Abstract
Wnt signaling increases bone mass by stimulating osteoblast lineage commitment and expansion and forms the basis for novel anabolic therapeutic strategies being developed for osteoporosis. These strategies include derepression of Wnt signaling by targeting secreted Wnt pathway antagonists, such as sclerostin. However, such therapies are associated with safety concerns regarding an increased risk of osteosarcoma, the most common primary malignancy of bone. Here, we analyzed 5 human osteosarcoma cell lines in a high-throughput screen for epigenetically silenced tumor suppressor genes and identified Wnt inhibitory factor 1 (WIF1), which encodes an endogenous secreted Wnt pathway antagonist, as a candidate tumor suppressor gene. In vitro, WIF1 suppressed β-catenin levels in human osteosarcoma cell lines, induced differentiation of human and mouse primary osteoblasts, and suppressed the growth of mouse and human osteosarcoma cell lines. Wif1 was highly expressed in the developing and mature mouse skeleton, and, although it was dispensable for normal development, targeted deletion of mouse Wif1 accelerated development of radiation-induced osteosarcomas in vivo. In primary human osteosarcomas, silencing of WIF1 by promoter hypermethylation was associated with loss of differentiation, increased β-catenin levels, and increased proliferation. These data lead us to suggest that derepression of Wnt signaling by targeting secreted Wnt antagonists in osteoblasts may increase susceptibility to osteosarcoma.
Introduction
Osteosarcoma is the most common primary malignancy of bone, and the third most common cancer in adolescents (1). Risk factors for osteosarcoma include states associated with increased osteoblast proliferation, such as chronic osteomyelitis, adolescence, Paget disease of bone, ionizing radiation, and various rare inherited syndromes (2). Osteosarcoma is characterized by morphologically abnormal osteoblastic cells producing aberrant osteoid. Loss of differentiation occurs in more than 80% of sarcomas, correlates with higher grade, and confers a 10%–15% decrease in survival (1, 3). Although the mechanisms that disrupt differentiation in osteosarcoma are poorly understood, strong evidence suggests that epigenetic processes are important (4). Implantation of even markedly aneuploid cancer genomes into blastocysts or enucleated zygotes appears compatible with more or less normal development of the derived embryos (5, 6). It has been suggested that these reversible events are epigenetic in character, since it is known that epigenetic templates are erased during early embryonic development (7). It is not clear which physiologic pathways responsible for differentiation are recurrently epigenetically inactivated during carcinogenesis.
Wnt signaling coordinates osteoblast proliferation and differentiation (8), and disruptions in various components of the Wnt pathway result in disordered bone development and homeostasis (9–12). The Wnt pathway is tightly controlled by secreted antagonists that either directly bind Wnts, exemplified by Wnt inhibitory factor 1 (Wif1), the secreted frizzled-related protein (Sfrp) family, and Cerberus (13), or bind proteins that directly bind Wnt receptors, exemplified by the Dickkopf (Dkk) family (Dkk1–Dkk4; ref. 14) and sclerostin (Sost; refs. 15, 16). Wnt signaling is also strongly linked to cancer, with oncogenic mutations reported in β-catenin, E-cadherin, adenomatous polyposis coli (APC), Wnt1, axis inhibition protein 1 (AXIN), and T cell factor 4 (TCF4) (17). Osteosarcomas frequently exhibit high levels of cytoplasmic and/or nuclear β-catenin (18), which is also associated with metastasis (19, 20). Canonically, β-catenin is stabilized after binding of Wnts to coreceptors Frizzled and LRP5/6 and then enters the nucleus, where it cooperates with TCF/lymphoid enhancer–binding factor (TCF/LEF) to transcriptionally activate oncogenes, including C-MYC, MYB, CCND, and CCNE (21). Epigenetic silencing of secreted Wnt pathway antagonists, including WIF1, DKK1, DKK3, SFRP1, SFRP2, SFRP4, and SFRP5, is a strikingly common event in many cancers (22–26), although the causal relationship between loss of expression and carcinogenesis is less well established. We undertook a high-throughput screen to identify novel epigenetically silenced tumor suppressor genes linking loss of differentiation to transformation in osteosarcoma.
Using this high-throughput strategy, we identified WIF1 to be epigenetically silenced in human osteosarcoma cell lines. Reexpression of WIF1 in cell lines suppressed β-catenin levels and cellular proliferation and induced expression of osteoblastic differentiation. In mice, Wif1 was not required for normal skeletal development, but loss of Wif1 increased susceptibility to radiation-induced osteosarcomas. WIF1 was silenced in primary human osteosarcoma samples by promoter hypermethylation, with a corresponding loss in WIF1 protein expression, and was associated with increased β-catenin levels and increased proliferation. The results from our studies represent a significant step forward in understanding the role of WIF1 in bone development and tumorigenesis.
Results
Epigenetic screen for genes linking differentiation and transformation in osteosarcoma.
A panel of 5 osteosarcoma cell lines (B143, G292, HOS, SAOS2, and SJSA) was treated with individually titrated doses of the demethylating agent 5-aza-2-deoxycytidine (dAC; 5–10 μM) for 3 d (Figure 1A). This treatment resulted in growth arrest and differentiation, as measured by alkaline phosphatase (ALP) activity (Figure 1B) and mineralization (mean increase of 2.2-fold across 5 cell lines). Next, we performed genome-wide transcriptional profiling of the dAC-treated cell lines to identify epigenetically silenced genes using cDNA microarrays containing 9,386 probes (27). Expression of genes involved in osteoblast differentiation, including the master osteoblast transcription factor RUNX2, osterix (OSX), collagen type 1A1 (COL1A1), osteopontin (OP), bone morphogenetic protein 2 (BMP2), BMP4, osteoprotegerin (OPG), osteoglycin, and ALP, increased after demethylation (Figure 1, C and D, and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI37175DS1). No significant increase in muscle-related gene expression was observed, which suggests that demethylation specifically reactivated the osteoblast transcription program. Transcripts of putative oncogenes, including cyclin B1, cyclin D1, cyclin E2, CDK2, CDK4, MDM2, and the RNA subunit of telomerase, were repressed. These data suggest that epigenetic mechanisms integrate transformation and disruption of differentiation in osteosarcoma lines.
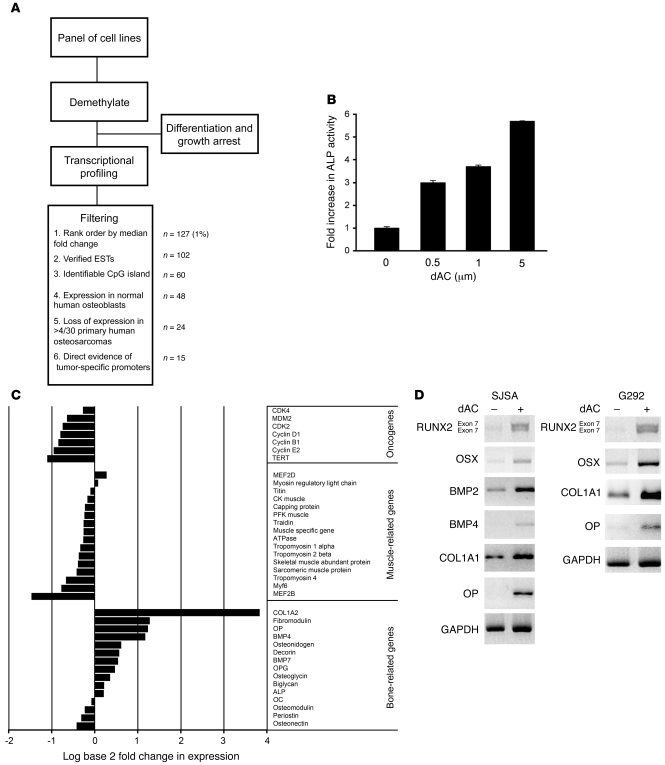
Figure 1. A high-throughput screen for epigenetically silenced tumor suppressor genes linking transformation with loss of differentiation in osteosarcoma.
(A) Design of high-throughput screen and filtering strategy for identification of epigenetically silenced tumor suppressor genes linking differentiation and transformation in osteosarcoma. (B) Dose-dependent induction of ALP activity in G292 cells treated with dAC for 5 d. Values are mean ± SEM of 3 separate determinations. (C) Effect of demethylation on expression of osteoblast- and muscle-specific genes, as well as a set of known oncogenes. Data are from transcriptional profiling, and are the median fold change across all 6 cell lines after treatment for 3 d with 5–10 μM dAC. The experiment was performed independently twice. (D) RT-PCR confirmation of changes in gene expression in SJSA and G292 cells. OSX, osterix.
