generated mutation. The PTGF-β-expressing vector along with the empty vector was transfected into DLD-1 colon carcinoma cells by Lipofectamine, followed by G418 selection and ring-cloning (27). To determine PTGF-β expression in stable lines, confluent cells were serum-starved for 48 hr, and conditioned medium was collected, concentrated by Centricon 10 (Ambion), and assayed by Western blotting (28) for PTGF-β expression using Flag-tag antibody (Sigma). To prepare conditioned medium for growth inhibition assay, confluent PTGF-β-expressing DLD-Cl7 cells as well as Dn1-3 control cells were serum-starved for 72 hr. The conditioned medium was collected and concentrated, and protein concentration was measured (Bio-Rad). The conditioned medium was then aliquoted and stored at −70°C until use.
Antibody Generation, Affinity Purification, and Blockage of PTGF-β-Induced Growth Inhibition.
A polyclonal antibody against PTGF-β protein was generated using standard methods by Zymed. The antigen used is a 20-aa peptide [PTGF-l: (C)QKTDTGVSLQTYDDLLAKD-COOH] located in the C terminus of PTGF-β protein. The antibody was affinity-purified with the antigen peptide, using the SulfoLink Kit (Pierce). For neutralization of PTGF-β, affinity-purified antibody was preincubated with PTGF-β-containing conditioned medium (20 μg of antibody protein added into 1 ml of conditioned medium containing 100 μg of protein) for 1 hr at 4°C before being added to growth medium for cell proliferation assay.
BrdUrd Incorporation Assay for Cell Proliferation.
Subconfluent cells (Mv1Lu, Du145, R1B/L17, RKO, or MDA-MB486) were seeded at 5,000 cells/well in 96-well flat-bottomed microtiter plates in medium containing 10% FCS. Next day, pure TGF-β1 protein (BMB) or serum-starved conditioned media from DLD-Cl7 and Dn1-3 cells were added, respectively, with various protein concentrations. The plates were subsequently cultured for 24 hr, followed by incubation with BrdUrd for 6 hr. Incorporation of BrdUrd into DNA is measured by immunoassay as specified in the kit (Boehringer Mannheim).
Results
Identification of Two p53 Binding Sites in the Promoter of the PTGF-β Gene.
We have recently used Affymetrix GeneChip arrays (HUM6000) in an attempt to identify genes responsible for apoptosis induced by etoposide, a p53 activator (18). We compared potential p53-inducible genes identified by our chip screening (18) and serial analysis of gene expression (SAGE) technology (8). TGF-β type II receptor was identified by the chip (18), and PTGF-β, a TGF-β superfamily member (13–17) was identified by SAGE (8). Computer searching of the gene promoters for p53 consensus binding sequence revealed no binding site for TGF-β receptor but two putative binding sites for PTGF-β. The first binding site is PTGF-FBS01:5′-CATCTTGCCCAGACTTGTCT-3′ containing two mismatches (underlined) to the consensus sequence. The second binding site is PTGF-SBS01: 5′-AGCCATGCCCGGGCAAGAAC-3′ containing three mismatches (underlined). They are located, respectively, at −898 to −879 and −43 to −24 upstream from possible translation initiation site (14, 15).
p53 Specifically Binds to These Two Binding Sites in the PTGF-β Promoter.
