Abstract
The SQUAMOSA PROMOTER BINDING PROTEIN–LIKE (SPL) gene family is an SBP-box transcription family in Arabidopsis. While several physiological responses to SPL genes have been reported, their biological role remains elusive. Here, we use a combined analysis of expression correlation, the interactome, and promoter content to infer the biological role of the SPL genes in Arabidopsis thaliana. Analysis of the SPL-correlated gene network reveals multiple functions for SPL genes. Network analysis shows that SPL genes function by controlling other transcription factor families and have relatives with membrane protein transport activity. The interactome analysis of the correlation genes suggests that SPL genes also take part in metabolism of glucose, inorganic salts, and ATP production. Furthermore, the promoters of the correlated genes contain a core binding cis-element (GTAC). All of these analyses suggest that SPL genes have varied functions in Arabidopsis.
Keywords: Arabidopsis, SPL, co-expression, network, promoter
1. Introduction
Transcription factors (TFs) are DNA-binding proteins that regulate gene expression at the level of mRNA transcription. In plants, as in all living organisms, transcription factors are an important level of gene regulation. Many families of transcription factors have been identified in plants. Analysis of the Arabidopsis genome reveals 29 classes of transcription factors, 16 of which appear to be unique to plants [1]. One of these contains a DNA binding domain referred to as the SQUAMOSA PROMOTER BINDING PROTEIN (SBP) domain and is encoded by the SBP-box, a feature characteristic of the Arabidopsis SQUAMOSA PROMOTER BINDING PROTEIN–LIKE (SPL) gene family [2, 3].
The first SBP-domain proteins, which were isolated from snapdragon [4], showed in vitro binding to a sequence motif in the promoter region of the floral meristem identity gene SQUAMOSA [5]. Since then, SBP-box genes have been identified in many plants. For example, the Arabidopsis genome alone contains 16 SPL genes [3].
A number of physiological and biochemical studies have implicated the SBP-box gene in the regulation of plant tissue development. SPL3 may interact in vivo with promoters of the AP1 or CAL MADS-box genes of the SQUAMOSA family to act as a positive transcriptional regulator modulating floral development [2]. An SPL gene is involved in the early stages of microsporogenesis and megasporogenesis [6] and as well as the development of normal plant architecture [7]. This family of genes was also associated with maize kernel development [8] and tomato fruit ripeness [9]. Very recently, a study showed that the SBP-box genes SPL9 and SPL15, which are regulated by microRNAs, control shoot maturation in Arabidopsis [10].
Despite an increasing body of physiological and biochemical data, the biological role of the SBP-box transcription family remains elusive. In order to infer a biological role for it, we took advantage of the large repositories of Arabidopsis thaliana microarray data to study the co-expression gene with SPL. We identified 112 genes whose expression most highly correlated with them in response to various treatments and during development. Furthermore, we analyzed the promoters of these genes in order to identify the regulatory motifs. The results of our study predict that the SBP-box transcription family is involved in plant tissue development, responses to abiotic and biotic stresses, and the activation of other transcription factors and membrane proteins. Furthermore, we demonstrate how computational analyses linking expression data with regulatory information from studies of promoter elements can provide novel insights into the function of a transcription family whose function is unclear.
2. Materials and Methods
2.1. Identification of correlated genes and GO analysis
A total of 1,779 Affymetrix raw data Cel files were downloaded from the AtGenExpress and GEO sites [11–13]. All of them were derived from the Affymetrix ATH1 gene GeneChip microarray for Arabidopsis, and the samples contained at least two replicate chips. The required probe set-to-locus mappings for the ATH chip were obtained from TAIR (ftp://ftp.arabidopsis.org/home/tair/Microarrays/Affymetrix, version 2-5-2007).
Normalization of the raw data Cel files was performed in R using the MAS5 algorithms, which are implemented in the affy package available from the BioConductor project [14, 15]. R scripts were used to calculate non-parametric correlation coefficients (Spearman’s rho) between the expression of SPL genes and the expression of each of the ~22,000 genes represented on the Affymetrix array used to generate this dataset. We ranked the genes according to the correlation coefficients and reported genes that were most positively correlated with SPL gene expression. The p-values were calculated using bivariate normal distribution, with p representing the probability of observing an equal or larger positive or negative correlation by chance. For details of the method, see the article by Wei [16].
To characterize the correlated genes, the web-based ‘Classification SuperViewer’ program (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) was used to search for differential distributions of gene ontology (GO) and biological terms within the correlation genes.
