Abstract
The transcription factor serum response factor (SRF) plays a crucial role in the development of several organs. However, its role in the skin has not been explored. Here, we show that keratinocytes in normal human and mouse skin expressed high levels of SRF but that SRF expression was strongly downregulated in the hyperproliferative epidermis of wounded and psoriatic skin. Keratinocyte-specific deletion within the mouse SRF locus during embryonic development caused edema and skin blistering, and all animals died in utero. Postnatal loss of mouse SRF in keratinocytes resulted in the development of psoriasis-like skin lesions. These lesions were characterized by inflammation, hyperproliferation, and abnormal differentiation of keratinocytes as well as by disruption of the actin cytoskeleton. Ultrastructural analysis revealed markedly reduced cell-cell and cell-matrix contacts and loss of cell compaction in all epidermal layers. siRNA-mediated knockdown of SRF in primary human keratinocytes revealed that the cytoskeletal abnormalities and adhesion defects were a direct consequence of the loss of SRF. In contrast, the hyperproliferation observed in vivo was an indirect effect that was most likely a consequence of the inflammation. These results reveal that loss of SRF disrupts epidermal homeostasis and strongly suggest its involvement in the pathogenesis of hyperproliferative skin diseases, including psoriasis.
Introduction
The outermost layer of the skin is formed by the epidermis, a stratified squamous epithelium mainly consisting of keratinocytes. A remarkable property of the epidermis is its self-renewing capacity. Keratinocytes of the basal layer proliferate to generate new cells that differentiate and move upward. Terminally differentiated and metabolically inactive cells form a tightly linked sheet; they are continuously sloughed off from the surface and replaced by cells from the underlying layers (1). To maintain the balance between proliferation and differentiation, a tight spatial and temporal regulation of gene expression is essential. Therefore, it is of particular interest to identify and functionally characterize the transcription factors involved in the control of epidermal homeostasis.
A promising new candidate in this regulation is serum response factor (SRF), a member of the highly conserved MADS box family of transcription factors (2, 3). SRF regulates the expression of immediate early genes (3–5) and of muscle-specific genes (reviewed in ref. 6) after binding to its target sequence (CArG box), which is often located within a serum response element. In fibroblasts, SRF-dependent gene transcription is controlled by 2 types of cofactors that are regulated by different signaling pathways. Cofactors of the myocardin-related transcription factor (MRTF) family can respond to Rho family GTPases and monomeric actin. The second class of SRF coactivators includes the ternary complex factors, which are activated by the MAPK pathway and involve SRF in response to mitogenic and stress stimuli (reviewed in ref. 7). SRF target genes identified in ES cells functionally relate to cell-cycle control, apoptosis, and wound healing, and a substantial portion encode proteins of the cytoskeleton, including several keratins and focal adhesion proteins (8, 9). These findings point to a role of SRF in skin morphogenesis and homeostasis. Surprisingly, however, the expression and function of SRF in keratinocytes has not been investigated so far.
In this study, we analyzed the expression of SRF in keratinocytes of normal, wounded, and diseased skin, and we explored the consequences of SRF downregulation in epidermal and hair follicle keratinocytes of mice. We show that inflammatory skin conditions that are characterized by keratinocyte hyperproliferation are associated with downregulation of SRF expression in the epidermis and that SRF is essential for the maintenance of epidermal homeostasis in mice.
Results
SRF is expressed in keratinocytes of normal skin but downregulated in hyperproliferative epidermis.