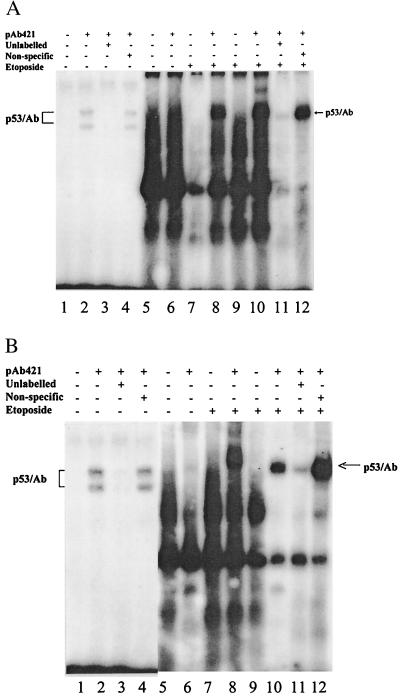
To examine whether p53 binds to these putative p53 binding sites, we used both partially purified recombinant p53 protein from baculovirus (23) and nuclear extract from p53-positive U2-OS cells treated with etoposide (25 μM). As shown in Fig. 1, consistent with the previous observations of many other p53 binding oligonucleotides (22–24, 29), neither PTGF-FBS01 (A) nor PTGF-SBS01 (B) bound to p53 in the absence of p53 antibody (lanes 1, 5, 7, and 9). The binding resumed in the presence of p53 antibody, pAb421, known to enhance and stabilize p53-DNA binding (30) (Fig. 1 A and B, lanes 2, 8, and 10). Binding of either oligonucleotide to purified p53 revealed two binding bands (Fig. 1 A and B, lane 2), possibly reflecting a different protein-DNA conformation (23). Both p53 binding sites were also found to bind to endogenous p53 in nuclear extract treated with etoposide for 6 and 24 hr, respectively (Fig. 1 A and B, lanes 8 and 10), but not in untreated samples (Fig. 1 A and B, lane 6). The binding is sequence-specific because it can be largely competed away by 100-fold excess of unlabeled oligonucleotide (Fig. 1 A and B, lanes 3 and 11), but not by a sequence-nonspecific competitor (Fig. 1 A and B, lanes 4 and 12). Thus, p53 binds to both p53 binding sites found in the PTGF-β promoter.
Figure 1.
p53 binds to putative p53 binding sites in the promoter of the PTGF-β gene. Synthetic oligonucleotides (PTGF-FBS01 and PTGF-SBS01) were labeled with [γ-32P]ATP and were used as probes in gel shift assays. (A) The oligonucleotide PTGF-FBS01. (B) The oligonucleotide PTGF-SBS01. Lanes: 1–4, partially purified p53 protein (3 μg) with or without pAb421 antibody; 5–12, nuclear extracts (8.5 μg) prepared from U2-OS cells treated with etoposide (25 μM) for 0 hr (lanes 5 and 6), 6 hr (lanes 7 and 8), and 24 hr (lanes 9–12). Competition was performed with a 100-fold excess of unlabeled specific oligonucleotides, PTGF-FBS01 or PTGF-SBS01, respectively, or nonspecific oligonucleotide mT3SF (5′-GGGGTTGCTTGAAGAGCGTC-3′) (29).
p53 Dose- as well as p53 Binding Site-Dependent Transactivation of the PTGF-β Promoter.
To examine whether p53 transactivates the PTGF-β promoter and whether activation is p53 dose-dependent, a luciferase reporter construct driven by a 923-bp promoter fragment containing both p53 binding sites (PTGFβW/p53BS) was made. This luciferase reporter was cotransfected with various amounts of p53-expressing plasmid into p53-negative human Saos-2 cells. p53 transactivated the PTGF-β promoter in a dose-dependent manner. The luciferase activity was increased by 12-, 18-, or 23-fold with an amount of input p53 DNA of 0.1, 0.3, and 0.7 μg, respectively (data not shown).
To examine whether transactivation of the promoter by p53 depends on the p53 binding sites, three additional luciferase reporter constructs were made. As presented in Fig. 2A, four constructs are PTGF-β W/p53BS (containing two p53 binding sites); PTGF-β W/Fp53BS (the first site); PTGF-β W/Sp53BS (the second site); and PTGF-β W/O p53BS (no p53 site). Each of these constructs was individually cotransfected with a p53 expressing plasmid into Saos-2 cells. As shown in Fig. 2B, in the absence of p53 (vector control), these four luciferase reporters give rise to a similar level of luciferase activity. However, in the presence of p53, a p53 binding site-dependent transactivation was revealed. The first p53 binding site induced a 3.5-fold activation whereas the second binding site contributed an 18-fold activation. With both binding sites, a 29-fold transactivation was achieved. Thus, transactivation of the PTGF-β promoter is p53-binding site-dependent, and the second binding site appears to be more active.
Lack of Transactivation of the PTGF-β Promoter by Most p53 Mutants.