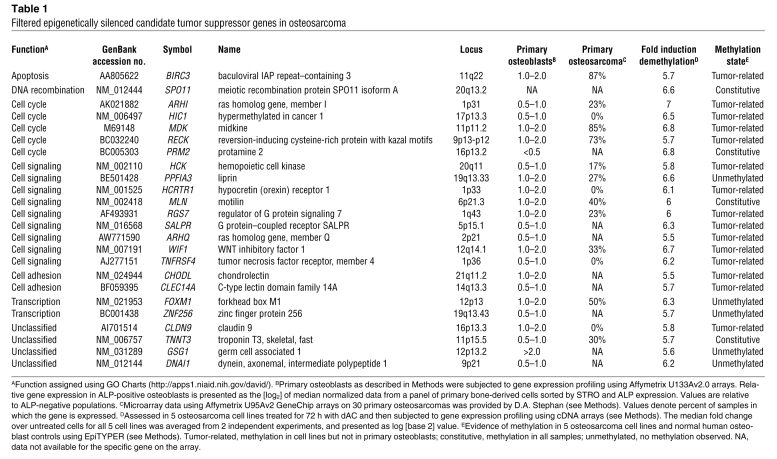
We next filtered the array data for candidate tumor suppressor genes (Figure 1A). Probes were first ranked by fold induction after treatment with dAC (Supplemental Table 1). The top-ranked 1% of genes were filtered for: (a) corresponding expressed sequence tags (hg17, May 2004 build), (b) presence of CpG island within 5 kb of the transcriptional start site, (c) loss of expression in a data set of human osteosarcoma samples arrayed using Affymetrix U95Av2 (28), and (d) expression in Affymetrix U133Av2.0 arrays of ALP-expressing primary human osteoblasts. The latter 2 filters were used to exclude genes epigenetically silenced after commitment to the osteoblast lineage rather than after transformation. The fold change in expression of 24 candidate genes after dAC treatment is shown in Supplemental Table 2. These 24 genes were screened for direct and selective promoter methylation (see Methods and ref. 29) in osteosarcoma cell lines, but not primary human osteoblasts (Table 1). We found that 19 of 24 (79%) candidates demonstrated promoter methylation, and 15 were methylated selectively in osteosarcoma cell lines. The failure to identify methylation in 5 candidates may be due to the restricted promoter regions analyzed in these studies. Notably, hypermethylated in cancer 1 (HIC1) was recently identified as an osteosarcoma-specific epigenetically silenced tumor suppressor (30).
Table 1 .
Filtered epigenetically silenced candidate tumor suppressor genes in osteosarcoma
We focused on WIF1 because of the known importance of Wnt signaling in coordinating osteoblast proliferation and differentiation (8). WIF1 is a highly conserved gene located on chromosome 12q14 and encodes a secreted 379–amino acid protein, which binds Wnt proteins in the extracellular space and inhibits their ability to bind to their receptors (31). Tumor-associated epigenetic silencing of secreted Wnt pathway antagonists (22–24), including Wif1 (25, 32, 33), has been widely reported. While compelling, it is unknown whether silencing of Wif1 is a cause or effect of tumorigenesis.
Epigenetic silencing of WIF1 activates Wnt signaling.
Treatment of the osteosarcoma cell lines with dAC resulted in suppression of β-catenin levels (Figure 2A) and in TCF/LEF-dependent transcriptional reporter activity (data not shown). As predicted by the array data, WIF1 transcript expression was absent in the osteosarcoma cell lines and expressed after demethylation (Figure 2B). As assessed by semiquantitative immunocytochemistry, treatment of B143 cells with recombinant WIF1 significantly reduced total β-catenin levels by 45% (P = 0.029; n = 45 [control]; 54 [WIF1]) and cytoplasmic β-catenin levels by 52% (P = 0.001; n = 20 [control]; 20 [Wif1]; Figure 2C) compared with control. As expected, Wif1 also downregulated TCF/LEF activity (Figure 2D). Taken together, these data suggest that epigenetic silencing of WIF1 may lead to derepression of Wnt signaling in osteosarcoma cell lines.
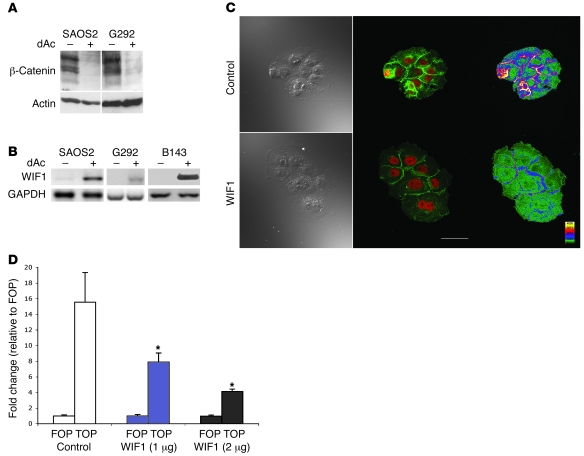
Figure 2. Epigenetic silencing of Wif1 derepresses Wnt signaling.
(A) Western blot showing marked suppression of β-catenin levels after treatment of osteosarcoma cell lines with 5–10 μM dAC for 3 d. (B) RT-PCR showing induction of WIF1 transcript after demethylation of B143 cells for 72 h. (C) B143 cells were treated with 3 μg/ml recombinant GST-WIF1 or GST alone (control) for 24 h, then fixed and stained for β-catenin (green) and counterstained with propidium iodide (red). Shown are brightfield (left), coregistered (middle), and β-catenin intensity heatmap (right) images. Scale bar: 100 μm. The experiment was performed independently 3 times with similar results. (D) Effect of WIF1 expression on TCF/LEF reporter activity. TOP-flash and FOP-flash luciferase reporter constructs were cotransfected with constant amounts of LacZ vector control (10:1 ratio) into B143 cells with varying amounts of WIF1 expression vector. Cells were harvested within 24 h, and luciferase activity was measured. Data are normalized to β-galactosidase expression. Values represent independent transfections done in triplicate and are presented as fold change relative to FOP-flash (mean ± SEM). *P < 0.01 versus control, Tukey test with 1-way ANOVA.
WIF1 regulates osteoblast differentiation and suppresses tumor cell growth in vitro.
Consistent with previously published data (35), we observed Wif1 expression after differentiation of murine preosteoblastic MC3T3E1 cells and in primary human bone–derived cultures differentiated with BMP2 and vitamin D3. We detected expression of WIF1 mRNA and protein in primary human osteoblasts flow sorted for ALP expression, but could not detect WIF1 expression (mRNA or protein) in cells negative for ALP but positive for STRO1, a mesenchymal progenitor marker (Supplemental Figure 1, B and C). To our knowledge, the role of WIF1 in osteoblast differentiation has not yet been established. Primary osteoblasts were treated with recombinant WIF1 protein, inducing expression of ALP (Figure 3A). ALP-positive primary osteoblasts (sorted by fluorescence-activated cell sorting) that were treated with WIF1 protein increased mRNA levels of several markers of osteoblast differentiation, including RUNX2, OPG, OP, osteonectin, and COL1A1, but not WIF1 (Figure 3B). Treatment of primary osteoblasts and the osteosarcoma cell lines with recombinant WIF1 protein induced ALP in every cell type except SAOS2, in which ALP expression is constitutively high (Supplemental Figure 2D). To determine whether Wif1 was required for the osteoblast phenotype, shRNAs were designed targeting Wif1 expression in MC3T3E1 cells. Knockdown of Wif1 mRNA and protein was confirmed and resulted in decreased Runx2 transcript (Figure 3C) and protein levels (Figure 3D) in MC3T3E1 cells. Differentiating conditions resulted in reduced mineralization of Wif1 knockdown MC3T3E1 cells compared with control cells (Figure 3E). These data confirm that Wif1 is highly expressed during osteoblast differentiation; moreover, Wif1 appears to be required for osteoblast differentiation in vitro.
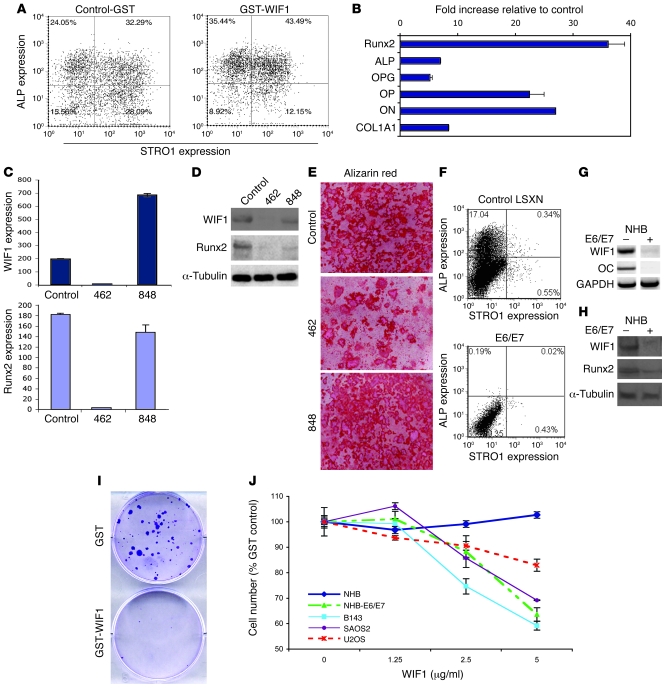
Figure 3. WIF1 regulates osteoblast differentiation and suppresses growth of osteosarcoma cell lines in vitro.