2.2. Arabidopsis interactome network and clustering analysis
The Arabidopsis interactome network was built by the AtPID. The AtPID (Arabidopsis thaliana Protein Interactome Database) represents a centralized platform for depicting and integrating information pertaining to protein-protein interaction networks, domain architecture, ortholog information, and GO annotation in the Arabidopsis thaliana proteome. We mapped the correlated genes to the network and also selected the first neighbors of the genes. To identify the function of SPL-correlated proteins in network clusters, the latest versions of the Biological Network Gene Ontology (BiNGO) [17] and GOlorize tools [18] were used for the statistical evaluation of groups of proteins. This evaluation was carried out using existing annotations of the Gene Ontology Consortium (http://www.geneontology.org).
2.3. Promoter analysis
Applications in the web-based promoter analysis program Promomer [19] were used to analyze the promoters (−1 kb upstream of the predicted translation start site) of the 112 genes showing the strongest correlations to SPL genes. The Promomer program aims to provide a user-friendly, web-based interface to accomplish two goals: (1) to identify statistically over-represented elements in a gene or a group of genes in Arabidopsis using the enumerative method, and (2) to find the number and position of occurrences of an element in genes across the Arabidopsis genome or in a subgroup of genes.
3. Results
3.1. Expression correlation and gene ontology (GO) analyses
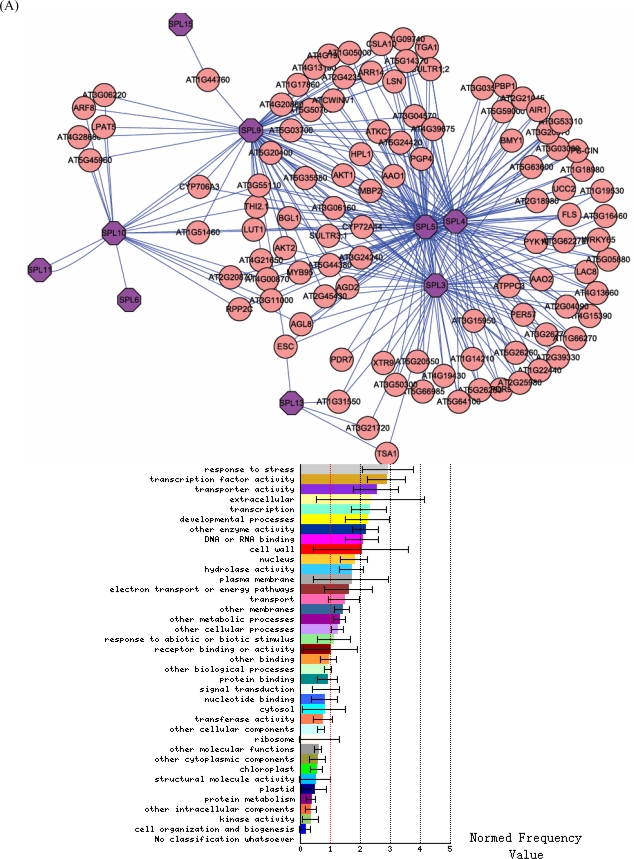
In the first step of the analyses, we extracted and ranked the 112 genes whose expression most tightly correlates with that of SPL genes (Table 1). We took advantage of a visualization tool called Cytoscape to view expression data and analysis results. Using the available data regarding correlations between different Arabidopsis genes, we constructed a network composed of the SPL genes and their co-expressed genes. This network contains 112 Arabidopsis genes (nodes) and 294 correlations (edges) (Figure 1A). Three interesting facts are observed from the co-expression network. First, some SPL genes are absent from the network, including SPL1, SPL2, SPL6, SPL7, SPL8, SPL12, SPL14 and SPL16. This reflects the current lack of knowledge regarding correlations between these SPL genes and other genes. Second, many genes correlate with SPL3, SPL4, SPL5, and SPL9; some genes correlate with SPL10 and SPL13; while SPL11 and SPL15 have only one co-expression gene. Third, within the co-expression network, there is a region comprising four SPL genes that contain 20 Arabidopsis genes (nodes) and 79 correlations (edges) (Figure 1B).
Table 1.