We next examined whether the PTGF-β promoter was also subjected to transactivation by p53 mutants. Plasmid DNAs encoding p53 mutants were individually co-transfected into Saos-2 cells with two luciferase reporters, PTGF-β W/p53BS or PTGF-β W/Sp53BS. The p53 mutants used are p53-143A, p53-175H, p53-248W, p53-273H, and p53-281G, five p53 mutants most commonly found in human cancers (31), and p53-280T, a dominant negative p53 mutant found in nasopharyngeal carcinomas (32, 33). As shown in Fig. 2C, compared with the vector control and wild-type p53, all of the p53 mutants, except p53-281G, do not induce any significant transactivation of the promoter. Transactivation by p53-281G was very striking, with a level comparable to that of the wild-type p53. However, activation by p53-281G was found to be p53 binding site-independent. All four reporters produced a similar fold activation (Fig. 2C; data not shown). Furthermore, expression of p53-281G protein is much higher than that of the wild type (19). If normalized with the p53 expression level, wild-type p53 should have a much higher activity in transactivation of the PTGF-β promoter.
p53-Dependent Transactivation Can Be Enhanced by Etoposide.
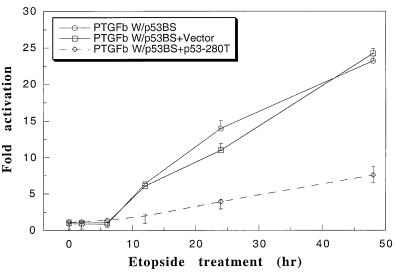
We have shown that etoposide can activate p53 in U2-OS cells (24). We therefore examined whether etoposide would induce p53-dependent transactivation of the PTGF-β promoter. As shown in Fig. 3, transactivation of the luciferase reporter driven by PTGF-β W/p53BS was induced by etoposide in an incubation time-dependent manner. A 25-fold activation was achieved at 48 hr. To confirm that the etoposide-induced transactivation is p53-dependent, we transfected p53-280T, a known dominant negative p53 mutant (19, 32, 33), into U2-OS cells, followed by etoposide treatment and luciferase assay. As a control, the empty vector was used. Etoposide-induced activation was dramatically reduced by p53-280T (PTGFβW/p53BS + p53-280T), but not at all by the vector control (PTGFβW/p53BS + Vector). The results demonstrate that transactivation of the PTGF-β promoter by etoposide is largely p53-dependent.
Figure 3.
Induction of p53-dependent transactivation of the PTGF-β promoter by etoposide. Subconfluent U2-OS cells were co-transfected with β-galactosidase-expressing construct and PTGF-β W/p53BS luciferase reporters, individually, or in combination with a construct expressing a dominant negative p53 mutant (p53-280T) or the vector control by the Lipofectamine method. Cells were treated with etoposide (25 μM) 24 hr after transfection for 0, 2, 6, 12, 24, and 48 hr, respectively, followed by luciferase activity measurements. Three independent transfections, each run in duplicate, were performed, and results are presented as fold activation ± standard error after normalization with β-galactosidase activity for transfection efficiency. To calculate the fold activation, the luciferase activity from PTGF-β W/p53BS construct after 0 hr of etoposide treatment was arbitrarily set as 1.
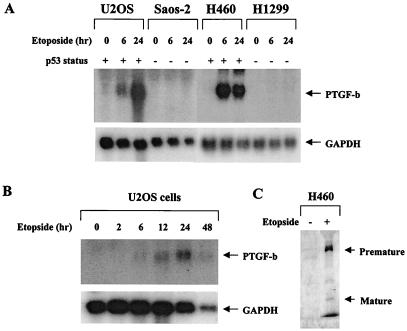
Increased PTGF-β mRNA Expression by Etoposide in p53-Positive but not p53-Negative Cells and Induction of PTGF-β Protein by Etoposide.