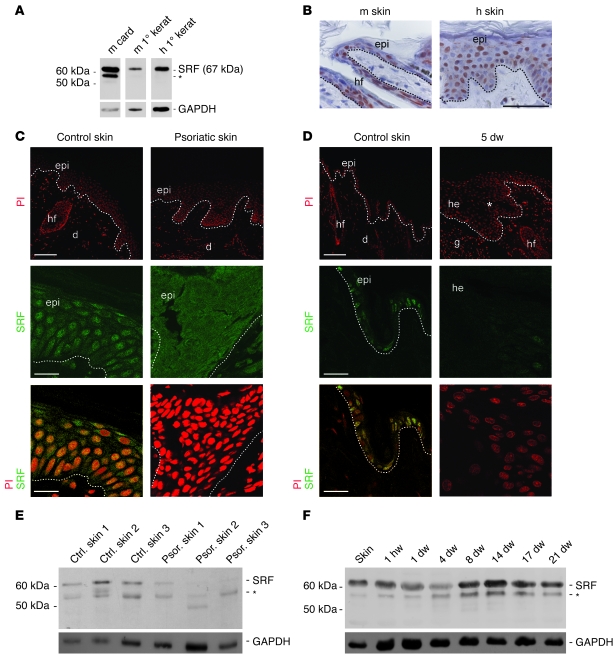
We first analyzed the expression of SRF in keratinocytes of mouse and human skin using a newly developed rat anti-SRF mAb (2C5; this study). A protein of the expected molecular weight (67 kDa) was detected by Western blot analysis in lysates from mouse and human primary keratinocytes (Figure 1A). In lysates from mouse cardiac muscle, we detected an additional protein between 50 and 60 kDa that was also present in total murine skin and murine wound lysates and in lysates from normal and psoriatic human skin (Figure 1, A, E, and F). This band was not present in lysates of primary keratinocytes (Figure 1A), indicating that this putative SRF variant is mainly present in stromal cells in skin. Immunohistochemical stainings of mouse and human skin sections with the SRF mAb revealed a nuclear localization of SRF throughout all epidermal layers (Figure 1, B, C, and D). A similar staining pattern was obtained with a commercially available rabbit polyclonal antibody (Santa Cruz Biotechnology Inc.; data not shown). In contrast, no specific staining was obtained with the secondary antibodies (data not shown).
Figure 1. Downregulation of SRF expression in hyperproliferative epidermis.
(A) SRF expression in murine (m) and human (h) primary (1°) keratinocytes (kerat) and in murine cardiac muscle (card) shown by Western blotting using a rat anti-SRF mAb (2C5). (B) Immunohistochemical analysis of sections from mouse and human skin using the 2C5 antibody shows nuclear localization of SRF in the epidermis (epi). hf, hair follicle. Scale bar: 50 μm. (C) Immunofluorescence stainings of SRF (green, middle and lower panels) show nuclear localization of SRF in epidermal keratinocytes of a healthy patient. Nuclear SRF was not detected in lesional skin of a psoriatic patient. PI-stained sections are shown at low magnification in the upper panels. Representative biopsies (from 5 healthy and 5 psoriatic patients) are shown. Control skin, yellow from double staining with propidium iodide (PI), red. d, dermis. Scale bars: 100 μm (upper panels); 20 μm (middle and lower panels). (D) Immunofluorescence stainings of SRF (middle panels and, in combination with PI, lower panels) show nuclear localization in keratinocytes of normal mouse skin but reduced staining in the hyperproliferative epithelium of 5-day wounds (5 dw). PI-stained sections are shown at low magnification in the upper panels. Asterisk indicates area of hyperproliferative epithelium at the wound edge shown at higher magnification in the lower panels. g, granulation tissue; he, hyperproliferative epithelium. Scale bars: 100 μm (upper panels); 20 μm (middle and lower panels). Dotted lines indicate basement membrane. (E and F) Western blot analysis of SRF using lysates from 3 healthy patients (ctrl) and affected skin of 3 psoriatic (psor) patients (E) or lysates of normal mouse skin and excisional wounds (F). (A, E, and F) Asterisks denote an additional band that is recognized by the anti-SRF antibody. GAPDH was used as a loading control.
SRF affects migration and proliferation of several cell types (8, 10–12). To explore the role of SRF in keratinocyte biology, we analyzed SRF expression in skin conditions that are associated with increased migration and/or proliferation of keratinocytes. First, we analyzed the expression of SRF in psoriatic skin, which is characterized by hyperproliferation of keratinocytes (13). Whereas SRF was abundant in nuclei of keratinocytes in normal human skin, its expression was strongly reduced in affected skin of all psoriasis patients analyzed (Figure 1C). Remarkably, there was an almost complete loss of nuclear SRF staining, and only a weak cytoplasmic signal was seen in some areas (Figure 1C and data not shown). Western blot analysis confirmed the reduction in SRF protein levels in the skin of psoriatic patients compared with that of healthy control patients (Figure 1E). Reduced nuclear SRF staining was also observed in biopsies from patients with lichen planus, an inflammatory skin disease associated with keratinocyte hyperproliferation, and to a much lesser extent in patients showing lichenification as a result of chronic eczematous dermatitis (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI37771DS1). In both situations, the extent of SRF downregulation correlated with the extent of acanthosis. However, the complete loss of SRF that we observed in psoriatic epidermis was not observed in patients with lichen planus or chronic eczematous dermatitis.