(A) WIF1 induced markers of bone differentiation. Primary human bone–derived NHB osteoblasts were flow sorted using antibodies to STRO1 and ALP. Cultures were treated with GST-WIF1 or equimolar amounts of GST for 24 h. The percentage of cells in each quadrant is indicated. (B) Quantitative RT-PCR analysis of osteoblast gene expression in flow-sorted ALP-positive NHB cells after treatment for 3 d with GST-WIF1. Values show fold change relative to GST treated control. The experiment was performed in triplicate. ON, osteonectin. (C–E) MC3T3E1 cells were infected with empty vector control or with shRNAmir hairpin to Wif1 (462) or nonsense hairpin (848) and selected using puromycin. (C) Quantitative RT-PCR analysis and (D) Western blot of Wif1 and Runx2 in control and Wif1 knockdown cells. (E) Mineralization of control and Wif1 knockdown cells after treatment with β-glycerophosphate and ascorbic acid for 2 wk. Cells were stained with alizarin red. (F) NHB cells were immortalized with LXSN-HPV16-E6/E7 (E6/E7) or control LXSN vector, and stably transfected cells were analyzed by flow cytometry for STRO1 and ALP expression. The percentage of cells in each quadrant is indicated. (G) RT-PCR for Wif1 and OC and (H) Western blot for WIF1 and Runx2 in immortalized cells compared with control. (I) Effect of WIF1 and control GST on B143 cells. Cells were stained with Crystal violet. (J) Cell lines and primary NHB cells were plated at 30% confluence and treated with 3 μg/ml GST-WIF1 or GST alone for 3 d followed by an MTT assay. Data (mean ± SEM for triplicate wells) are expressed as percentages relative to GST control.
Primary human osteoblastic cultures were immortalized by infection with retrovirus encoding human papilloma virus 16 (HPV16) E6/E7, whereas LXSN-infected control NHB cells underwent classical senescence at passage 10–15 after transduction (data not shown). Immortalization of primary osteoblasts was associated with loss of ALP expression, as assessed by flow cytometry (Figure 3F), as well as reduced WIF1 and osteocalcin (OC) mRNA expression (Figure 3G) and decreased WIF1 and RUNX2 protein expression (Figure 3H). The effects of restoring WIF1 expression on cell growth and viability were examined. Treatment of B143 cells with recombinant WIF1 protein profoundly suppressed colony formation (Figure 3I). Identical results were seen after transfection with a mammalian expression vector expressing WIF1 (data not shown). Two independent clones of HPV16 E6/E7–immortalized osteoblasts and osteosarcoma cell lines treated with WIF1 for 72 h demonstrated growth inhibition (Figure 3J), which was caused by both cell death and G1 cell cycle arrest (data not shown). No effect on cell number or morphology was seen in primary osteoblasts. Moreover, we did not observe differences in cell death in MC3T3E1 cells after knockdown of Wif1 (data not shown). Wif1 expression is lost early during immortalization, while reexpression of Wif1 suppresses the growth and viability of transformed human osteoblasts.
Wif1 is highly expressed in the developing and adult skeleton in vivo, but is not required for normal development.
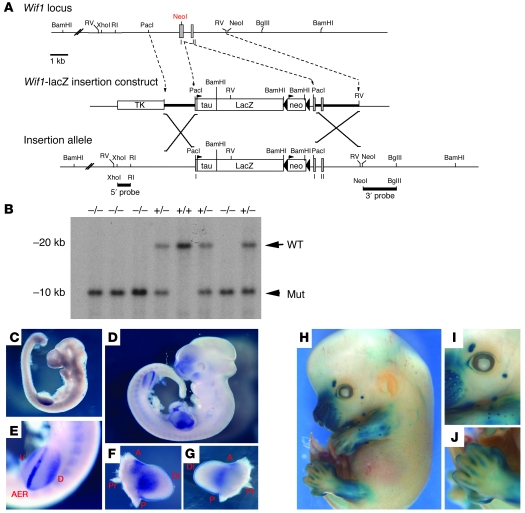
To study the developmental and tumor suppressor functions of Wif1 in vivo, transgenic mice were generated in which the Wif1 coding sequence was disrupted by insertion of a tau-LacZ reporter cassette (see Methods and Figure 4A). The insertion of the transgene disrupted transcription from the native Wif1 locus and encoded multiple in-frame stop codons. The presence of the inserted transgene was confirmed by Southern blot in ES cells and derived mice (Figure 4B). LacZ expression in Wif1+/+ mice recapitulated patterns seen using in situ hybridization (Figure 4, C–J) and demonstrated highly tissue-specific patterns similar to that described for wif1 in Xenopus and zebrafish embryos (31). Although initially undetectable, by E9.0 Wif1 expression was noted within the fore limb and somites. By E11.5, Wif1 was readily detected within the craniofacial regions, developing limb buds, and somites (Figure 4, C and D). The limb buds showed distinctly stronger dorsal expression compared with the ventral domain, and expression was also noted throughout the apical ectodermal ridge in both fore and hind limbs (Figure 4, E–G). At E14.5, LacZ expression was seen within the retina, hair follicles, cartilage, and bone in developing limbs at E14.5 (Figure 4, H–J).
Figure 4. Gene targeting Wif1 in vivo.
(A) Gene targeting strategy. Mice were generated containing a knockin allele of tau/LacZ in the first exon of Wif1, resulting in loss of expression of the Wif1 gene. (B) Southern blot of derived mice tails, demonstrating WT, heterozygous, and homozygous alleles. (C–G) Expression of Wif1 during mouse embryogenesis was detected by in situ hybridization. (C) Wif1 expression was initiated at E9.0 within the fore limb and weakly in the developing somites. (D) At E11.5, Wif1 expression was detected in the branchial arches, limb buds, and somites. (E) Wif1 was expressed in the apical ectodermal ridge (AER) and in the dorsal (D) and ventral (V) limb buds at E11.5. (F and G) Expression within the fore-limb bud at E11.5 was stronger on the dorsal (F) than the ventral (G) side. A, anterior; P, posterior; Pr, proximal; Di, distal. (H–J) LacZ expression under the control of the native Wif1 promoter in the developing embryo at E14.5. Patterns of β-galactosidase expression reflect endogenous patterns of Wif1 expression. (H) LacZ activity was detected in the craniofacial region, hair cells, limbs, and retina. (I) Expression of LacZ within the eyelid and retina. (J) Expression of LacZ within the digits of the hind limb.
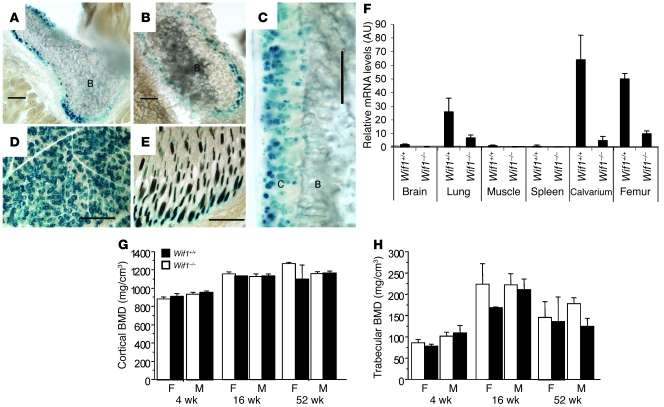
In the adult mouse, Wif1 was highly expressed in the skeleton and a limited range of additional tissues. LacZ expression was identified in sternum and ribs (Figure 5, A and B), cartilage (Figure 5C), hair follicles (Figure 5D), salivary glands (Figure 5E), and small intestinal mucosa (data not shown). Wif1 transcripts from all tissues studied from adult homozygous knockin mice (Wif1–/–) were markedly reduced, as determined by real-time RT-PCR, with the highest levels identified in bone and lung (Figure 5F). Wif1–/– mice were born at the expected Mendelian ratios, and adult mice lacked obvious phenotype in any tissues in which Wif1 was expressed, gained weight normally, and were able to breed successfully on a homozygous knockin background. Wif1–/– animals were compared with littermate controls at 4, 16, and 52 wk of age using high-resolution peripheral quantitative CT. These analyses revealed a lack of significant changes in femoral cortical bone mineral density at all time points and a nonsignificant trend to decreased trabecular bone mineral density between Wif1–/– and WT mice at 16 wk (Figure 5, G and H). In independent experiments, while gross femoral length and width and histomorphometric analysis of tibial bone structure were largely unremarkable, there was again a nonsignificant trend to decreased trabecular bone at 16 wk (Supplemental Figure 2A), consistent with the radiologic data. Consistent with these observations, expression of markers of osteoblast differentiation also demonstrated a nonsignificant trend to decreased expression of osteoblast-related genes (Supplemental Figure 2B). While these data may suggest a possible role for Wif1 in osteoblast function in vivo, the skeletal consequences of Wif1 loss are subtle at best.