To examine whether endogenous PTGF-β is subjected to p53 regulation, we treated four human cell lines with etoposide and measured mRNA expression by Northern blot analysis. The four cell lines used included osteogenic sarcoma lines, U2-OS (p53-positive), Saos-2 (p53-negative), and lung carcinoma lines NCI-H460 (p53-positive) and H1299 (p53-negative). As shown in Fig. 4A, no endogenous basal level of PTGF-β mRNA could be detected in any of four cell lines. Upon etoposide treatment, PTGF-β mRNA was induced only in p53-positive U2-OS and H460 cells, but not in p53-negative Saos-2 or H1299 cells. We further examined the time course of PTGF-β induction by etoposide in U2-OS. Induction of PTGF-β mRNA started to occur at 6 hr post-treatment and then gradually increased and reached maximal level at 24–48 hr (Fig. 4B). The induction pattern agreed well with p53 activation by etoposide (ref. 24; also see Fig. 1). Finally, we examined whether etoposide induced PTGF-β protein expression in p53-positive H460 cells. As shown in Fig. 4C, both premature and mature forms of PTGF-β protein were detected in conditioned medium collected from cells treated with etoposide, but not from DMSO control cells. These results clearly demonstrated a p53-dependent induction of endogenous PTGF-β expression and indicate that PTGF-β is an endogenous p53 target gene.
Figure 4.
Induction of endogenous PTGF-β expression by etoposide in p53 positive cells. (A) Subconfluent U2-OS, Saos-2, H460, and H1299 cells were subjected to etoposide (25 μM) treatment for various times up to 48 hr, followed by total RNA isolation and Northern blot analysis, using PTGF-β cDNA as a probe. (B) The time course of PTGF-β mRNA induction by etoposide in U2OS cells. (C) PTGF-β protein induction by etoposide. Subconfluent H460 cells were subjected to DMSO or etoposide (25 μM) treatment for 24 hr under serum-free condition. The media were collected, and proteins were TCA-precipitated and subjected to Western blot analysis using rabbit anti-PTGFβ antibody.
Suppression of Tumor Cell Growth by PTGFβ.
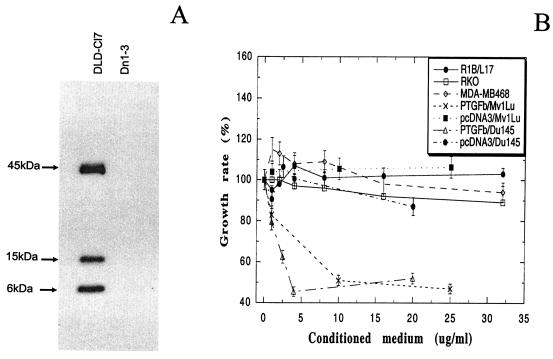
It has been recently shown that p53, upon activation, can induce the secretion of growth inhibitory cytokines that exert an antiproliferative effect on neighboring tumor cells (21). We hypothesized that PTGF-β could be one of these secretory proteins that are activated by p53 and mediate p53-induced growth inhibition. To test potential growth inhibitory activity of PTGF-β, we generated stable clones overexpressing flag-tagged PTGF-β in DLD-1 colon carcinoma cells (27). Because PTGF-β is a secretory protein, expression of PTGF-β was examined from the conditioned media collected from these stable lines by Western blot analysis using anti-flag antibody. As shown in Fig. 5A, three bands with the sizes of 6, 15, and 45 kDa, respectively, were revealed in the conditioned medium from a PTGF-β-transfected clone (DLD-Cl7), but not from the vector control (Dn1-3). The 15-kDa band corresponds to the mature form of PTGF-β whereas the 45-kDa band is the premature form (16). The nature of the 6-kDa band is not clear. It might be a degradation product of a mature form. This band was not detectable when the same PTGF-β-encoding plasmid was transiently transfected into 293 cells (data not shown). The conditioned medium was collected from DLD-Cl7 and Dn1-3 control and was applied to Mv1Lu mink lung epithelial cells, a common TGF-β target cells, along with pure TGF-β1 protein (BMB) as a positive control. While pure TGF-β1 induced a dose-dependent growth inhibition (data not shown), the conditioned medium collected from DLD-Cl7 (PTGF-β), but not from Dn1-3 control (pcDNA3), inhibited Mv1Lu cell growth, also in a dose-dependent manner. The IC50, (a concentration that caused half-maximal growth inhibition) was ≈3.5 μg/ml (Fig. 5B). Thus, PTGF-β inhibits growth of cells that are sensitive to TGF-β1.
Figure 5.
