Abstract
Possession of a variant Cytochrome P450 2C9 (CYP2C9) genotype has been associated with a higher risk of hemorrhagic complications among warfarin users. [1; 2] Although the influence of the common variant alleles (CYP2C9*2, CYP2C9*3) on warfarin response is well documented that of other rare defective alleles, CYP2C9*5, the null allele CYP2C9*6 and CYP2C9*11 found in African-Americans [3; 4] has not. The presence of these alleles may represent the higher genomic sequence diversity in populations of African descent. Herein we describe discovery of a new putative deleterious CYP2C9 polymorphism identified in an African American participant from an ongoing prospective study during routine testing.[5] Analysis of the patient’s genotype identified a new CYP2C9 polymorphism G1078A coding for a D360N in the coding region of exon 7 one codon downstream from the I359L coding change seen in CYP2C9*3.
JJ, a 63-year old African American man (68″ 198 lbs) with a history of coronary artery disease, hyperlipidemia, hypertension, diabetes mellitus, anemia, osteoarthritis, glaucoma, and prostate cancer was initiated on warfarin for atrial fibrillation. The patient is a non-smoker, does not consume alcohol and consumes two-three servings of vitamin K rich foods per week. Sixteen months after initiation of warfarin, he was admitted with report of dark stools over the preceding four days. Medications on admission included: sotolol 120mg twice daily, digoxin 0.125 mg/day, atorvastatin 10 mg/day, warfarin 42 mg/week, clopidogrel 75 mg/day, isosorbide mononitrate 30mg/day, Oscal-D 500mg bid, docusate 100mg as needed, and acetaminophen 500mg as needed. On admission his hematocrit was 30gm/dl (prior 40gm/dl), INR 2.5, BUN 28 and creatinine 0.9 and heme-positive stool. Colonoscopy revealed old clotted blood in the colon. Endoscopy did not reveal any tumor or any actively bleeding loci. During the course of hospitalization, the patient received five units of blood and clopidogrel and warfarin were discontinued and ferrous sulfate 325mg twice daily and pantoprazole 40mg twice daily added to his medication regimen. The hemorrhagic event was adjudicated by an independent expert without knowledge of the patients’ genotype as dictated by the study protocol. [2]
One week after discharge the patient reported no further signs of gastrointestinal bleeding (hematocrit 40gm/dl, heme-negative stool). In light of his medical history and incessant paroxysmal arrhythmia warfarin therapy was re-started (37.5 mg/week) with an INR range of 2–2.5. Patient was maintained on warfarin for the following nine months with no further problems (hematocrit 40 to 42gm/dl).
Analysis of the patient’s genotype was determined without knowledge of the case history. CYP2C9 and VKORC1-1173C/T (rs9934438) genotyping methodology has been detailed previously. [2; 5] All exons and intron exon junctions of this individual were amplified by PCR using Amplitaq Gold (Applied Biosystems, Foster, CA). The amplification products were therefore sequenced directly in both directions using Big Dye Terminator Cyler Sequencing Kit (Applied Biosystems) and analyzed on an ABI 3100 Genetic Analyzer.