Figure 5. Mice lacking expression of Wif1 undergo normal skeletal development.
(A–E) Patterns of LacZ expression in the adult mouse. Mice were sectioned and stained for β-galactosidase activity. (A) Sternum. (B) Cross section through rib. (C) Cartilage surface of hip joint. (D) Salivary gland. (E) Hair follicles in dermis. Scale bars: 100 μm. C, cartilage; B, bone. (F) The knockin allele disrupted expression of endogenous Wif1. Real-time RT-PCR for Wif1 expression in tissues from WT and Wif1–/– mice. Data (mean ± SEM for 3 primer sets spanning exons 1–2, 3–4, and 7–8) were repeated twice. (G) Femoral cortical bone mineral density (BMD), measured at 25% of bone length distal to the growth plate, in female (F) and male (M) WT and WIF1 knockout mice. (H) Trabecular bone mineral density, measured at 5% of bone length distal to the growth plate. n = 4–11 for all groups, except 52-wk-old females of both genotypes and 16-wk-old WT females (n = 2). No statistically significant differences were detected.
Wif1–/– mice are susceptible to spontaneous and radiation-induced osteosarcoma.
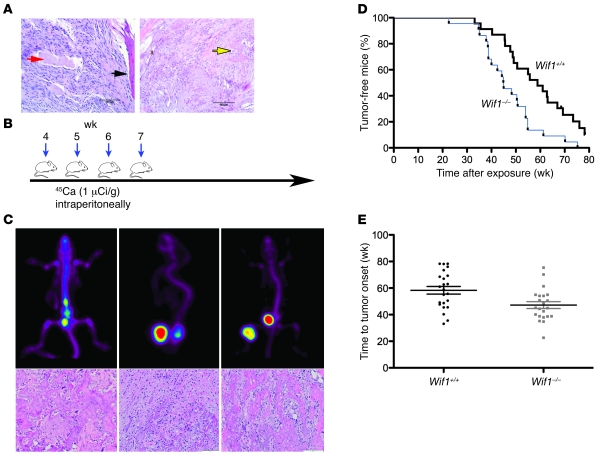
Of 13 Wif1–/– mice, 2 developed spontaneous sarcomas (1 limb sarcoma arising adjacent to bone at 89 wk, and 1 osteosarcoma at 56 wk), which suggests an in vivo tumor suppressor role for Wif1 (Figure 6A). No tumors were observed in 30 littermate control mice. To quantitate the tumor suppressor role of Wif1 in the development of osteosarcoma, we used a radiocarcinogen model (35), since irradiation is a known risk factor for osteosarcoma. 45Ca is a long-lived, low-energy β-emitter that is incorporated into the skeleton and induces osteosarcomas in mice with 100% penetrance. Cohorts of Wif1–/– mice and littermate controls were injected with 45Ca from 4 to 8 wk of age (Figure 6B). Tumor emergence was monitored clinically and by the use of 18F-PET, which provided a sensitive marker for sites of osteoblast activity (our unpublished observations). The first osteosarcomas were seen approximately 26 wk after 45Ca exposure and developed in the spine, hind and fore limbs, pelvis, and cranium (Figure 6C). Histologically, the tumors resembled human osteosarcoma, with malignant osteoid production (Figure 6A, left). We did not observe any nonosteosarcoma tumors. As previously reported for this radiocarcinogen mouse model (35), female mice were more susceptible to the development of osteosarcomas than were male mice (P < 0.001). Wif1–/– mice were predisposed to the development of radiocarcinogen-induced osteosarcoma. The median latency of tumor onset (measured as a visible growth, palpable tumor, limping, or paralysis and verified by 18F-PET scan) in 45Ca-treated Wif1–/– mice was shortened by 14 wk compared with littermate controls (P = 0.0032, Mantel-Cox; Figure 6D). Mean time to onset was 58.32 ± 2.9 wk in WT mice (n = 23) and 47.12 ± 2.5 wk in Wif1–/– mice (n = 22; Figure 6E). Median survival was shortened by 8.3 wk in Wif1–/– compared with WT mice as assessed by Kaplan-Meier curves (P = 0.0382, Mantel-Cox; data not shown).
Figure 6. Wif1–/– mice are susceptible to spontaneous and radiation-induced osteosarcoma.
(A) Left: Sarcoma from the hind left limb of an 89-wk-old male Wif1–/– mouse, arising adjacent to periosteum, and infiltrating skeletal muscle. The adjacent cortical bone (black arrow) and skeletal muscle fiber surrounded by sarcoma cells (red arrow) are indicated. Right: Osteosarcoma from the lumbar spine of a 56-wk-old female Wif1–/– mouse, showing a malignant osteoid (yellow arrow). Scale bars: 100 μm. (B) Radiation-induced osteosarcoma model schematic. (C) Typical imaging appearance of osteosarcomas. Top: 18F-PET scan demonstrating intense avidity of osteoblastic tumors from the same mouse. Bottom: Histology of mouse radiation-induced osteosarcoma. Original magnification, ×20. (D and E) Kaplan-Meier plots of tumor onset (D; P = 0.0032, Mantel-Cox) and survival (E; P = 0.0382, Mantel-Cox) of Wif1–/– and WT mice after exposure to 45Ca (n = 23 [WT]; 22 [Wif1–/–]). (E) Data points represent individual mice; horizontal bars and error bars denote mean ± SEM.
WIF1 expression inversely correlates with proliferation and is silenced by promoter hypermethylation in human osteosarcomas.
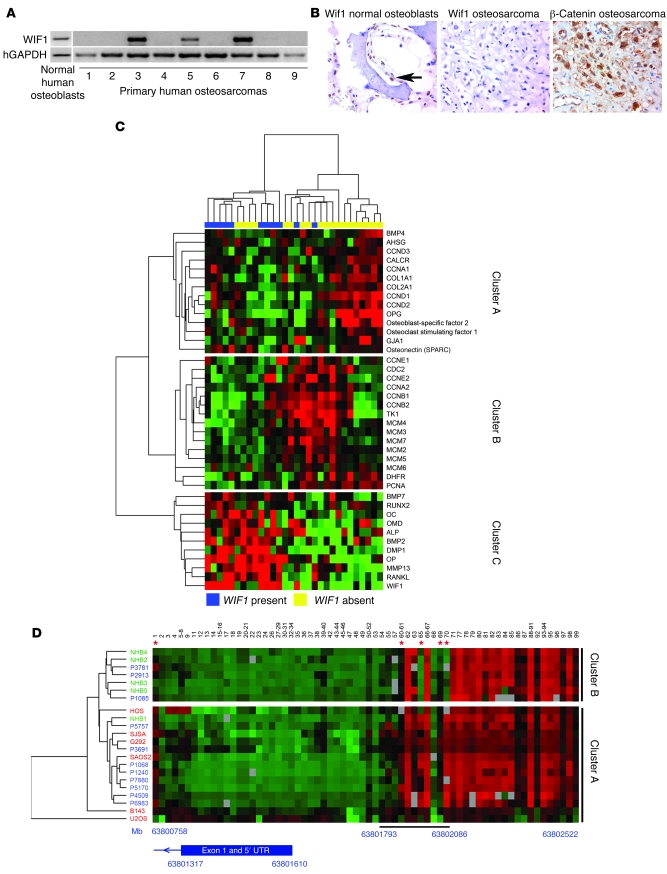
WIF1 transcript was readily detected in human primary osteoblast cultures, but not in 15 of 20 primary human osteosarcomas examined (6 of the 9 samples shown in Figure 7A). WIF1 protein was expressed in osteoblasts, but undetectable in 93% (71 of 76) of osteosarcomas, by immunohistochemistry (Figure 7B). In contrast, strong staining (grade 2 or 3; see Methods) for β-catenin was observed in 48% of tumors (Figure 7B), and β-catenin was associated with increased expression of proliferating cell nuclear antigen (P < 0.05, χ2 test), a marker of proliferation.
Figure 7. Expression patterns and epigenetic silencing of WIF1 in human osteosarcoma.