Expression of PTGF-β in conditioned medium and requirement of intact TGFβ signaling pathway for growth inhibition. (A) Expression of PTGF-β in the conditioned medium collected from PTGF-β-expressing DLD-Cl7 cells, but not from Dn1-3 vector control. Conditioned medium was collected and subjected (85 μg) to Western blot analysis using Flag-tag antibody. (B) PTGF-β-induced growth inhibition of TGF-β-sensitive but not TGF-β-resistant cells. The target cells used are TGF-β-sensitive Mv1Lu or Du145 cells as well as TGF-β-resistant cells, including RKO (TβRII mutation), R1B/L17 (TβRI mutation), and MDA-MB468 (Smad4-null). Conditioned medium was collected from PTGF-β expressing clone, DLD-Cl7 or the vector control, and Dn1-3 cells and was concentrated and added into the target cells at various protein concentrations, followed by BrdUrd incorporation assay. All results were presented as percent of growth rate ± standard error of the mean from three independent experiments, each run in quadruplet. The growth rate was calculated with the control group (without any addition of conditioned medium) setting as 100%.
To determine potential tumor cell growth inhibition, the conditioned medium was applied to Du145 prostate carcinoma cells. As also shown in Fig. 5B, a dose-dependent growth inhibition with an IC50 of 2 μg/ml was achieved by using the medium from DLD-Cl7 cells, but not from Dn1-3 cells. Similarly, pure TGF-β1 induced a dose-dependent growth inhibition (data not shown). Thus, like TGF-β, PTGF-β has growth inhibitory activity against human tumor cells.
To determine whether observed growth inhibition was largely attributable to PTGF-β, the conditioned medium was applied to an anti-FLAG M2 affinity gel (Sigma), and partially purified PTGF-β (Flag-tagged) was then used for cell proliferation assay. Growth inhibitory activity against both MvlLu and Du145 cells was significantly increased, with an IC50 of 0.8 μg/ml for Mv1Lu cells and 0.4 μg/ml for Du145 cells (data not shown). Furthermore, PTGF-β-induced growth inhibition of Mv1Lu cells was completely blocked by preincubation of conditioned medium with anti-PTGF-β antibody (data not shown). Thus, PTGF-β is the major protein in conditioned medium that confers growth inhibition.
Growth Suppression by PTGF-β Requires an Intact TGF-β/Smad4 Signaling Pathway.
Last, we determined whether PTGF-β-mediated growth suppression was via TGF-β signaling pathway. It is known that the TGF-β signaling pathway is mediated by type II (TβRII) and type I (TβRI) TGF-β receptors as well as receptor-activated Smad4 (12). Mutations in either of these genes would render cells resistant to TGF-β as well as PTGF-β if it acts through TGF-β signaling pathway. Indeed, neither TGF-β1 (data not shown) nor condition medium from PTGF-β-expressing cells (Fig. 5B) had any growth inhibitory activity against TβRII mutant RKO cells (34), TβRI mutant R1B/L17 cells (35), and Smad4 null MDA-MB468 cells (36). These results indicate that PTGF-β-induced growth inhibition requires an intact TGF-β receptor/Smad4 signaling pathway.
Discussion
The finding reported here indicates that PTGF-β, a TGF-β family member, is subjected to p53 activation and is a p53 target gene. This conclusion is supported by the following: (i) p53 binds to the p53 binding sites in the promoter of the PTGF-β gene; (ii) p53 transactivates the PTGF-β promoter in p53 dose- and p53 binding site-dependent manner; (iii) the p53 binding and transactivation of the PTGF-β promoter was enhanced by etoposide, a p53 activator; (iv) etoposide/p53-induced transactivation of the PTGF-β promoter can be largely blocked by a dominant negative p53 mutant; and (v) expression of endogenous PTGF-β was remarkably induced by etoposide only in p53-positive cells. The finding made a linkage between p53 tumor suppressor and a TGF-β family member and indicated a vertical signal transduction pathway from p53 to PTGF-β.