List of genes that are expression correlated with SPL in our analysis.
| AGI ID | Annotation | AGI ID | Annotation |
|---|---|---|---|
| AT5G20960 | ALDEHYDE OXIDASE 1 | AT3G18850 | LPAT5__LPAT5 |
| AT1G52030 | MYROSINASE-BINDING PROTEIN 2 | AT4G00870 | basic helix-loop-helix (bHLH) family protein |
| AT2G26650 | ARABIDOPSIS K TRANSPORTER 1 | AT5G37020 | ARF8 |
| AT3G55110 | ABC transporter family protein | AT3G09260 | phosphate starvation-response 3.1 oxidoreductase, 2OG-Fe(II) oxygenase family |
| AT4G15440 | HYDROPEROXIDE LYASE 1 | AT5G20400 | protein |
| AT3G06160 | transcriptional factor B3 family protein | AT4G19430 | unknown protein |
| AT5G44380 | FAD-binding domain-containing protein | AT5G17820 | peroxidase 57 (PER57) (P57) (PRXR10) |
| AT5G60910 | FUL | AT3G06220 | DNA binding / transcription factor |
| AT2G42200 | SPL9 | AT5G45960 | GDSL-motif lipase/hydrolase family protein |
| AT2G45430 | DNA-binding protein-related | AT4G21650 | subtilase family protein UCC2__UCC2 (UCLACYANIN 2); copper |
| AT3G04570 | DNA-binding protein-related | AT2G44790 | ion binding |
| AT4G32650 | ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1 | AT3G28500 | 60S acidic ribosomal protein P2 (RPP2C) |
| AT3G14680 | CYP72A14 | AT4G12550 | Auxin-Induced in Root cultures 1 |
| AT1G20900 | ESCAROLA | AT4G20860 | FAD-binding domain-containing protein |
| AT1G60680 | ARF-GAP DOMAIN 2 | AT4G28680 | tyrosine decarboxylase, putative |
| AT3G53130 | LUTEIN DEFICIENT 1 | AT5G63600 | flavonol synthase, putative |
| AT3G51895 | SULFATE TRANSPORTER 1 | AT2G01760 | ARABIDOPSIS RESPONSE REGULATOR 14 |
| AT2G47000 | P-GLYCOPROTEIN 4 | AT5G59000 | zinc finger (C3HC4-type RING finger) family protein |
| AT4G39675 | unknown protein | AT5G03700 | PAN domain-containing protein |
| AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | AT2G21045 | similar to unknown protein |
| AT3G24240 | leucine-rich repeat transmembrane protein kinase, putative | AT5G50760 | auxin-responsive family protein |
| AT1G72260 | toxin receptor binding | AT3G15950 | (TSA1-LIKE); unknown protein |
| AT3G15270 | SPL5 | AT1G17860 | trypsin and protease inhibitor family protein |
| AT2G33810 | SPL3 | AT2G20870 | cell wall protein precursor, putative |
| AT1G53160 | SPL4 | AT5G20550 | oxidoreductase |
| AT5G05880 | UDP-glucosyl transferase family protein | AT1G05000 | tyrosine specific protein phosphatase family protein |
| AT1G24070 | Cellulose synthase-like A10 | AT3G43600 | AAO2 |
| AT3G20370 | MATH domain-containing protein | AT4G22200 | Arabidopsis K+ transporter 2 |
| AT5G26280 | MATH domain-containing protein | AT3G21720 | isocitrate lyase, putative |
| AT1G51460 | ABC transporter family protein | AT1G52410 | TSK-ASSOCIATING PROTEIN 1 |
| AT3G16460 | jacalin lectin family protein | AT4G19880 | similar to unknown protein |
| AT3G53480 | PLEIOTROPIC DRUG RESISTANCE 9 | AT2G18980 | peroxidase, putative |
| AT5G44620 | CYP706A3 | AT1G18980 | germin-like protein, putative |
| AT5G01040 | laccase 8 | AT4G13180 | short-chain dehydrogenase |
| AT5G65210 | TGA1 | AT5G63590 | Flavonol synthase |
| AT1G52400 | BGL1 | AT3G03090 | ATVGT1__sugar transporter family protein |
| AT5G14370 | similar to CIL | AT3G25820 | TERPENE SYNTHASE-LIKE SEQUENCE-1,8-CINEOLE |
| AT1G66270 | beta-glucosidase | AT4G15210 | BETA-AMYLASE |
| AT5G02030 | HB-6_RPL_LSN_BLH9_BLR_PNY_RPL_VAN__LSN | AT2G42350 | zinc finger family protein |
| AT4G13660 | pinoresinol-lariciresinol reductase, putative | AT3G50300 | transferase family protein |
| AT4G15390 | transferase family protein | AT3G03520 | phosphoesterase family protein |
| AT5G64100 | peroxidase, putative | AT1G15210 | PLEIOTROPIC DRUG RESISTANCE 7 |
| AT1G78000 | SULFATE TRANSPORTER 1;2 | AT3G16420 | PYK10-BINDING PROTEIN 1 |
| AT1G29280 | WRKY DNA-binding protein 65 | AT3G13790 | ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
| AT2G39330 | jacalin lectin family protein | AT3G62270 | anion exchange family protein |
| AT1G09740 | ethylene-responsive protein, putative | AT4G25820 | XYLOGLUCAN ENDOTRANSGLYCOSYLASE 9 |
| AT1G22440 | alcohol dehydrogenase, putative | AT1G27370 | SPL10 |
| AT5G26260 | MATH domain-containing protein | AT5G66985 | unknown protein |
| AT2G25980 | jacalin lectin family protein | AT3G26770 | short-chain dehydrogenase |
| AT1G31550 | carboxylic ester hydrolase/lipase | AT3G11000 | similar to kelch repeat-containing protein |
| AT1G19530 | unknown protein | AT3G14940 | PHOSPHOENOLPYRUVATE CARBOXYLASE 3 |
| AT1G74430 | AtMYB95 | AT1G44760 | universal stress protein (USP) family protein |
| AT2G04090 | MATE efflux family protein | AT1G14210 | ribonuclease T2 family protein |
| AT5G35580 | kinase | AT1G27360 | SPL11 |
| AT3G53310 | transcriptional factor B3 family protein | AT1G69170 | SPL6 |
| AT5G50570 | SPL13 | AT3G57920 | SPL15 |
Figure 1.