We next analyzed murine SRF expression during the healing process of full-thickness excisional mouse wounds. In this wound model, strong keratinocyte migration and proliferation occurs within the first few days after injury (reviewed in ref. 14). We found a transient reduction in SRF protein levels at day 4 after wounding (Figure 1F), when keratinocyte proliferation is strongly increased, followed by a spike in SRF expression during the tissue remodeling phase. This kinetics of SRF expression was observed in 2 independent experiments using wound lysates from different animals (data not shown). SRF regulation was not seen on the mRNA level (data not shown), suggesting posttranscriptional regulation of SRF during reepithelialization, possibly involving microRNA. In accordance with lower SRF protein levels, immunofluorescence revealed strongly reduced nuclear staining in the hyperproliferative epithelium at the edge of 5-day-old wounds compared with nonwounded mouse skin (Figure 1D).
Keratinocyte-specific depletion of SRF in mice.
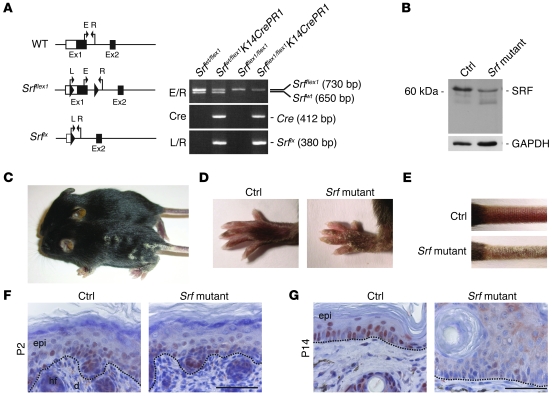
To determine whether the observed downregulation of SRF expression in hyperproliferative epidermis is functionally important, we attempted to generate mice with a keratinocyte-specific knockout of the Srf gene. Mice carrying Srf alleles flanked by loxP sites (floxed; Srfflex1/flex1; ref. 15) were mated with heterozygous floxed Srf mice that also express Cre recombinase under the control of the keratin (K) 5 promoter (K5Cre; ref. 16). The latter is active in basal cells of stratified epithelia, in particular in keratinocytes of the skin (ref. 16 and Supplemental Figure 2, A–C). From these breedings, Srfflex1/flex1K5Cre offspring could not be obtained at postnatal stages. None of the animals that we genotyped at P28 was SRF deficient. Since we did not observe postnatal death, it seems most likely that the mutant mice die before birth. In timed matings, we could only detect them until E16.5, further indicating embryonic lethality of mice with 2 recombined Srf alleles (Supplemental Figure 3A). Some of the Srfflex1/flex1K5Cre embryos that we analyzed at days E14.5 and E15.5 showed severe edema, skin blistering (Supplemental Figure 3, B–D), and hemorrhage (Supplemental Figure 3B).
To circumvent the embryonic lethality, we aimed to generate an inducible conditional mutation of Srf using K14CrePR1 mice (17). These animals allow the activation of Cre recombinase by treatment with RU486 (18). Srfflex1/flex1K14CrePR1 mice (Srf mutant mice) were obtained at the expected Mendelian ratio by crossing Srfflex1/flex1 with SrfWT/flex1K14CrePR1 mice. The homozygous mutant mice appeared normal at birth. However, skin lesions of variable severity appeared spontaneously around day 7 after birth on all of the more than 40 animals that we obtained. Genotyping showed recombination of the Srf allele in newborn mice (Figure 2A), but at that stage Srf mutant mice were indistinguishable from their littermates. Thus, activation of Cre recombinase obviously occurred to a certain extent, although we did not treat the animals with RU486. Histology of back skin at P2 did not reveal any obvious abnormalities, and SRF expression was similar in control and Srf mutant mice (Figure 2F). Skin abnormalities of Srf mutant mice were first visible at 1 week after birth and progressed rapidly. At 2 weeks after birth (P14), severe scales were observed on the paws (Figure 2D) and the tail (Figure 2E), and mutant mice often showed several scaling plaques on the back (Figure 2C). The ears as well as the eyelid epithelium were also affected, but the severity of the phenotype diverged markedly between different litters. Srf mutant mice had to be sacrificed 3–6 weeks after birth to avoid suffering of the animals. The general weakness of the animals also precluded further activation of the inducible Cre protein by RU486 treatment.