PTGF-β is a newly identified secretory protein (13–17). Although it only shares 25% overall sequence identity to TGF-β family members, PTGF-β has characteristics of TGF-β superfamily, including a signal peptide, a consensus RXXRA/S cleavage signal for processing to mature form, and seven conserved cysteine residues in carboxyl terminal (mature form) (13–17). It is known (for review, see ref. 12) that TGF-β-mediated signaling pathway is initiated by the binding of TGF-β to type II TGF-β receptor that leads to activation of type I receptor via phosphorylation. Activated type I receptor then phosphorylates receptor-regulated Smads (such as Smad3) that forms heterodimer or trimer (37) with common Smad4 and translocates into nucleus. In nucleus, heterotrimerized Smads interact with nuclear transcription factors such as FAST-1 (38) to regulate expression of target genes such as plasminogen activator inhibitor-type 1 (39). Other TGF-β target genes include p21/Waf1 (40), c-myc (41), p15 (42), and cdc25A (43). Smad4 is a Mad-related protein independently identified as a candidate tumor suppressor gene dpc4 (44). It plays a central role in mediating signaling pathways involving multiple TGF-β family ligands (45). We have shown here that, like TGF-β1, PTGF-β suppresses growth of cells that contain intact TβR/Smad4 signal, but not the cells that lack functional TGF-β receptors or Smad4. Although it is unknown whether PTGF-β binds to one of TGF-β receptors, it appears that PTGF-β acts through TGF-β signaling pathway. Thus, PTGF-β transmits p53 signal to TGF-β signal transduction pathway.
The promoter sequence of the PTGF-β gene has been recently cloned and characterized (15). There are several transcription factor binding sites, including SP1, AP-1, AP-4, and several GR elements. Indeed, PTGF-β was found to be regulated by androgens (16). We showed here that, in addition, PTGF-β is strongly subject to p53 up-regulation. This p53 activation is mediated through two p53 binding sites in the promoter of PTGF-β, and the second p53 binding site appears to play the major role. It is noteworthy that the second site is located downstream from the TATA box and the transcriptional start (15), but immediately upstream from the ORF (14, 15). It is well known that p53-induced transactivation of its downstream target genes is mediated by p53 binding sites located either in the promoter (e.g., Waf-1, Bax, MMP2) (3, 5, 24) or in the introns (e.g., Mdm2, Gadd45) (46, 47). We showed here an example that the p53 binding site fell downstream from the transcriptional start site but is still active in mediating p53 activation.
In addition to wild-type p53, one p53 mutant, p53-281G, but not other p53 mutants, also up-regulates the PTGF-β gene. p53-281G is a DNA contact mutant (48) that fails to bind to p53 consensus sequence (49). However, in addition to activating the PTGF-β gene, p53-281G also transactivates several other genes, including EGFR, MDR-1, PCNA, GPX, and c-myc (19, 50–53). The fact that p53-281G transactivates these gene promoters in a p53-binding site-independent manner indicates that transactivation must be mediated through other cis-element(s) in the promoter, as demonstrated recently in the c-myc gene (53). Alternatively, this p53 mutant-induced transactivation is merely a secondary effect. Of interest will be defining whether the transactivation of the PTGF-β promoter by p53-281G is mediated through the cis-element(s) in the promoter and, if so, mapping the element(s).
Biological significance of our finding is implicated in the role of p53 played in cancer and inflammation. First of all, it is well known that p53-induced growth suppression is mediated mainly by transactivation of p21/Waf-1, a universal inhibitor of cyclin-dependent kinase that induces G1 arrest (3, 54). Our data shown here indicate that, in addition, PTGF-β, a TGF-β family member, can also mediate this growth inhibitory activity. Importantly, unlike p21/Waf-1, which is a cellular protein and only functions in the cells that express it, PTGF-β is a secretory protein that can function in the cells secreting it (autocrine) as well as in their neighboring cells (paracrine). Thus, p53 antiproliferating signal can be transmitted from its responder cells to nonresponder cells through this “paracrine” effect. It has been previously reported that insulin-like growth factor binding protein 3 is a p53 target gene (55). However, p53-enhanced secretion of insulin-like growth factor binding protein 3 is much limited to inhibiting mitogenic signaling induced by insulin-like growth factor 1 (55). Thus, p53-induced growth inhibition on the neighboring cells can be mediated more widely by PTGF-β. It is most likely that PTGF-β is one of growth inhibitory cytokines secreted from cells after p53 activation by ionizing radiation (21).