(A) RT-PCR showing expression of WIF1 transcript in primary human osteoblasts, and lost in primary human osteosarcoma samples. hGAPDH, human GAPDH. (B) Immunohistochemical staining for WIF1 and β-catenin in normal bone and osteosarcomas. A tissue microarray was probed with antibodies to WIF1. Arrow indicates normal osteoblast. Original magnification, ×20. (C) Heatmap clustering of 30 primary osteosarcomas using differentiation and proliferation gene cassettes. Cluster A is a gene cassette of intermediate osteoblastic lineage, cluster B contains proliferation markers, and cluster C is a gene cassette of an osteoblastic phenotype. (D) Heatmap demonstrating unsupervised hierarchical clustering of CpG methylation in primary human osteoblast cultures (green), osteosarcoma cell lines (red), and primary osteosarcoma samples (blue). Individual CpGs are identified by numbers above, and the approximate relationship between CpG location and the WIF1 coding region is identified below. Cluster A represents the tumor-associated methylation group, and cluster B represents the normal osteoblast methylation group. The region underlined below the heatmap, from 63,801,793 to 63,802,086, appeared to be differentially methylated. Green denotes no methylation; red denotes 100% methylation. Asterisks indicate individual CpGs whose degree of methylation significantly correlated with expression of WIF1 transcript (all P < 0.05, ANOVA). The schematic below shows the position of Wif1 exon 1 and the untranslated region (UTR) in relation to regions of methylation.
We used an array data set on 30 primary osteosarcomas to quantitate the relationship between WIF1 expression and tumor differentiation and proliferation (28). Loss of differentiation correlates with higher grade in osteosarcoma, while increased proliferation correlates with tumor grade and an adverse prognosis (1, 36). Unsupervised hierarchical clustering was applied to transcriptional profiling data using gene sets enriched for osteoblast- and proliferation-related genes (Supplemental Table 3). On gene clustering, osteoblastic genes formed a distinct cluster, cluster C, which included markers of the osteoblast phenotype (ALP, OC, OP, RUNX2, dentine matrix protein, RANKL, osteomodulin, MMP13 [collagenase 3], BMP2, and BMP7). Cluster B was enriched in genes implicated in S-phase progression of the cell cycle (including PCNA, thymidine kinase 1, dihydrofolate reductase, minichromosome maintenance proteins 3–7, and a range of S-phase and G2/M-phase cyclins). Finally a third cluster, cluster A, was observed, representing less mature markers of the osteoblast lineage, presumably intermediate with respect to proliferative status (including COL1A1, COL1A2, OPG, and G1/S-phase cyclin D1–cyclin D3).
Tumor clustering identified 3 distinct groups. Group 1 contained 9 WIF1-positive tumors of 13, group 2 contained 2 WIF1-positive tumors of 10, and group 3 contained 0 WIF1-positive tumors of 7. Group 1 was characterized by high expression of osteoblastic cluster C, and group 2 was associated with the expression of proliferation cluster B, whereas group 3 expressed elements of clusters A and B. Expression of cluster C genes significantly correlated with WIF1 expression (P = 0.002, χ2, Pearson uncorrected 2-tailed), and only 2 of 17 tumors with proliferation markers expressed WIF1, compared to 9 of 13 well-differentiated tumors (P < 0.001, χ2, Pearson uncorrected). These data suggest that WIF1 is lost in poorly differentiated osteosarcomas, which are associated with increased proliferation.
The human WIF1 promoter has 105 CpGs located within 1.5 kb of the 5′ transcriptional start site of human WIF1 (25). WIF1 promoter CpG methylation patterns were mapped in detail in normal human bone samples (n = 5), osteosarcoma cell lines (n = 6), and primary human osteosarcomas (n = 11). Osteosarcoma-specific differences in methylation patterns were observed with unsupervised hierarchical clustering using Spearman rank correlation followed by average clustering method (Figure 7D). In all samples, including normal human osteoblasts, there was dense methylation of CpGs between –476 bp and –912 bp from the transcriptional start site of the WIF1 gene. Tumor-specific methylation (cluster A) correlated with loss of Wif1 expression in 14 of 17 samples, including all osteosarcoma cell lines and 8 of 11 primary tumors. Cluster B correlated with retention of expression in 5 of 7 samples, including 4 of the 5 normal osteoblast samples along with P3781, which retained Wif1 expression. Clusters A and B were distinguished by differential methylation of 4 of 16 CpGs within a 293-bp region from –183 to –476 located approximately 150 bp from the transcription start site: CpG 60–61, CpG 65, CpG 69, and CpG 70. The distribution of primary osteoblasts to tumors and the cell lines was significantly different (P = 0.022, χ2, Pearson uncorrected 2-tailed). Tumor-specific differential methylation of the WIF1 promoter was also confirmed using bisulphite restriction analysis (data not shown). Tumor-specific methylation patterns were seen in both osteosarcoma cell lines and primary osteosarcomas, but not in primary cultured osteoblasts or lymphocytes (data not shown). Microsatellite analysis revealed that the WIF1 locus (12q14) showed evidence of loss of heterozygosity in 3 of 6 cell lines, but we did not detect any loss of heterozygosity in any primary osteosarcomas examined.
Discussion
The present work causally links epigenetic silencing of WIF1 to spontaneous and radiation-induced osteosarcoma in vivo, providing key support to mounting evidence that WIF1 is frequently methylated in cancer (25, 32, 33, 37). Wif1 expression correlated with markers of differentiation in primary osteosarcomas and inversely correlated with proliferation. It is important to note that about 30% of tumors expressed WIF1 transcript, although expression of WIF1 protein was seen in fewer than 10% of osteosarcomas. It is possible that activating mutations in β-catenin or other intracellular mediators of Wnt signaling may bypass the inhibitory effects of secreted Wnt pathway antagonists.
Epigenetic processes are important in the loss of differentiation in cancer (4–6, 38). Detailed mapping of the WIF1 promoter revealed an approximately 300-bp differentially methylated region, located within 200 bp of the coding region and found to be rich in stimulatory protein 1 (Sp1) sites. This region separates a constitutively heavily methylated region located 450–900 bp upstream of the coding region from the transcriptional start site. We identified 4 CpGs that were differentially methylated in a tumor-specific manner and in general correlated with WIF1 transcript expression. Differential methylation of quite small stretches of DNA (less than 300 bp) has previously been shown to affect gene transcription (39–41). Wif1 resembles E-cadherin (42), in which Sp1 sites may function as insulators that limit spreading of methylation from Alu-rich distal regions (43, 44). However, it also remains formally possible that the extremely focal methylation patterns observed in the WIF1 promoter of osteosarcoma cell lines and primary tumors may reflect unknown upstream methylation events affecting other genes. While we did not observe genetic events in our study, a recent report identified the first genetic rearrangement disrupting WIF1 in salivary gland tumors (45), which is interesting given the high levels of expression of Wif1 in the murine salivary gland. Whether direct or indirect, epigenetic templates associated with less mature states of differentiation may favor tumorigenesis and, when erased during embryogenesis, may reset the cancer genome, permitting normal cell development, as predicted by Hochedlinger and colleagues (5).
The frequency with which secreted negative regulators of Wnt signaling, including multiple members of the Dkk and Sfrp families, are silenced by promoter methylation in cancer is remarkable. Why the Wnt pathway, why bone, and why Wif1? The Wnt pathway is critical to cancer as well as to bone development and homeostasis. Oncogenic mutations have been reported in β-catenin, E-cadherin, APC, Wnt1, AXIN, and TCF4 (18). Inactivating mutations in LRP5, the Wnt coreceptor, are associated with osteoporosis-pseudoglioma syndrome, a condition associated with low bone mass (46), while activating mutations lead to increased bone mass (10). Mice with targeted deletions of β-catenin in mesenchymal precursors have impaired bone development (47), while later inactivation of β-catenin affects both osteoblast and osteoclast functions (9). Wnt signaling is under the control of numerous secreted negative regulators. Inactivating mutations in SOST, which binds and inhibits LRP5, are associated with increased bone mass (48), while deletions in Dkk1 (12) and Sfrp1 resulted in anabolic effects (11). These effects are mediated by increased osteoblast commitment and lineage expansion. On the other hand, Dkk2-null mice had increased numbers of nonmineralizing osteoids, and while exogenous expression of Dkk2 early in osteoblast cultures in vitro abolished osteogenesis, late expression (after peak Wnt7b expression) induced mineralization (49).
The results of the present study show that Wif1 was highly expressed in the murine skeleton during development, that mouse and human osteoblasts expressed high levels of Wif1, and, importantly, that Wif1 was required for osteoblast differentiation in vitro. Terminal osteoblast differentiation is associated with expression of negative regulators of Wnt signaling, including Dkk2 and Wif1, in vitro (present study and refs. 34, 49). In vitro, exogenous Wif1 suppressed β-catenin levels and TCF/LEF-dependent transcriptional activity (data not shown), consistent with previous studies showing that an activated allele of β-catenin represses Runx2-dependent transcriptional activity (50). Wif1 and Runx2 were coexpressed in cluster C in primary osteosarcomas. Like Dkk2, Wif1 may initiate terminal cell cycle exit partly by shutting down Wnt signaling, thereby removing a stimulus to proliferation and derepressing functions of Runx2 relevant to terminal osteoblast differentiation. The absence of an obvious developmental phenotype in the Wif1–/– mice may be caused by functional redundancy in vivo, since a recent report linked loss of WIF1 expression to human craniosynostosis, the premature fusion of calvarial sutures (51). Recognizing the limitations of the assays we used as biomarkers of Wif1 activity, we acknowledge the possibility that Wif1 may play a role in other processes relevant to tumorigenesis, including invasion, metastasis, and bypassing of senescence.