Figure 2. Spontaneous recombination of the floxed Srf allele in Srfflex1/flex1K14CrePR1 mice.
(A) Schematic illustration of Srf alleles and genotyping of Srfflex1/flex1 × SrfWT/flex1K14CrePR1 offspring: Srfflex1/flex1K14CrePR1 (Srf mutant) mice were identified by a 730-bp fragment (Srfflex1), a 412-bp fragment (Cre recombinase), and the absence of the 650-bp fragment that identifies the WT allele (SrfWT). A 380-bp fragment was amplified from the recombined Srf allele (Srflx). E/R and L/R indicate primer pairs described in ref. 15. (B) A representative Western blot of total back skin lysates showing reduced SRF protein levels in the skin of Srfflex1/flex1K14CrePR1 mice at P14 compared with skin of control littermates. GAPDH was used as a loading control. (C–E) Macroscopic appearance of control and Srf mutant mice at P14. Note the scaly lesions on the back (C), the paws (D), and the tail (E). (F) Immunohistochemical analysis of SRF in back skin from Srf mutant and control mice at P2 shows comparable SRF expression patterns. (G) At P14, nuclear SRF staining is markedly reduced in the epidermis of Srfflex1/flex1K14CrePR1 mice. (F and G) Dotted lines indicate the basement membrane. Scale bars: 50 μm. Sections were counterstained with hematoxylin.
Western blot analysis of total skin lysates revealed reduced SRF protein levels in skin of mutant mice at P14 (Figure 2B). As expected, this was due to loss of SRF in keratinocytes as determined by immunohistochemical analysis of skin sections from Srf mutant mice (Figure 2G). Nuclear SRF staining was completely absent in lesional epidermis, and only a weak cytoplasmic staining was detected in some areas.
Epidermal hyperplasia and increased keratinocyte proliferation in skin of Srf mutant mice.
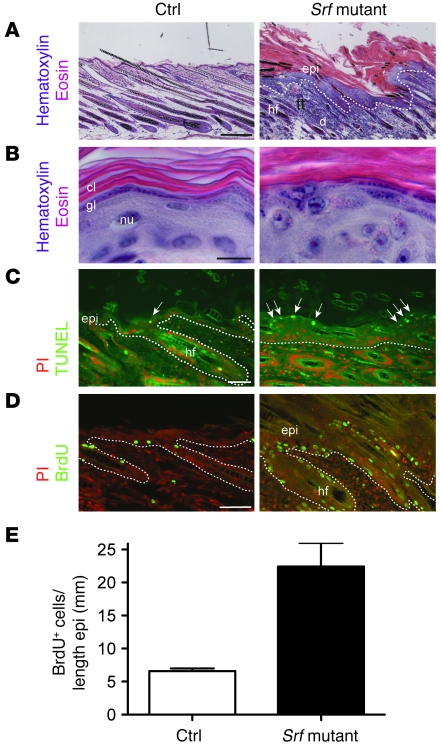
To examine the consequences of reduced SRF expression in keratinocytes, we analyzed the skin of Srf mutant mice between P11 and P15. Histology of back and tail skin revealed hyperthickening of most parts of the epidermis. Lesions of back and tail skin showed strongly thickened epidermis (acanthosis) with a hyperthickened stratum corneum (hyperkeratosis) containing nucleated cells (parakeratosis) (Figure 3, A and B, and Figure 2G). A granular layer was still present in most of the lesions (Figure 3B), although it was hardly detectable in severely affected skin (data not shown). These abnormalities were confirmed by ultrastructural analysis (see below).
Figure 3. Loss of SRF expression in keratinocytes leads to hyperproliferative epidermis.