Second, although p53 has been well documented as a tumor suppressor, the potential role of p53 in inflammation is not well understood. In fact, increased p53 expression has been found in number of inflammatory tissues, including (i) coronary restenosis lesions (56), (ii) atherosclerotic lesions (57, 58); (iii) pancreatitis (59); and (iv) ulcerative colitis and Crohn's disease (60). Importantly, increased p53 in inflammatory tissues, such as atherosclerotic lesions, was found to be functional active, as demonstrated by an induction of expression of Waf-1/p21 and Mdm2, two known p53 target genes (57, 58). Because PTGF-β is a potent p53 target gene (this report) and has anti-inflammatory activity (17), increased p53 expression in inflammatory tissues may reflect a tissue defense mechanism that triggers signaling pathway, leading to activation of PTGF-β and, finally, the inhibition of inflammation. Thus, p53-PTGF-β signaling pathway could play an important role in inflammation modulation, which is an interesting subject for future investigation.
In summary, we have identified PTGF-β, a member of TGF-β superfamily, as a p53 target gene. Because PTGF-β, as a secretory ligand, inhibits tumor cell growth and suppresses inflammation, the finding reveals a mechanism by which p53 acts as a tumor suppressor and implies a little-known but potentially important role of p53 in inflammation regulation.
Abbreviations
- TGF-β
type β transforming growth factor
- TβRI
type I TGF-β receptor
References
- 1.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2.el Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 3.el Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 5.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 6.Knudson C M, Tung K S, Tourtellotte W G, Brown G A, Korsmeyer S J. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 7.Wu G S, Burns T F, McDonald E R, III, Jiang W, Meng R, Krantz I D, Kao G, Gan D D, Zhou J Y, Muschel R, et al. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 8.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 9.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Cold Spring Harbor Symp Quant Biol. 1994;59:483–489. doi: 10.1101/sqb.1994.059.01.053. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, Wicha M, Leopold W R. Mol Carcinog. 1999;24:25–28. [PubMed] [Google Scholar]
- 11.Derynck R, Feng X H. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 12.Whitman M. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama Kobayashi M, Saeki M, Sekine S, Kato S. J Biochem (Tokyo) 1997;122:622–626. doi: 10.1093/oxfordjournals.jbchem.a021798. [DOI] [PubMed] [Google Scholar]
- 14.Hromas R, Hufford M, Sutton J, Xu D, Li Y, Lu L. Biochim Biophys Acta. 1997;1354:40–44. doi: 10.1016/s0167-4781(97)00122-x. [DOI] [PubMed] [Google Scholar]
- 15.Lawton L N, Bonaldo M F, Jelenc P C, Qiu L, Baumes S A, Marcelino R A, de Jesus G M, Wellington S, Knowles J A, Warburton D, et al. Gene. 1997;203:17–26. doi: 10.1016/s0378-1119(97)00485-x. [DOI] [PubMed] [Google Scholar]
- 16.Paralkar V M, Vail A L, Grasser W A, Brown T A, Xu H, Vukicevic S, Ke H Z, Qi H, Owen T A, Thompson D D. J Biol Chem. 1998;273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 17.Bootcov M R, Bauskin A R, Valenzuela S M, Moore A G, Bansal M, He X Y, Zhang H P, Donnellan M, Mahler S, Pryor K, et al. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Rea T, Bian J, Gray S, Sun Y. FEBS Lett. 1999;445:269–273. doi: 10.1016/s0014-5793(99)00136-2. [DOI] [PubMed] [Google Scholar]
- 19.Tan M, Li S, Swaroop M, Guan K, Oberley L W, Sun Y. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 20.Tan M, Heizmann C W, Guan K, Schafer B W, Sun Y. FEBS Lett. 1999;445:265–268. doi: 10.1016/s0014-5793(99)00135-0. [DOI] [PubMed] [Google Scholar]
- 21.Komarova E A, Diatchenko L, Rokhlin O W, Hill J E, Wang Z J, Krivokrysenko V I, Feinstein E, Gudkov A V. Oncogene. 1998;17:1089–1096. doi: 10.1038/sj.onc.1202303. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Bian J, Wang Y, Jacobs C. Oncogene. 1997;14:385–393. doi: 10.1038/sj.onc.1200834. [DOI] [PubMed] [Google Scholar]