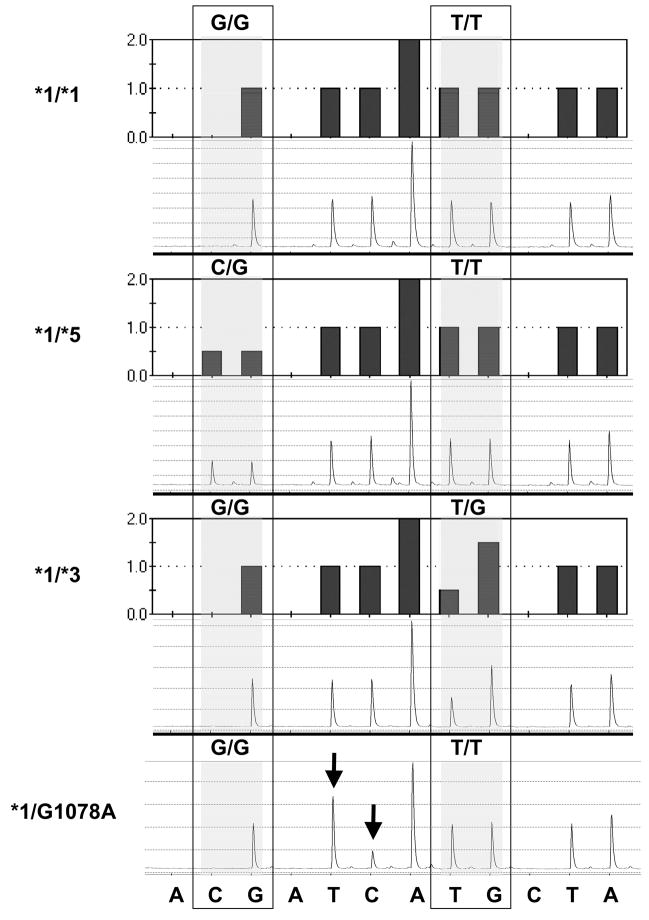
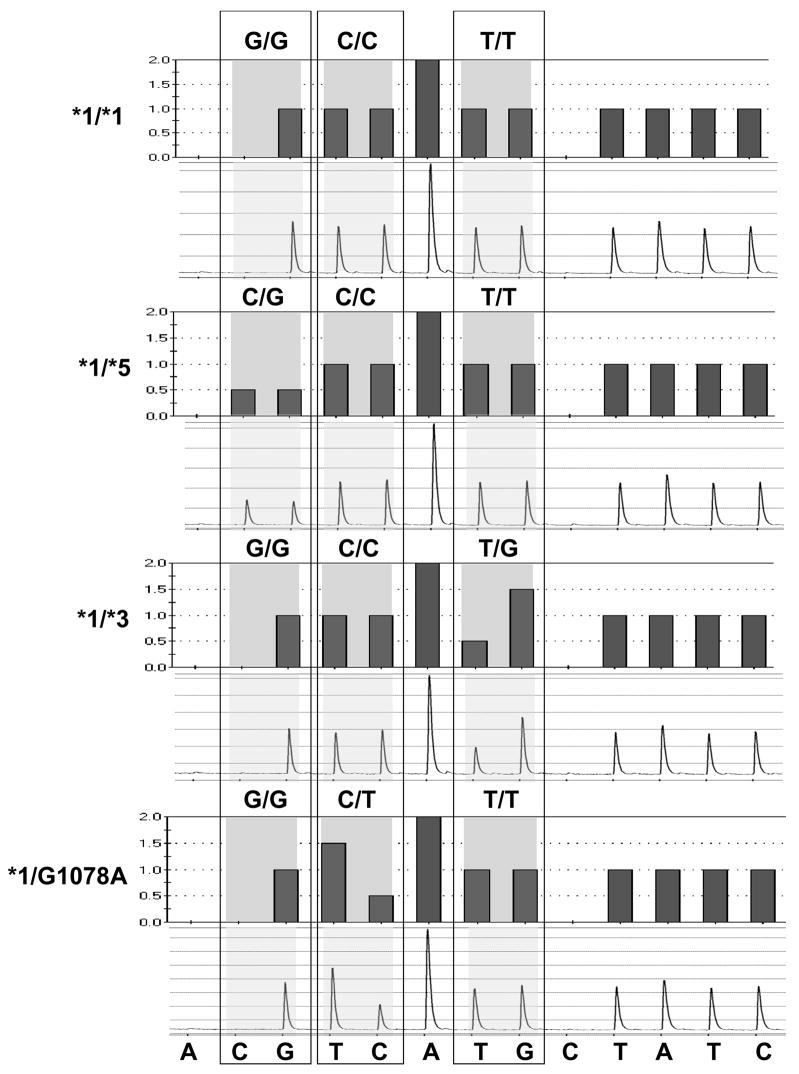
The patient was homozygous for the wild-type allele for the VKORC1 polymorphism (VKORC1-1173C/C, rs9934438) assessed. During the pyrosequencing tests for the CYP2C9*3 and CYP2C9*5 alleles an abnormal pyrogram was observed showing an extra T peak at 1078 and the disappearance of a C on the antisense strand of exon 7 in one African-American patient (Figure 1, Panel 4).[6] This change is between the A1075C expected in individuals carrying the CYP2C9*3 allele and the C1080G expected in individuals carrying the CYP2C9*5 allele. DNA sequencing confirmed the presence of a heterozygous G1078A change in the cDNA (g.42617) coding for D360N in exon 7 of CYP2C9 consistent with the abnormal pyrogram, indicating the patient was heterozygous for CYP2C9*1/D360N. The sequence of the coding strand of part of exon 7 for this patient and the wild-type CYP2C9 allele are compared in Figure 2. Since this patient carried a coding SNP indicative of a new allele, all 9 exons were sequenced to better predict the complete haplotype of the new allele. The patient was also heterozygous for three other mutations. In exon 1, a C8A change codes for a S3Y change at amino acid three (The genomic numbering is consistent with the access number of NT_029381.4 for the CYP2C9 gene). A second new noncoding mutation was detected in intron 1 (IVS1 +83) at g.215 of the CYP2C9 genome. A third known noncoding mutation was noted at IVS2 +73 (g.3411). The third mutation has previously been found in predicted haplotypes E and L (which differ by an additional upstream mutation in haplotype L), both noncoding variants of the CYP2C9*1 allele which were reported by Blaisdell et al [7] in African-Americans. Due to the length of the 2C9 gene and the fact that the changes were all heterozygous and identified in only one patient, the haplotype cannot be definitely determined from the present information. However, it is reasonable to infer that the new CYP2C9 allele may also contain the S3Y change, the new change in intron 