(A and B) H&E staining of back skin sections of control and Srf mutant mice shows epidermal hyperthickening and parakeratosis in skin lesions of Srf mutant mice. At high magnification, the presence of a granular layer was still seen in most of the mutant mice (B). Scale bar: 10 μm. cl, cornified layer; gl, granular layer; nu, nucleus. (C) TUNEL staining in combination with PI shows an increased number of apoptotic keratinocytes in lesional skin of Srf mutant mice (indicated by arrows). (D) Immunofluorescence analysis in combination with PI shows an increased number of proliferating keratinocytes by BrdU staining. Dotted lines indicate the basement membrane. (E) SRF mutant epidermis shows an increased number of BrdU-positive cells compared with control epidermis. n = 3 per genotype. Error bars indicate mean ± SEM. Scale bars: 200 μm (A); 10 μm (B); 20 μm (C); 100 μm (D).
TUNEL staining revealed only very few apoptotic keratinocytes in control epidermis. In contrast, an increased number of TUNEL-positive cells were detected in lesional skin of Srf mutant mice (Figure 3C). This was further confirmed by ultrastructural analysis (data not shown). Labeling with BrdU showed an increase in the number of proliferating keratinocytes in hair follicles and in the interfollicular epidermis, and BrdU-positive cells were no longer restricted to the basal layer (Figure 3D). Even in mild lesions, the number of BrdU-positive keratinocytes in the epidermis was 3-fold higher than in control mice (Figure 3E), and the hyperproliferation was even more pronounced in severe lesions (data not shown). We also detected suprabasal expression of p63, a key regulator of epidermal development and differentiation (19) that is predominantly expressed in keratinocytes of the basal layer of normal skin (data not shown).
Disruption of the actin cytoskeleton and of cell-cell contacts in Srf mutant epidermis.
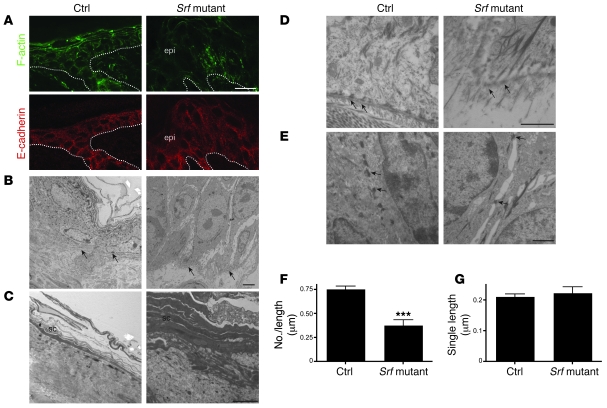
We next characterized the organization of F-actin in Srf mutant keratinocytes, since SRF is a direct regulator of the actin cytoskeleton (20). Phalloidin staining was irregular in most areas of Srf mutant epidermis, revealing alterations in actin polymerization. In severe lesions, phalloidin staining was strongly reduced (Figure 4A) and there was also a loss of cell compaction in all epidermal layers, as determined by ultrastructural analysis (see below). E-cadherin, a major component of adherens junctions, was present in Srf mutant keratinocytes, and we could not observe a difference in the staining pattern between control and mutant mice by immunofluorescence (Figure 4A).
Figure 4. Disruption of the actin cytoskeleton and reduced cell-cell and cell-matrix contacts in Srf mutant mice.
(A) Phalloidin-FITC staining of skin sections of control and Srf mutant mice at P14 shows markedly reduced levels of polymerized actin (F-actin) in lesions of Srf mutant skin (upper panels). Immunofluorescence staining for E-cadherin shows membrane localization in skin of control and Srf mutant mice (lower panels). Dotted lines indicate the basement membrane. Scale bar: 20 μm. (B–E) Ultrastructural analysis of control and Srf mutant epidermis at P14. (B) Keratinocytes in the basal cell layer of lesional Srf mutant epidermis are strongly enlarged and lack most cell-cell contacts compared with control skin, in which basal keratinocytes are small and densely packed. Arrows indicate the basement membrane. (C) Stratum corneum (sc) of lesional Srf mutant epidermis is thicker and less compact than stratum corneum of control epidermis. (D) Epidermal lesions of Srf mutant skin show areas with defective hemidesmosomes (arrows). Note that collagen bundles below the basement membrane are absent in Srf mutant skin. (E) Desmosomes (arrows) are markedly reduced in Srf mutant skin, and cell-cell contacts are largely absent. (F and G) Morphometric analysis confirmed a distinct reduction of the number of desmosomes (F), while no change of the mean length of single desmosomes could be found (G). Error bars indicate SD. Scale bars: 5 μm (B and C); 1 μm (D and E). ***P ≤ 0.001.