- 23.Bian J, Jacobs C, Wang Y, Sun Y. Carcinogenesis. 1996;17:2559–2562. doi: 10.1093/carcin/17.12.2559. [DOI] [PubMed] [Google Scholar]
- 24.Bian J, Sun Y. Mol Cell Biol. 1997;17:6330–6338. doi: 10.1128/mcb.17.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Pommier Y, Colburn N H. Cancer Res. 1992;52:1907–1915. [PubMed] [Google Scholar]
- 26.Sun Y, Nakamura K, Hegamyer G, Dong Z, Colburn N. Mol Carcinog. 1993;8:49–57. doi: 10.1002/mc.2940080111. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Hildesheim A, Li H, Li Y, Chen J Y, Cheng Y J, Hayes R B, Rothman N, Bi W F, Cao Y, et al. Cancer Epidemiol Biomarkers Prev. 1995;4:261–267. [PubMed] [Google Scholar]
- 28.Duan H, Wang Y, Aviram M, Swaroop M, Loo J A, Bian J, Tian Y, Mueller T, Bisgaier C L, Sun Y. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian J, Sun Y. Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hupp T R, Meek D W, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 31.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Dong Z, Nakamura K, Colburn N H. FASEB J. 1993;7:944–950. doi: 10.1096/fasebj.7.10.8344492. [DOI] [PubMed] [Google Scholar]
- 33.Wallingford J B, Seufert D W, Virta V C, Vize P D. Curr Biol. 1997;7:747–757. doi: 10.1016/s0960-9822(06)00333-2. [DOI] [PubMed] [Google Scholar]
- 34.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 35.Attisano L, Carcamo J, Ventura F, Weis F M, Massague J, Wrana J L. Cell. 1993;75:671–680. doi: 10.1016/0092-8674(93)90488-c. [DOI] [PubMed] [Google Scholar]
- 36.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 37.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 39.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datto M B, Yu Y, Wang X F. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 41.Malliri A, Yeudall W A, Nikolic M, Crouch D H, Parkinson E K, Ozanne B. Cell Growth Differ. 1996;7:1291–1304. [PubMed] [Google Scholar]
- 42.Hannon G J, Beach D. Nature (London) 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 43.Iavarone A, Massague J. Nature (London) 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 44.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 45.Lagna G, Hata A, Hemmati Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 46.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kastan M B, Zhan Q, el Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 48.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Funk W D, Wright W E, Shay J W, Deisseroth A B. Oncogene. 1993;8:2555–2559. [PubMed] [Google Scholar]
- 50.Ludes Meyers J H, Subler M A, Shivakumar C V, Munoz R M, Jiang P, Bigger J E, Brown D R, Deb S P, Deb S. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin J, Teresky A K, Levine A J. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 52.Deb S, Jackson C T, Subler M A, Martin D W. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frazier M W, He X, Wang J, Gu Z, Cleveland J L, Zambetti G P. Mol Cell Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 55.Buckbinder L, Talbott R, Velasco Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Nature (London) 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 56.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 57.Ihling C, Menzel G, Wellens E, Monting J S, Schaefer H E, Zeiher A M. Arterioscler Thromb Vasc Biol. 1997;17:2218–2224. doi: 10.1161/01.atv.17.10.2218. [DOI] [PubMed] [Google Scholar]
- 58.Ihling C, Haendeler J, Menzel G, Hess R D, Fraedrich G, Schaefer H E, Zeiher A M. J Pathol. 1998;185:303–312. doi: 10.1002/(SICI)1096-9896(199807)185:3<303::AID-PATH106>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 59.Maacke H, Kessler A, Schmiegel W, Roeder C, Vogel I, Deppert W, Kalthoff H. Br J Cancer. 1997;75:1501–1504. doi: 10.1038/bjc.1997.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krishna M, Woda B, Savas L, Baker S, Banner B. Mod Pathol. 1995;8:654–657. [PubMed] [Google Scholar]