Co-expression network and GO analysis of SPL genes. (A) A network composed of SPL and correlated genes. GO analysis shows that the network is involved in extensive functions. (B) A complex within the co-expression network and its function.
In order to identify a functional role for SPL genes, the correlated genes were analyzed in a web-based gene ontology (GO) analysis tool to identify any bias in GO functional annotation terms in the correlated genes compared to the remainder of the A. thaliana genome (Supplementary Table 1). When GO clustering was applied to the 112 co-expression genes, some GO biological categories were retrieved. Most of these related to stress response, transporter activity, and transcription factor activity (Figure 1A). Other biological processes identified were associated with extracellular activities, development processes, and enzyme activity (Figure 1A). Thus, given previous work [3] suggesting that SPL genes play a role in controlling plant development, we can infer that SPL genes may not only do this but also take part in many other biological processes. In the co-expression network, a complex related to development and the plasma membrane was found (Figure 1B). We presume that the complex is very important for SPL gene function of development (Figure 1B), and SPL proteins may form heterodimers or multimers to carry out their functions.
3.2. Interactome analysis
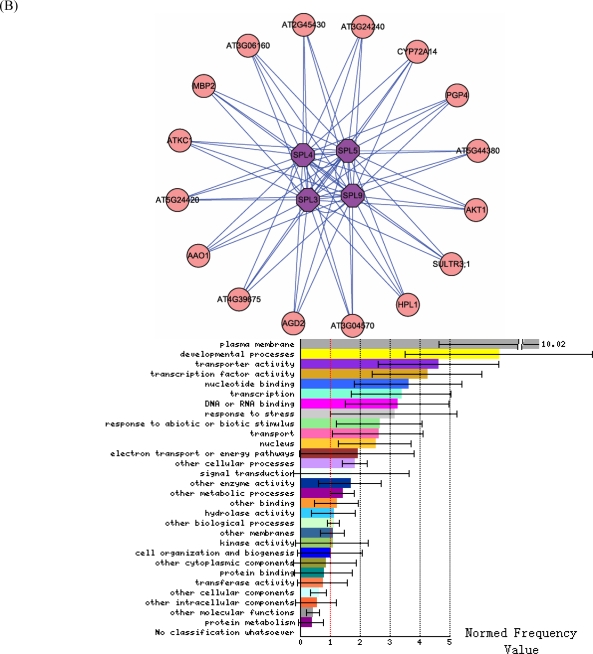
To evaluate functional associations between co-expressed proteins, we mapped the genes whose expression was correlated with SPL genes using the Arabidopsis thaliana interactome network (Figure 2A). Since pairs of interacting proteins are more likely to contain similar GO annotations than pairs of unrelated proteins, annotations of neighbor proteins in the network can be used to make predictions of functions. In particular, the GO annotations allow functional assignment of uncharacterized gene products, such when At3g62270 was identified in a large-scale interactome mapping study of anion transport (Figure 2B).
Figure 2.
Network analysis of the Arabidopsis interactome showing direct interactions between co-expressed gene products and their first neighbors. (A) Direct interactions between correlated gene products. (B) Direct interactions between correlated gene products and their first neighbors. Some sub-networks show enrichment in GO terms (functional modules). Arrows indicate the SPL-correlated gene nodes present in these functional modules.