Ultrastructural analysis of Srf mutant epidermis revealed markedly enlarged keratinocytes in the basal layer (Figure 4B), and some of the keratinocytes in severe lesions revealed features of apoptosis, including chromatin condensation (data not shown). Cells of the stratum corneum were stratified but were significantly thicker and less densely packed (Figure 4C). The basement membrane was not obviously affected, but the ultrastructural analysis revealed areas of lesional skin with severe defects in hemidesmosomes, reduction in hemidesmosome number, and absence of collagen bundles below the basement membrane (Figure 4D). There was also a severe reduction of intact desmosomes (Figure 4E). This was verified by morphometric analysis of 10 randomly selected epidermal areas from 2 mice per genotype. The number of desmosomes between keratinocytes was significantly lower in mutant mice (0.379 ± 0.241 desmosomes/μm cell membrane) compared with control mice (0.755 ± 0.148 desmosomes/μm cell membrane; Figure 4F), while no change of the mean length of single desmosomes was detected (control: 0.212 ± 0.081 μm; Srf mutant: 0.224 ± 0.129 μm; Figure 4G). Cell-cell contacts between desmosomes were largely absent, resulting in extended intercellular gaps (Figure 4E). These results indicate that impaired actin polymerization prevents the formation of anchoring junctions in lesional epidermis of Srf mutant mice.
Impaired keratinocyte differentiation in Srf mutant epidermis.
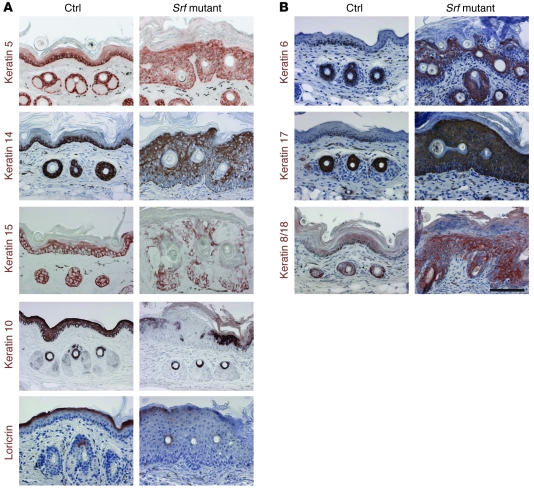
As expected from the histological abnormalities seen in the epidermis of Srf mutant mice, keratinocyte differentiation was impaired. Staining of lesional tail skin with antibodies against the basal keratins K14, K15, and K5 revealed strongly reduced levels of these keratins in basal keratinocytes of Srf mutant mice compared with control animals (Figure 5A). Cells with positive staining for K14 and K15 were scattered throughout the epidermis, and K5 staining was seen in all epidermal layers. The expression of the early differentiation marker K10 was markedly reduced and delayed. In some areas of particularly severe lesions, expression of K10 almost completely disappeared. These regions also lacked expression of loricrin, a protein of the cornified envelope used as a late differentiation marker (Figure 5A). Impaired keratinocyte differentiation of Srf mutant epidermis was also indicated by defective lamellar bodies that were characterized by large vacuoles (data not shown). Thus, both the actin cytoskeleton and the intermediate filament network were distorted. Consistent with the hyperproliferative skin phenotype, K6 and K17 were strongly expressed in interfollicular epidermis of mutant animals, whereas expression of these keratins was restricted to hair follicles in control mice (Figure 5B). Finally, keratinocytes in skin lesions aberrantly expressed K8 and K18, which are normally restricted to single-layered or simple epithelia (Figure 5B).
Figure 5. Keratinocyte differentiation is impaired in Srf mutant epidermis.
(A and B) Immunohistochemical analysis of tail-skin sections from control and Srf mutant mice (P11–P15) for the expression of epidermal differentiation markers. (A) Expression of K14, K15, and K5 was reduced and their localization was irregular in Srf mutant tail skin. Expression of the differentiation markers K10 and loricrin was delayed and reduced compared with that in control skin. (B) Srf mutant mice show abnormal interfollicular expression of K6, K17, and K8/K18. Sections were counterstained with hematoxylin. Scale bar: 100 μm.