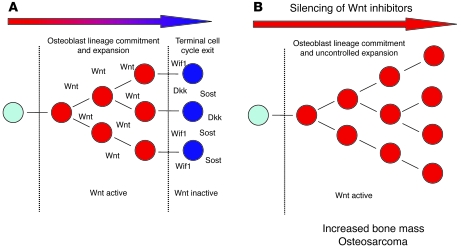
We propose the following model (Figure 8), which may account for the epigenetic inactivation of Wnt inhibitors during tumorigenesis. Under normal circumstances, Wnt signaling is required not only for osteoblast lineage commitment, but also for expansion. Together, these processes contribute to increasing bone mass. In a temporospatially restricted fashion, expression of Wnt antagonists, such as WIF1, SOST, and DKK1, initiates terminal cell cycle exit. Interestingly, in some cases (e.g., SOST), these inhibitors constrain bone mass, and in other cases, they stimulate bone formation (e.g., DKK2), while WIF1 appears dispensable for bone formation (Figure 8A). These effects may reflect subtle differences in timing or function that we do not yet understand. While we have here focused on tumor-related epigenetic silencing of WIF1, demethylation was associated with reexpression of additional inhibitors noted in the original screen: Wnt11, Sfrp4, and Sfrp1. Collectively, these data suggest that epigenetic silencing of Wnt pathway antagonists derepresses Wnt signaling, thereby removing a physiologic constraint on osteoblast expansion (Figure 8B). At least in the case of Wif1, loss of expression may increase susceptibility to osteosarcoma, although this also represents a therapeutic strategy to increase bone mass.
Figure 8. Role of Wnt signaling in osteoblast development and osteosarcoma.
(A) Physiologic control by Wnts of osteoblast lineage commitment and expansion is terminated by a range of secreted antagonists, including WIF1, SOST, and DKKs. (B) Therapeutic targeting of Wnt antagonists such as SOST increases bone mass by increasing the commitment and number of osteoblasts. Epigenetic silencing of WIF1 leads to unopposed activity of Wnts, resulting in failure to undergo terminal cell cycle exit and increasing the likelihood of malignant transformation.
Osteoporosis is a major human health problem, affecting between 5 and 8 million people worldwide. Until recently, treatment focused on inhibiting bone resorption, which stabilized bone loss but could not restore lost mass. Achieving peak bone mass is as much a function of osteoblast lineage expansion as is precisely controlled cellular differentiation. The discovery that intermittent parathyroid hormone (PTH) increases bone mass, in part through promoting osteoblast proliferation, represented a major advance in the treatment of osteoporosis (52). Wnt signaling is critical to achieving peak bone mass and has therefore been an attractive therapeutic strategy for osteoporosis. PTH itself may act by downregulating SOST, a negative regulator of Wnt signaling, thereby activating Wnt signaling (53). Drugs that activate Wnt signaling in bone by directly blocking the actions of SOST are in late phase clinical development (16). The strategy of targeting negative regulators of Wnt signaling to stimulate bone formation may be freighted with unwanted consequences. Osteosarcomas frequently exhibit high levels of β-catenin (18), which is associated with metastasis (19). States in which osteoblast proliferation is stimulated, including skeletal growth during adolescence, Paget disease, and chronic osteomyelitis, are associated with osteosarcoma. Intermittent PTH has caused osteosarcoma in preclinical models (54, 55), and a recent case report of osteosarcoma in a patient being treated with intermittent PTH has caused concern (56), while a number of additional reports are currently under review. Our present findings suggest that activation of Wnt signaling, in this case by loss of Wif1, may increase the risk of osteosarcomas in vivo. Caution is warranted as Wnt pathway agonists enter clinical trials for disorders of bone loss.
Methods
Cell lines and tissue samples.
B143, G292, HOS, SAOS2, SJSA, U2OS, and MC3T3E1 cell lines were obtained from ATCC and maintained in DMEM-HEPES (Gibco-BRL, Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin, and 10 mg/ml streptomycin. The mouse preosteoblast cell line MC3T3E1 was cultured in α-MEM supplemented with 10% FBS. Human osteosarcoma samples were obtained from the Peter MacCallum Cancer Centre tissue bank following patient consent. All procedures using human tissue were reviewed and approved by the Ethics Committee of Peter MacCallum Cancer Centre. Trabecular bone specimens were obtained from patients undergoing routine hip replacement surgery and processed as previously described (57). Osteoblast differentiation was induced with 100 ng/ml BMP2, or 10 mM β-glycerophosphate and 25 μg/ml ascorbic acid, for 21 d. ALP activity and mineralization using Alizarin red were assayed as previously described (58). Primary human osteoblasts were immortalized by infection with retrovirus containing the HPV16 E6 and E7 oncogenes (LXSN-HPV16-E6/E7) or control vector as described previously (59). After transduction, cells were selected in media containing G418 (50 μg/ml). Colony-forming assays were carried out as described previously (60).
Transcriptional profiling.
The high-throughput screen for epigenetically silenced genes linking differentiation and transformation in osteosarcoma was undertaken as follows. Osteosarcoma cell lines B143B, G292, HOS, SAOS2, and SJSA were cultured until 70% confluent and treated for 3 d with 5–10 μM dAC (Sigma-Aldrich). The concentration of dAC used was empirically determined in each line to induced cell cycle arrest but not apoptosis. Total RNA was then isolated, and molecular profiling was performed using 2-color cDNA arrays as described previously (27). All array experiments were performed independently twice. Data were normalized with LOWESS and analyzed using GeneSpring GX software (version 7.3.1; Silicon Genetics; ref. 61). Levels in dAC-treated samples were normalized to those of untreated cells, data from replicate experiments were averaged for each line, and median values derived from all 6 cell lines were used for analyses (Supplemental Table 1).
The array data set on 30 primary osteosarcomas (28) was provided by D.A. Stephan (Translational Genomics Research Institute, Phoenix, Arizona, USA). This data set was generated on Affymetrix U95Av2 arrays. The gene cassettes described were generated from genes selected for their role in osteoblast differentiation or proliferation based on exhaustive literature review (Supplemental Table 3). Wif1 expression was defined as present or absent by Affymetrix calls on each array. Finally, profiling of primary human osteoblasts was undertaken using the Affymetrix U133Av2.0 arrays. Data from the methylation analyses and the primary osteosarcoma data sets were clustered after log transformation and median normalization using Spearman’s rank correlation and centroid linkage in Cluster (version 3.1) and presented graphically using Treeview (version 1.1.1; both available at http://rana.lbl.gov). This analysis segregated the samples into 3 distinct clades (clusters). The correlation coefficient for cluster C by this analysis was 0.31, for cluster B, 0.34; and for cluster A, 0.31 (Figure 7C).
Methylation analyses.
CpG island methylation of the WIF1 promoter region was analyzed using quantitative high-throughput mass spectrometry on SEQUENOM’s EpiTYPER platform (29). The details of primers used to amplify the proximal human WIF1 promoter and first exon (chromosome 12, 63,800,758–63,802,522 bp) are given in Supplemental Table 4. The median amplicon length was 419 bp (min, 314; max, 467), with a median of 22 CpGs per amplicon (min, 13; max, 46). To estimate the differences between groups in mean scaled methylation level for each CpG, linear models were fitted to the data. An ANOVA was then performed on each model to determine which of these differences were significant at the 5% level. Bisulfite restriction analysis was undertaken as previously described (62), using methylation-specific and methylation-independent primers (Supplemental Table 4).
Fluorescence-activated cell sorting.
NHB cells were stained for STRO1 and ALP clone B4-78 (R&D systems) as described previously (63). NHB cells were treated with 1 ng/ml PDGF for 3 d to optimize the STRO1+ population and sorted on a BD LSRII flow cytometry system (BD Biosciences). For cell sorting, 107–108 NHB cells were stained, and STRO1+ALP– or STRO1–ALP+ cells were sorted and collected using a FACS Diva flow cytometry system (BD Biosciences).
Reporter assays.
Osteosarcoma cell lines were transfected with TOP-Flash, control FOP-Flash (gifts of R. Moon, University of Washington, Seattle, Washington, USA), or a reporter containing 6 tandem runx2 binding sites from the OC promoter 6OSE2-luciferase (60) using FuGENE 6 (Roche). Constructs were cotransfected with Wif1 vector (hWIF1PJCH76; provided by J.C. Hsieh, SUNY Stony Brook, Stony Brook, New York, USA), and β-galactosidase–expressing vector was used as a control. After 24 h, luciferase and β-galactosidase assays were performed according to the manufacturers’ instructions (Promega and Roche).