Cluster analysis of this network suggested the molecular mechanisms underlying the function of SPL genes. Analysis of the connected sub-graphs and their GO terms identified functional modules enriched for carbohydrate transport, electron transport, anion transport, flavonoid biosynthetic processes, diaminopimelate metabolic processes, response to hypoxia and stress, and root development (Figure 2B). Many genes and proteins co-expressed with SPL genes are located in these modules, which supports their role in a diversity of physiology processes. These observations support the theory that the network modeled here constitutes a framework that can guide in-depth experimental study of genes and proteins related to SPL gene function in Arabidopsis.
3.3. Promoter analysis
The shared expression profiles of the 112 correlated genes in response to development and stress suggests that these genes are under the same regulatory control and are thus likely to share cis-elements in their promoter regions. Since these genes are co-expressed with SPL genes, we hypothesized that they might be directly regulated by SPL genes. The cis-element of the SBP-box transcription factor is unclear. To reveal which DNA motif can bind to SPL genes, we analyzed the promoter regions of these genes 1 kb upstream of the predicted transcription start site. We also analyzed aspects of shared transcriptional activation sites on these genes for the presence of known plant cis-elements.
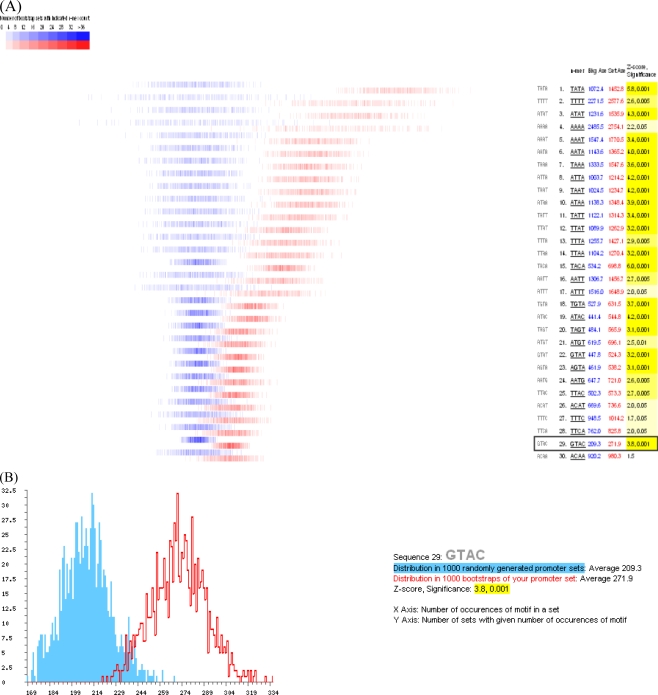
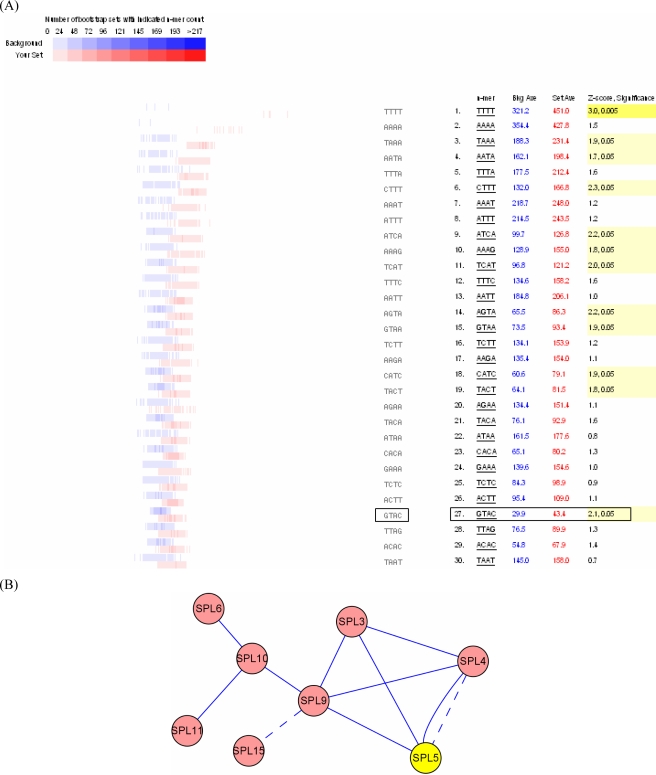
Based on previous research [3, 20–22], we hypothesized that the GTAC motif was the cis-element common to the SBP-box genes. Indeed, our analysis showed the GTAC motif to be a statistically over-represented element (Figure 3). Two distributions were created from 1,000 bootstrapped sets, each containing the same number of genes. The first was obtained by sampling for the frequency of occurrence of an element from the correlated genes cluster promoter set bootstrapped 1,000 times; the second was obtained from the same bootstrapping process carried out on 1,000 whole genome promoter data sets containing the same number of sequences as the cluster set. The distribution of occurrence of a given element in both data sets was then obtained and plotted [23] and significant differences were highlighted (Supplementary Figure 1). For comparison, significant tetramers whose expression is unrelated to that of SPL genes are also shown.