Skin lesions of Srf mutant mice show features of inflammatory human skin disease.
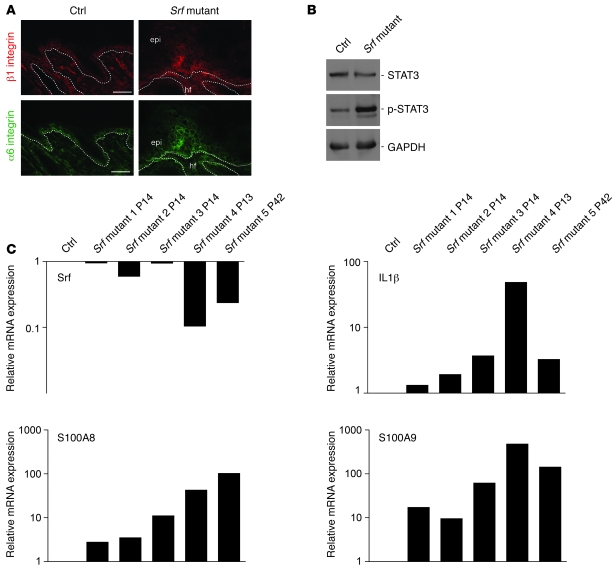
Expression of the β1 integrin subunit is restricted to the basal layer in normal epidermis. Its suprabasal expression is associated with hyperproliferation and perturbed differentiation of keratinocytes and skin inflammation, as seen in psoriasis (21). In the skin lesions of Srf mutant mice, we reproducibly found suprabasal expression of the β1 integrin subunit in the interfollicular epidermis (Figure 6A). Staining with an antibody against the α6 integrin subunit indicated that hemidesmosomes were still present in mutant mice, although electron microscopy revealed that their number was reduced (see above). However, similar to the staining pattern seen with the β1 integrin antibody, α6 integrin staining was extended and was also seen in suprabasal layers of lesional epidermis (Figure 6A). In contrast, suprabasal integrin expression was never seen in control skin.
Figure 6. Skin lesions of Srf mutant mice reveal markers of inflammatory skin disease.
(A) Immunofluorescence analysis of skin sections shows high suprabasal expression of the β1 integrin (upper panels) and α-integrin (lower panels) subunits in Srf mutant skin. Scale bars: 20 μm. Dotted lines indicate the basement membrane. (B) p-STAT3 levels are markedly increased in total skin lysates from Srf mutant mice. GAPDH was used as a loading control. (C) Relative mRNA levels of Srf, IL-1β, S100A8, and S100A9 in 5 individual Srf mutant mice are shown (1–3, P14; 4, P13: severe scaling phenotype; 5, P42: slowly progressing phenotype). Reduced Srf expression correlates with increased IL-1β, S100A8 and S100A9 mRNA levels. RNA levels of a control littermate are set as 1. mRNA of the housekeeping gene Gapdh was used for normalization. PCR analysis was performed in duplicate, and the average of both values is shown. Each result was reproduced in an independent experiment.
Epidermal hyperthickening, infiltration of immune cells, expression of proinflammatory cytokines, and activation of the signaling protein STAT3 are additional features of inflammatory human skin diseases, in particular of psoriasis (13, 22). Therefore, we determined whether such abnormalities are also present in Srf mutant mice. Indeed, Western blot analysis revealed strongly elevated levels of phosphorylated (activated) STAT3. The levels of total STAT3 differed markedly among individual mice, but increased activation of STAT3 in Srf mutant skin was consistently observed in all samples analyzed (Figure 6B).
Factors that activate STAT3 are frequently produced by inflammatory cells. Indeed, we found infiltrating leukocytes in lesional skin by ultrastructural analysis (Supplemental Figure 4A). Staining with antibodies against different inflammatory cells confirmed the presence of an infiltrate. Thus, hyperthickened areas of mutant skin showed an increased number of CD3-positive cells in the dermis and epidermis compared with control skin (Supplemental Figure 4B). Staining for the neutrophil-specific marker Ly6G revealed that the lesions are also infiltrated with a high number of neutrophils (data not shown). In Srf mutant skin, we also detected more MHC class II–positive (MHCII-positive) cells. Most of them are obviously macrophages, since they were stained with an antibody against a macrophage-specific lectin. Additionally, more dermal mast cells were present in Srf mutant skin, as shown by toluidine blue staining (Supplemental Figure 4B). The dermis of lesional skin was also highly vascularized, and the vessels below the hyperproliferative skin were enlarged (Supplemental Figure 4B).