RNA analyses.
RNA was prepared and converted to cDNA using standard techniques. Semiquantitative PCR was carried out using Hotstar Taq (Qiagen) according to the manufacturer’s instructions. Real-time RT-PCR was carried out using SyBr Green (Applied Biosystems) according to the manufacturer’s instructions using an ABI-Prism 7000 Sequence Detection System. All primer sequences are listed in Supplemental Table 4.
WIF1 siRNA knockdown studies.
To knock down Wif1 in MC3T3E1 cells, sequences from mouse Wif1 (Supplemental Table 4) were cloned into pSUPER.retro.puro (64). Ecotropic retroviral supernatants produced using Phoenix packaging cells were used to infect MC3T3E1 cells, which were then selected using 2 μg/ml puromycin.
Western blot analysis.
Western blots were carried out using whole cell extracts, separated on 10% SDS-PAGE genes, and transferred to PVDF (Millipore). Blots were stained with anti-mouse Wif1 mAb (clone 134714; R&D Systems), anti-human WIF1 mAb (clone 133015; R&D Systems), β-catenin (clone 14; BD Biosciences), or rabbit anti-Runx2 (human/rodent; CeMines). Western blots were developed using enhanced chemiluminescence (Amersham Biosciences).
Immunohistochemistry and immunofluorescence studies.
The osteosarcoma tissue microarray was a gift from B. Rubin (Cleveland Clinic Lerner College of Medicine, Cleveland, Ohio, USA). Each tissue microarray contained a section of normal bone and 96 osteosarcomas. Sections were stained with H&E and antibodies against anti-human Wif1 mAb (clone 133015; R&D Systems), β-catenin (clone 14; Dako), and PCNA (clone PC10; Dako). H&E staining was graded as follows: 0, no staining; <2, less than 50% staining; 2, 50%–90% staining; 3, greater than 90% staining of sections. Cells were treated with dAC or wheat germ–derived human recombinant WIF1 (Abnova) for 2 d or 8–16 h, respectively. Cells were fixed in 4% freshly prepared paraformaldehyde for 20 min and permeabilized with 0.2 % Triton X-100 for 10 min. After blocking, cells were incubated with β-catenin antibody (diluted 1:100) for 2 h. After washing, cells were incubated with Alexa Fluor 488–conjugated goat anti-mouse IgG (diluted 1:1,000; Invitrogen), and with 2 μg/ml propidium iodide for 10 min. Immunostaining was analyzed by laser scanning confocal microscopy, and intensity was semiquantitatively assessed using Metamorph software (version 7.5; Molecular Devices).
Wif1 LacZ reporter knockin mice.
All procedures using animals were reviewed and approved by the NICHD, NIH, Animal Care and Use Committee. To generate the Wif1 LacZ knockin construct, a BAC clone (Genome Systems) containing the complete Wif1 gene locus was used. A restriction fragment (approximately 5 kb) containing the first 2 Wif1 exons with extensive upstream and intron sequences was subcloned into pBluescript II vector and subjected to site-directed mutagenesis (Quikchange; Stratagene). To disrupt the Wif1 coding sequence and to insert a tau-LacZ reporter gene, the initiation methionine (NcoI site) was mutated to create a PacI restriction site (see Supplemental Table 4 for primer details). The tau-LacZ/neomycin cassette inserted into the PacI site was under the control of endogenous Wif1 promoter and enhancers, and at the same time the insertion disrupted normal Wif1 transcription and translation because it introduced multiple in-frame stop codons. The viral thymidine kinase gene was subcloned into the 5′ end of the Wif1 genomic sequence to allow for secondary selection of targeted ES cells. The knockin construct was linearized, electroporated into 129 ES cells, and selected by standard protocols. After selection, more than 300 ES clones were isolated and expanded. We screened 100 individual clones using Southern blots, and at least 20% exhibited correct insertion of the tau-LacZ cassette into the Wif1 locus. ES clone no. 58 was used to generate chimeric animals (65), which were outcrossed to obtain heterozygous animals. The background of these mice, referred to herein as Wif1–/–, was C57BL/6×129/SV. Mice were genotyped using a PCR-based method using primers listed in Supplemental Table 4. In E12.5 heterozygous embryos, β-galactosidase activity was detected by whole-mount staining with X-gal (66).
Quantitative CT and bone histomorphometry.
Wif1–/– and control littermates (n = 3–6 per group, male and female) were analyzed for bone developmental defects at 4 wk ± 2 d and for remodeling defects at 16 wk ± 1 wk and 1 yr. Femora and tibiae were fixed in 10% buffered formalin. Femoral cortical and trabecular bone mineral density, femoral circumference, and cortical thickness were measured by peripheral quantitative CT (XCT Research SA+; Stratec Medizintechnik GmbH) as previously described (67). For histomorphometry, mice were injected with 20 mg/kg calcein at 10 d and 3 d prior to killing. Femora and tibiae were fixed and embedded in methylmethacrylate, and 5-μm sections were stained with toluidine blue or analyzed unstained for fluorochrome labels according to standard procedures in the proximal tibia using the Osteomeasure system (Osteometrics Inc.) (68).
Radiation-induced osteosarcoma model.
Wif1–/– mice and littermate controls were injected weekly with 1 μCi/g 45Ca intraperitoneally from 4 to 8 wk of age (Figure 7B), as described previously (35). Mice were monitored for tumorigenesis, limping, paralysis (caused by spinal metastasis), loss of condition, poor feeding or grooming, or greater than 20% weight loss. Mice were scanned using 18F-PET to localize osteoblastic lesions (69), and then euthanized and scanned using CT prior to dissection.
Statistics.
Where single comparisons were made, 2-tailed Student’s t test or standard χ2 test was used, with corrections as specified in the text. For multiple comparisons, standard ANOVA was used. For time-to-event and survival analyses, significance was estimated using Cox proportional hazard regression models. The statistical analyses associated with the array studies are described above. Significance was conventionally accepted at a P value less than 0.05.
Supplementary Material
Acknowledgments
The authors would like to thank colleagues in the Bowtell and Thomas laboratories for helpful discussions and Alexander Grinberg, Andreas Tomac, Heiner Westphal, Martha Rebbert, and Paul Love for help in generating gene-modified mice. We thank Carleen Cullinane and staff for PET scanning of the mice. This work was supported by grants 350432 and 508982 from the National Health and Medical Research Council, by the Cancer Council Victoria, by the Sarcoma Foundation of America, by and the Intramural Research Program of the NICHD, NIH. D.M. Thomas was supported by R.D. Wright Career Development Award 2003-7 and by Victorian Cancer Agency Clinician Researcher Fellowship 2008-10.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: ALP, alkaline phosphatase; BMP, bone morphogenetic protein; COL1A1, collagen type 1A1; dAC, 5-aza-2-deoxycytidine; Dkk, Dickkopf; HPV, human papilloma virus; LEF, lymphoid enhancer–binding factor; OC, osteocalcin; OP, osteopontin; OPG, osteoprotegerin; Sfrp, secreted frizzled-related protein; Sost, sclerostin; TCF, T cell factor; Wif1, Wnt inhibitory factor 1.
Citation for this article: J. Clin. Invest. 119:837–851 (2009). doi:10.1172/JCI37175
Michael Tsang’s present address is: Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Laurent Kodjabachian’s present address is: Institut de Biologie du Développement de Marseille-Luminy, UMR 6216, CNRS–Université de la Méditerranée, Marseille, France.
See the related Commentary beginning on page 758.