1 (IVS2+83), and the coding change G1078A in the cDNA (genomic position g.42617) resulting in a D360N substitution are part of one haplotype, while the previously reported IVS2+73 probably represents haplotype E or L of the CYP2C9*1 allele on the other chromosome. Due to this uncertainty, however, the Human Cytochrome P450 Allele Nomenclature committee ruled that the haplotype could not be named at this time. The distance between the coding changes in exon 1 and exon 7 is 42,609 bases. This distance is too great to allow amplification and sub-cloning of the separate alleles to determine whether the bases are on the same allele. Therefore, we designate the allele CYP2C9D360N until such time as additional patients carrying the D360N change are identified in the future and can be examined for the other substitutions. We also devised a new pyrosequencing test which will directly read the new G1078A change, to allow for ease of detection of the new allele. This new genotyping test is shown in Figure 3.
Figure 1.
Histograms and pyrograms showing CYP2C9*5 and CYP2C9*3 genotyping tests used in the present study. The sequence at the bottom refers to the nucleotide dispensation for the reverse strand. While the dispensation order is A(negative)[CG]A(negative)TCA[TG]C(negative)TA, the actual sequence (reverse strand) analyzed is [C/G]TCAA[T/G]GTA (variant nucleotides in brackets with mutant nucleotides in bold). The shaded boxes indicate the variable region in which two nucleotides were dispensed, with the base designation of each allele. Pyrograms shown are examples of individuals homozygous for CYP2C9*1/*1 or heterozygous for CYP2C9*1/*5 and CYP2C9*1/*3. The final pyrogram is from index case who shows an abnormal pyrogram (see arrows) with the appearance of an extra T at bp 1078 and the disappearance of a C (on the antisense strand), confirmed by sequencing of the genomic DNA as a G1078A mutation on the sense strand). Figure reproduced with permission from Limdi et al, Personalized Medicine. (2007) 4(2), 157–16916 with permission of Future Medicine Ltd.