Figure 3.
Promoter analysis of SPL co-expression genes. (A) The distribution of the occurrence of the GTAC motif in 1000 sets of SPL-correlated genes promoters randomly selected from the Arabidopsis genome. (B) Distribution in 1000 randomly generated promoter sets: Average 209.3. Distribution in 1000 bootstraps of our promoter set: Average 271.9. Z-score, Significance: 3.8,0.01.
The analysis (Supplementary Table 2) indicates that the invariant core GTAC motif is present in 100/112 of our correlated genes a total of 271.9 times, which is higher than the average of 209.3 times across 1,000 randomly-generated promoter sets (Figure 3). The promoter analysis indicates that multiple copies of the SBP-box elements are present in a high percentage of SPL-correlated genes. The core GTAC motifs are significantly enriched compared to expected frequencies, suggesting that they are important regulatory elements in these co-expressed genes.
4. Discussion
Although transcription factors are generally low expressed, microarray has been widely used for transcription factors expression research [24–26]. With recent interest in co-expression, transcription factors co-expression has emerged as a novel holistic approach for their function analysis. For example, MADS-box proteins form specific homo- and heterodimers and even higher order complexes to conduct their functions [27, 28]. Specific interactions among MADS proteins require that they are present in the same cells and tissues under the same developmental stages, and correspondingly it has been shown that transcripts with overlapping expression patterns are preferred as protein interaction partners [29, 30]. Roosa et al. used co-expression method identify correlation genes with MADS in Gerbera hybrida, they found that MADS proteins not only might correlate with themselves but also correlated with MYB factors [31]. In our analysis, we used microarray data to classify co-expression network of SPL genes. We think the network may help us to infer the function of SPL genes.
4.1. SPL genes may play a key role in the stress response
Transcriptional control of the expression of stress-responsive genes is a crucial part of plant responses to a range of abiotic and biotic stresses. Research carried out in recent years [23] has been productive in identifying transcription factors that are important for regulating plant responses to these stresses. SPL genes have not been shown to be affected by stress in Arabidopsis. Nevertheless, our GO analysis revealed that some genes correlated with SPL genes were involved in the stress response (Supplementary Table 1). These genes include P57 (GO: 0006979, response to oxidative stress), AKT1 (GO: 0009651, response to salt stress), At3g16460 (GO: 0009409, response to cold), ATPPC3 (GO: 0009414, response to water deprivation), and ATCWINV1 (GO: 0009611, response to wounding).
In addition, two transcription factors (TGA1 and WRKY65) that belong to the bZIP and WRKY families, respectively, are co-expressed with SPL genes. bZIPs are a large family of transcription factors in plants, and 75 members are present in Arabidopsis [32]. One class of bZIP proteins that is linked to stress responses comprises the TGA/octopine synthase (ocs)-element-binding factor (OBF) proteins. These bind to the activation sequence-1 (as-1)/ocs element, which regulates the expression of some stress-responsive genes such as PR-1 and GLUTATHIONE S-TRANSFERASE6 (GST6) [33, 34]. In Arabidopsis, seven members of the TGA/OBF family play a role in plant defense, xenobiotic stress responses, and development. WRKY proteins are a novel family of transcription factors that are unique to plants and that form a large, 74-member family in Arabidopsis. WRKY proteins contain either one or two WRKY domains, which is 60-residue region that contains the amino acid sequence WRKYGQK and a motif similar to zinc fingers. Certain WRKY family members show enhanced expression or DNA-binding activity following induction by a range of pathogens, defense signals, and wounding [35]. Since the two transcription families are very important for plant response to stress, and because SPL genes can control some stress-responsive genes, we speculate that SPL genes also play a key role in the stress response.