References
- 1.International Agency for Research on Cancer. 2002. Pathology and genetics of tumours of soft tissue and bone. C.D.M. Fletcher, K.K. Unni, and F. Mertens, editors. IARC Press. Lyon, France. 427 pp. [Google Scholar]
- 2.Kansara M., Thomas D.M. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 3.Dahlin, D.C. 1957. Bone tumors. Charles C. Thomas Publisher Ltd. Springfield, Illinois, USA. 224 pp. [Google Scholar]
- 4.Thomas D., Kansara M. Epigenetic modifications in osteogenic differentiation and transformation. J. Cell. Biochem. 2006;98:757–769. doi: 10.1002/jcb.20850. [DOI] [PubMed] [Google Scholar]
- 5.Hochedlinger K., et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mintz B., Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo P.E., Mann J.R. Allelle-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 8.Macsai C.E., Foster B.K., Xian C.J. Roles of Wnt signalling in bone growth, remodelling, skeletal disorders, and fracture repair. J. Cell. Physiol. 2008;215:578–587. doi: 10.1002/jcp.21342. [DOI] [PubMed] [Google Scholar]
- 9.Glass D.A., 2nd, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Boyden L.J., et al. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 11.Bodine P.V., Seestaller-Wehr L., Kharode Y.P., Bex F.J., Komm B.S. Bone anabolic effects of parathyroid hormone are blunted by deletion of the Wnt antagonist secreted frizzled-related protein-1. J. Cell. Physiol. 2007;210:352–357. doi: 10.1002/jcp.20834. [DOI] [PubMed] [Google Scholar]
- 12.Morvan F., et al. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J. Bone Miner. Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 13.Kawano Y., Kypta R. Secreted antagonist of the Wnt signalling pathway. J. Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 14.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 15.Baron R., Rawadi G. Targeting the Wnt/beta-catenin pathway to regulate bone formation in the adult skeleton. Endocrinology. 2007;148:2635–2643. doi: 10.1210/en.2007-0270. [DOI] [PubMed] [Google Scholar]
- 16.Canalis E., Giustina A., Bilezikian J.P. Mechanisms of anabolic therapies for osteoporosis. N. Engl. J. Med. 2007;357:905–916. doi: 10.1056/NEJMra067395. [DOI] [PubMed] [Google Scholar]
- 17.Polakis P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Haydon R.C., et al. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int. J. Cancer. 2002;102:338–342. doi: 10.1002/ijc.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwaya K., et al. Cytoplasmic and/or nuclear staining of beta-catenin is associated with lung metastasis. Clin. Exp. Metastasis. 2003;20:525–529. doi: 10.1023/A:1025821229013. [DOI] [PubMed] [Google Scholar]
- 20.Iwoa K., et al. Frequent beta-catenin abnormalities in bone and soft-tissue tumors. Jpn. J. Cancer Res. 1999;90:205–209. doi: 10.1111/j.1349-7006.1999.tb00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 22.Aguilera O., et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–4121. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 23.Caldwell G.M., et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–888. doi: 10.1158/0008-5472.CAN-03-1346. [DOI] [PubMed] [Google Scholar]
- 24.Elston M.S., et al. Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology. 2008;149:1235–1242. doi: 10.1210/en.2007-0542. [DOI] [PubMed] [Google Scholar]
- 25.Mazieres J., et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 26.Roman-Gomez J., et al. Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood. 2007;109:3462–3469. doi: 10.1182/blood-2006-09-047043. [DOI] [PubMed] [Google Scholar]
- 27.Morgan T., et al. Molecular profiling of giant cell tumor of bone and the osteoclastic localization of ligand for receptor activator of nuclear factor kappaB. Am. J. Pathol. 2005;167:117–128. doi: 10.1016/s0002-9440(10)62959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mintz M.B., et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65:1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 29.Ehrich M., et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W.Y., et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat. Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh J.C., et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 32.Ai L., et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi H., et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–7952. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- 34.Vaes B.L., et al. Comprehensive microarray analysis of bone morphogenetic protein 2-induced osteoblast differentiation resulting in the identification of novel markers for bone development. J. Bone Miner. Res. 2002;17:2106–2118. doi: 10.1359/jbmr.2002.17.12.2106. [DOI] [PubMed] [Google Scholar]
- 35.Finkel M.P., Jinkins P.B., Biskis B.O. Parameters of radiation dosage that influence production of osteogenic sarcomas in mice. Natl. Cancer Inst. Monogr. 1964;14:243–269. [PubMed] [Google Scholar]
- 36.Scotlandi K., et al. Clinical relevance of Ki-67 expression in bone tumors. Cancer. 1995;75:806–814. doi: 10.1002/1097-0142(19950201)75:3<806::AID-CNCR2820750310>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Urakami S., et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin. Cancer Res. 2006;12:383–391. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 38.Baylin S.B., Herman J.G. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- 39.DiNardo D.N., Butcher D.T., Robinson D.P., Archer T.K., Rodenhiser D.I. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20:5331–5340. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- 40.Mancini D.N., et al. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 41.Sadikovic B., et al. In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma. PLoS ONE. 2008;3:e2834. doi: 10.1371/journal.pone.0002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graff J.R., Herman J.G., Myohanen S., Baylin S.B., Vertino P.M. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 43.Brandeis M., et al. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 44.Siegfried Z., et al. DNA methylation represses transcription in vivo. Nat. Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 45.Queimado L., Lopes C.S., Reis A.M. WIF1, an inhibitor of the Wnt pathway, is rearranged in salivary gland tumors. Genes Chromosomes Cancer. 2007;46:215–225. doi: 10.1002/gcc.20402. [DOI] [PubMed] [Google Scholar]
- 46.Gong Y., et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/S0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 47.Besplug J., et al. Sex and tissue-specific differences in low-dose radiation-induced oncogenic signaling. Int. J. Radiat. Biol. 2005;81:157–168. doi: 10.1080/09553000500103512. [DOI] [PubMed] [Google Scholar]
- 48.Balemans W., Van Hul W.D. The genetics of low-density lipoprotein receptor-related protein 5 in bone: a story of extremes. Endocrinology. 2007;148:2622–2629. doi: 10.1210/en.2006-1352. [DOI] [PubMed] [Google Scholar]
- 49.Li X., et al. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat. Genet. 2005;37:945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- 50.Kahler R.A., Westendorf J.J. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J. Biol. Chem. 2003;278:11937–11944. doi: 10.1074/jbc.M211443200. [DOI] [PubMed] [Google Scholar]
- 51.Coussens A.K., et al. Identification of genes differentially expressed by prematurely fused human sutures using a novel in vivo - in vitro approach. Differentiation. 2007;76:531–545. doi: 10.1111/j.1432-0436.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt I.U., Dobnig H., Turner R.T. Intermittent parathyroid hormone treatment increases osteoblast number, steady state messenger ribonucleic acid levels for osteocalcin, and bone formation in tibial metaphysis of hypophysectomized female rats. Endocrinology. 1995;136:5127–5134. doi: 10.1210/en.136.11.5127. [DOI] [PubMed] [Google Scholar]
- 53.Bellido T., et al. Chronic elevation of parathyroid hormone in mice reduces expression of sclerostinby osteocytes: a novel mechanism of hormonal control of osteoblastogenesis. Endocrinology. 2005;146:4577–4583. doi: 10.1210/en.2005-0239. [DOI] [PubMed] [Google Scholar]
- 54.Jolette J., et al. Defining a noncarcinogenic dose of recombinant human parathyroid hormone 1-84 in a 2-year study in Fischer 344 rats. Toxicol. Pathol. 2006;34:929–940. doi: 10.1080/01926230601072301. [DOI] [PubMed] [Google Scholar]
- 55.Vahle J.L., et al. Bone neoplasms in F344 rats given teriparatide [rhPTH(1-34)] are dependent on duration of treatment and dose. Toxicol. Pathol. 2004;32:426–438. doi: 10.1080/01926230490462138. [DOI] [PubMed] [Google Scholar]
- 56.Harper K.D., Krege J.H., Marcus R., Mitlak B.H. Osteosarcoma and teriparatide. J. Bone Miner. Res. 2007;22:334. doi: 10.1359/jbmr.061111. [DOI] [PubMed] [Google Scholar]
- 57.Gronthos S., et al. Differential cell surface expression of the STRO-1 and alkaline phosphatase antigens on discrete developmental stages of primary cultures of human bone cells. J. Bone Miner. Res. 1999;14:47–56. doi: 10.1359/jbmr.1999.14.1.47. [DOI] [PubMed] [Google Scholar]
- 58.Sellers W.R., et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller A.D., Rosman G.J. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–982, 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas D.M., et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y.H., et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frommer M., et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simmons P.J., Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55–62. [PubMed] [Google Scholar]
- 64.Brummelkamp T.R., Bernards R., Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/S1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 65.Nagy A., et al. Derivation of completely cell-culture derived mice from early-passage embryonic stem cell. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss D.J., Liggitt D., Clark J.G.1997. . In situ histochemical detection of beta-galactosidase activity in lung: assessment of X-Gal reagent in distinguishing lacZ gene expression and endogenous beta-galactosidase activity. Hum. Gene Ther. 81545–1554. 10.1089/hum.1997.8.13-1545 [DOI] [PubMed] [Google Scholar]
- 67.Sims N.A., Brennan K., Spaliviero J., Handelsman D.J., Seibel M.J. Perinatal testosterone surge is required for normal adult bone size but not for normal bone remodeling. Am. J. Physiol. Endocrinol. Metab. 2006;290:E456–E462. doi: 10.1152/ajpendo.00311.2005. [DOI] [PubMed] [Google Scholar]
- 68.Sims N.A., et al. Bone homeostasis in growth hormone receptor-null mice is restored by IGF-I but independent of Stat5. J. Clin. Invest. 2000;106:1095–1103. doi: 10.1172/JCI10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cullinane C., et al. An in vivo tumor model exploiting metabolic response as a biomarker for targeted drug development. Cancer Res. 2005;65:9633–9636. doi: 10.1158/0008-5472.CAN-05-2285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.