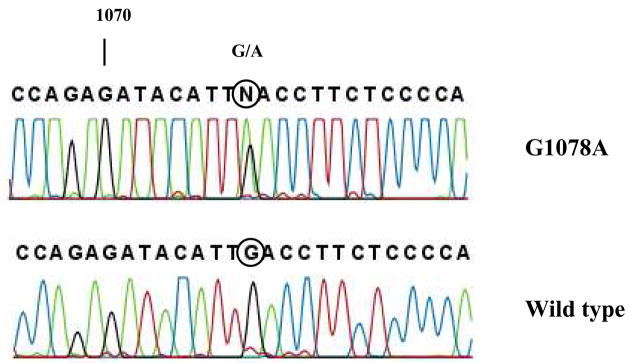
Figure 2.
Partial DNA sequence of exon 7 of African-American patient (top) and wild type CYP2C9*1/*1 (lower). Direct sequencing of the sense strand of exon 7 shows the patient is heterozygous for a G1078A mutation (numbering based on the cDNA). The circle designates base pair at 1078 which is heterozygous G/A in the top panel.
Figure 3.
Histograms and pyrograms showing a new pyrosequencing genotyping test which encompasses the CYP2C9*5, CYP2C9*3, and the new CYP2C9*G1078A(D360N) alleles. As in Fig. 1, the sequence at the bottom refers to the nucleotide dispensation for the reverse strand. The new dispensation order is A(negative)[CG][TC]A[TG]C(negative)TATC, the actual sequence (reverse strand) analyzed is [C/G]T[C/T]AA[T/G]GTATCT (variant nucleotides in brackets with mutant nucleotides in bold). (Note that the forward sense strand being analyzed is AGATC[A/C]1075TT[G/A]1078A[C/G]1080). The shaded boxes indicate the variable region in which two nucleotides were dispensed, with the base designation of each allele. Pyrograms shown (top to bottom) are examples of individuals homozygous for CYP2C9*1/*1 or heterozygous for CYP2C9*1/*5, CYP2C9*1/*3, or CYP2C9*1/*G1078A (CYP2C9*1/*D360N).
This polymorphism has not been previously reported and among the 448 study participants (215 African Americans) no other case of G1078A substitution has been encountered. Therefore from this one occurrence in 896 alleles, it was concluded that G1078A is a rare polymorphism possibly occurring primarily in African Americans with an allele frequency of 0.23% [95% CI: 0.01%–1.14%]. If the polymorphism is not specific to African Americans, the estimated allele frequency in the two populations would be 0.11% [95% CI: 0.005%–0.55%].
Although the new allele probably contains an S3Y coding change in addition to D360N, it is unlikely that the amino acid change in the N-terminus would change catalytic activity. In contrast, amino acids 359 and 360 lie within one of the substrate recognition sites of the CYP2 family of enzymes originally predicted by Gotoh and coworkers.[8] This area appears to be a “hot spot” for mutations in CYP2C9. Several defective alleles which arise from coding mutations in this region greatly affect the affinity, catalytic activity, and turnover number of the CYP2C9 enzyme resulting in large variability in dosage requirements of CYP2C9 substrates.[9] These alleles include CYP2C9*3 (I359L),[10] CYP2C9*4 (I359T, found in Japanese),[11] CYP2C9*5 (D360E) in African-Americans.[4] The new D360N coding change found in one African American in this study represents a new mutation within this substrate recognition site. An Y358C mutation has also been reported to the NCIdbSNP homepage (rs1057909). Since the prediction of the “substrate site” by Gotoh, recent crystal structures definitively show that the hydrophobic substrate pocket includes the adjacent amino acids Leu362 and Leu366. [12] Leu362 and Leu366 limit access for warfarin to the heme group.[13] I359 and D360 are in close proximity to these amino acids and substitutions in the region apparently affect the substrate pocket, explaining the reported effects of substitutions I359L [10], I359T [4] and D360N (present article) on turnover numbers of CYP2C9 for warfarin. Therefore D360N represents a new putative defective allele of CYP2C9 which merits further study and represents risk to individuals carrying this allele who are treated with anticoagulants.