4.2. A complex control network linking SPL genes and other transcription families
Our GO analysis shows transcription activity to be an important function for co-expressed genes. SPL genes can activate other transcription factor families, including B3, bZIP, WRKY, MYB, bHLH, and MADs-box. We believe that SPL genes function by controlling the expression of these transcription families. For example, the expression of AGL8 (FUL), a MADs-box gene, correlates with SPL gene expression. In another study, the MADs-box gene AP1 was also found to be activated by SBP-box genes [2]. The proteins AP1 and FUL may serve as hubs between the flower induction pathway, which is comprised of interacting proteins such as SVP, SOC1, and AGL24, and the floral organ identity proteins. Both AP1 and FUL have dual functions in floral meristem identity (early function) and floral organ determination (late function) [36, 37]. This is consistent with the fact that the flowering proteins and floral homeotic proteins come together to form dimers.
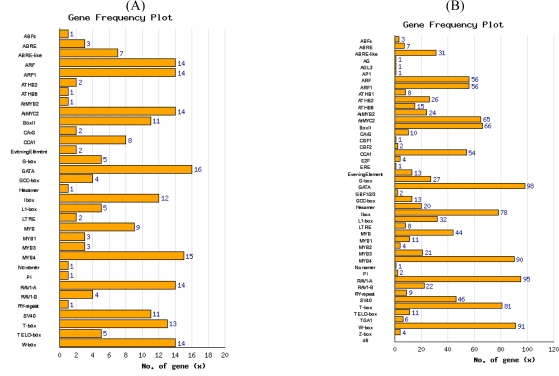
Analysis of cis-elements suggests an even more complex picture: (1) SPL genes may be activated by other transcription families (Figure 4A), and (2) the 112 genes correlated to SPL genes in our analysis may be controlled by other transcription families (Fig 4B). We therefore hypothesize a complex control network linking SPL genes with other transcription families.
Figure 4.
Known cis-element analysis of (A) SPL genes and (B) genes co-expressed with SPL genes.
4.3. SPL genes and integral membrane proteins
An integral membrane protein (IMP) is a protein molecule (or assembly of proteins) that is permanently attached to the biological membrane. In the co-expression network of SPL genes, some IMP genes correlate with SPL genes. These include ATK1 and ATK2 (ARABIDOPSIS K TRANSPORTER), ATVGT1 (sugar transporter family protein), PGP4 (P-GLYCOPROTEIN 4), SULTR1;2 (SULFATE TRANSPORTER 1;2), At3g55110 (ABC transporter family protein), and some genes encoding ligands of membrane proteins. The IMPs encoded by these genes include transporters, channels, receptors, enzymes, structural membrane-anchoring domains, proteins involved in accumulation and transduction of energy and sugar, and proteins responsible for the stress response. Our analysis of genes correlated with SPL genes leads us to speculate that SPL genes may participate in development, stress responses, and other biological processes by controlling the expression of IMPs. In addition, some SPL proteins have a transmembrane domain, which suggest that SPL may have interaction with IMPs.
4.4. SPL genes may regulate themselves
Our analysis indicates that the cis-element shared by SPL genes, the GTAC motif, is necessary for binding of SPL proteins to DNA. In the co-expression network, the expression of some SPL genes correlates with that of other SPL genes; these genes may form a complex within the network (Figure 1B). Therefore, we propose that SPL genes regulate themselves. Using promoter analysis, we found the GTAC motif to be present in the 1 kb upstream region in 13/16 of SPL genes a total of 56 times (Supplementary Table 3), which was significantly higher than the occurrence in tetramer sets of randomly selected promoters (Figure 5A). In the AtPID database [38], we found that SPL9 can interact with SPL15, and SPL4 can interact with SPL5. SPL genes may therefore regulate themselves using a feedback loop based on protein interactions and transcriptional regulation (Figure 5B).
Figure 5.
Promoter and network analysis of SPL genes. (A) The distribution of the occurrence of the GTAC motif in 1000 sets of SPL genes promoters randomly selected from the Arabidopsis genome. Distribution in 1,000 randomly generated promoter sets: Average 29.7. Distribution in 1,000 bootstraps of our promoter set: Average 42.9. Z-score, Significance: 2.2,0.05. (B) A putative network of SPL genes (a solid line indicates co-expression, and a dashed line indicates protein interactions).
4.5. SPL genes have varied functions
The SBP-box gene family belongs to a group of plant-specific zinc finger protein genes that encodes plant-specific transcription factors. The proteins encoded by SBP-box genes bind specifically to promoters of the floral meristem identity gene SQUAMOSA and its orthologous genes [2, 3, 6]. Except for the presence of a conserved SBP-box/domain, the SPL genes vary substantially in their genomic organization, transcript size, and the size and sequences of their encoded proteins. The position of the SBP domain within the protein also varies [39]. The different gene structures and the divergence of amino acid sequences among different SPL genes provide us with some hints that SBP transcription factors may have a variety of physiological functions. To date, several important and divergent biological processes regulated by SBP-box genes have been reported. These include flower and fruit development [2, 4, 8, 9], architecture formation [40], sporogenesis [6], response to copper and fungal toxin [7, 41], and control of GA level [42].
Our co-expression analysis has revealed that SPL genes may play a role in the stress response. Network analysis suggests that SPL genes function by controlling other transcription factor families, and that they have relatives encoding proteins involved in membrane protein transport. The interactome of the correlated genes has shown that SPL genes may also take part in glucose, inorganic salt, and ATP production. Notably, some SPL genes are not detected in our co-expression analysis. Among them, SPL8 can affect pollen sac development [6], and SPL14 plays a role in plant development and sensitivity to fumonisin B1 [7]. All of the available evidence suggests that SPL genes have varied functions in Arabidopsis.
5. Conclusion
The SPL gene family belongs to a group of plant-specific zinc finger protein genes that encodes plant-specific transcription factors. The expression of SPL genes are significantly correlated with that of genes involved in the defense response pathway in response to various biotic and abiotic stress. The expression of SPL genes are correlated with other transcription factors gene and some integral membrane proteins. Additionally, like the MADS-box genes, some SPL genes are correlated with themselves. Furthermore, the promoters of the correlated genes contain a core binding cis-element (GTAC). All of these analyses suggest that SPL genes have varied functions in Arabidopsis.
Appendix: Supplementary Material
Supplementary material is available from http://dx.doi.org/10.3390/ijms10010116
Acknowledgments
This study was supported by the National Natural Science Foundation of China (NO. 30600044, 30771463) and Natural Science Foundation of Chongqing of China (No. cstc, 2007BB1197, 2007BB1198).
References
- 1.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 2.Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12:367–77. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- 3.Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/s0378-1119(99)00308-x. [DOI] [PubMed] [Google Scholar]
- 4.Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- 5.Huijser P, Klein J, Lonnig WE, Meijer H, Saedler H, Sommer H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992;11:1239–49. doi: 10.1002/j.1460-2075.1992.tb05168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15:1009–1019. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone JM, Liang X, Nekl ER, Stiers JJ. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41:744–754. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008;67:183–95. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin LX, Beyer RP, Hudson FN, Linford NJ, Morris DE, Kerr KF. Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics. 2006;7:23. doi: 10.1186/1471-2105-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei H, Persson S, Mehta T, Srinivasasainagendra V, Chen L, Page GP, Somerville C, Loraine A. Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 2006;142:762–774. doi: 10.1104/pp.106.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 18.Garcia O, Saveanu C, Cline M, Fromont-Racine M, Jacquier A, Schwikowski B, Aittokallio T. GOlorize: A Cytoscape plug-in for network visualization with Gene Ontology-based layout and coloring. Bioinformatics. 2007;23:394–396. doi: 10.1093/bioinformatics/btl605. [DOI] [PubMed] [Google Scholar]
- 19.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 20.Kropat J, Tottey S, Birkenbihl RP, Depege N, Huijser P, Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Liang X, Nazarenus TJ, Stone JM. Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry. 2008;47:3645–3653. doi: 10.1021/bi701431y. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 29.Immink RG, Ferrario S, Busscher-Lange J, Kooiker M, Busscher M, Angenent GC. Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics. 2003;268:598–606. doi: 10.1007/s00438-002-0781-3. [DOI] [PubMed] [Google Scholar]
- 30.de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laitinen RA, Broholm S, Albert VA, Teeri TH, Elomaa P. Patterns of MADS-box gene expression mark flower-type development in Gerbera hybrida (Asteraceae) BMC Plant Biol. 2006;6:11. doi: 10.1186/1471-2229-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 33.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Singh KB. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 35.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 36.Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–237. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 37.Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 38.Cui J, Li P, Li G, Xu F, Zhao C, Li Y, Yang Z, Wang G, Yu Q, Shi T. AtPID: Arabidopsis thaliana protein interactome database--an integrative platform for plant systems biology. Nucleic Acids Res. 2008;36:D999–1008. doi: 10.1093/nar/gkm844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M. The liguleless-1 gene acts tissue specifically in maize leaf development. Dev. Biol. 1990;141:220–232. doi: 10.1016/0012-1606(90)90117-2. [DOI] [PubMed] [Google Scholar]
- 41.Eriksson M, Moseley JL, Tottey S, Del Campo JA, Quinn J, Kim Y, Merchant S. Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics. 2004;168:795–807. doi: 10.1534/genetics.104.030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Rong H, Pilbeam D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 2007;58:2329–2338. doi: 10.1093/jxb/erm114. [DOI] [PubMed] [Google Scholar]