Acknowledgments
The last author wishes to acknowledge Drs. Ronald T. Acton and Edward Faught for their support and mentorship. We are grateful to all the patients that participated in the study. We thank Janice Ware for her untiring efforts with patient recruitment and the staff of the Anticoagulation Clinic at The Kirklin Clinic, the Cooper Green Hospital and Jefferson Clinic P.C for their help with identification of potential participants. We also thank the physicians, especially Drs. Mark Wilson, and Melissa Baird; at the University of Alabama at Birmingham and the Health Service Foundation for their support of this research. Thanks to Steve Duncan and Darlene Green and the Office of Data Resources for their work with the POAT database and quality assurance.
Supported in part by a grant from the National Institute of Neurological Disorders and Stroke (Grant Number: K23NS45598-01) and in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
Footnotes
Financial & competing interests
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 2.Limdi N, McGwin G, Goldstein J, Beasley T, Arnett D, Adler B, Baird M, Acton R. Influence of CYP2C9 and VKORC1 1173C/T Genotype on the Risk of Hemorrhagic Complications in African-American and European-American Patients on Warfarin. Mol Ther. 2008;83:312–21. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidd RS, Curry TB, Gallagher S, Edeki T, Blaisdell J, Goldstein JA. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics. 2001;11:803–8. doi: 10.1097/00008571-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Dickmann LJ, Rettie AE, Kneller MB, Kim RB, Wood AJ, Stein CM, Wilkinson GR, Schwarz UI. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Molecular Pharmacology. 2001;60:382–7. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 5.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, ART Influence of CYP2C9 and VKORC1 polymorphisms on warfarin dose, anticoagulation attainment and maintenance among European American and African Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limdi NA, Goldstein JA, Blaisdell JA, Beasley TM, Rivers CA, Acton RT. Influence of CYP2C9 Genotype on warfarin dose among African American and European Americans Personalized Medicine. 2007;4:157–169. doi: 10.2217/17410541.4.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, Xi T, Mohrenweiser H, Ghanayem B, Goldstein JA. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–37. doi: 10.1097/01.fpc.0000114759.08559.51. [DOI] [PubMed] [Google Scholar]
- 8.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. Journal of Biological Chemistry. 1992;267:83–90. [PubMed] [Google Scholar]
- 9.Shintani M, Ieiri I, Inoue K, Mamiya K, Ninomiya H, Tashiro N, Higuchi S, Otsubo K. Genetic polymorphisms and functional characterization of the 5′-flanking region of the human CYP2C9 gene: in vitro and in vivo studies. Clinical Pharmacology & Therapeutics. 2001;70:175–82. doi: 10.1067/mcp.2001.117367. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Imai J, Ieiri I, Mamiya K, Miyahara S, Furuumi H, Nanba E, Yamane M, Fukumaki Y, Ninomiya H, Tashiro N, Otsubo K, Higuchi S. Polymorphism of the cytochrome P450 (CYP) 2C9 gene in Japanese epileptic patients: genetic analysis of the CYP2C9 locus. Pharmacogenetics. 2000;10:85–9. doi: 10.1097/00008571-200002000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–8. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 13.Seifert A, Tatzel S, Schmid RD, Pleiss J. Multiple molecular dynamics simulations of human p450 monooxygenase CYP2C9: the molecular basis of substrate binding and regioselectivity toward warfarin. Proteins. 2006;64:147–55. doi: 10.1002/prot.20951. [DOI] [PubMed] [Google Scholar]