Consistent with the presence of an inflammatory infiltrate, we found strongly increased mRNA levels of the proinflammatory cytokine IL-1β (Figure 6C). mRNA levels of the chemokines S100A8 and S100A9, which are not present in normal skin but overexpressed in wounded skin and in psoriasis (23), were also strongly increased in Srf mutant skin. The corresponding Srf mRNA levels in total skin of Srf mutant mice (which includes SRF-negative keratinocytes and SRF-positive stromal cells) are shown in the uppermost panel. Remarkably, the upregulation of cytokine expression correlated with the extent of SRF downregulation, indicating that the loss of SRF is indeed responsible for the inflammatory response.
SRF affects spreading and adhesion but not proliferation of cultured keratinocytes.
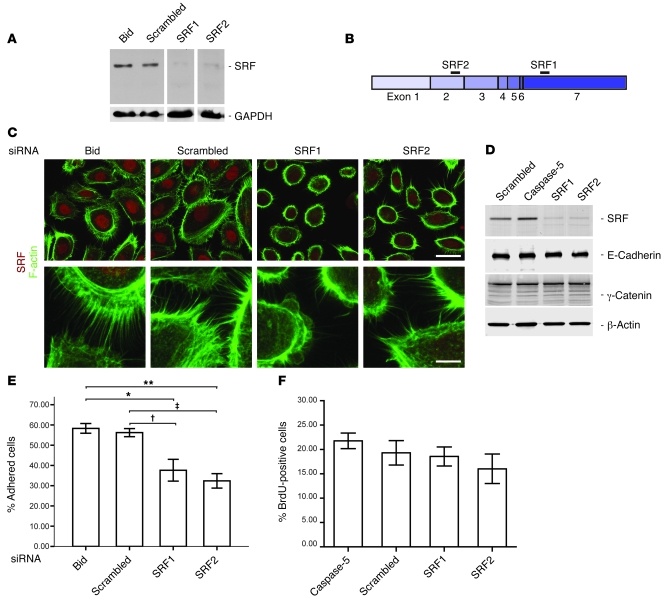
Since infiltrating inflammatory cells are known to influence epidermal homeostasis via secretion of growth factors and cytokines, we wondered whether the observed keratinocyte hyperproliferation seen in Srf mutant skin is a cell autonomous effect or is secondary to the severe inflammation in lesional skin. In addition, it is unclear whether the defect in cell adhesion directly results from the loss of SRF in keratinocytes. To address these issues, we used siRNA-mediated knockdown of SRF in human primary keratinocytes to characterize SRF-mediated cellular changes. Downregulation of SRF was efficiently achieved with 2 different SRF siRNAs (Figure 7A). Binding sites of the used siRNAs were located in exon 2 (SRF1) or in the 3′ UTR of exon 7 (SRF2) of the SRF mRNA, respectively, and are depicted in Figure 7B. Scrambled siRNA and siRNAs against mRNAs of unrelated proteins (Bid and caspase-5) were used as controls. Substantial nuclear SRF expression was seen in the control cultures, but nuclear staining was much weaker after transfection with SRF siRNAs (Figure 7C). To analyze the effect of SRF downregulation on the actin cytoskeleton, we stained keratinocyte cultures with FITC-conjugated phalloidin. Although polymerized actin was still present in SRF siRNA–treated cells, the area covered by polymerized actin was reduced in cells with an SRF knockdown. Concomitantly, there were fewer filopodia and the remaining filopodia were shorter and less organized (Figure 7C). Cell-cell contacts were reduced, and most cells were small and round, indicating impaired cell spreading and/or adhesion.
Figure 7. siRNA-mediated downregulation of SRF in primary human keratinocytes disrupts the actin cytoskeleton and affects cell adhesion but not proliferation.
(A) SRF protein is efficiently downregulated in primary human keratinocytes as shown by Western blot analysis. Scrambled siRNA and siRNA against Bid served as controls. GAPDH was used as a loading control. (B) Schematic representation of SRF mRNA. Black bars indicate siRNA-binding sites. (C) Immunofluorescence analysis of SRF (red) shows reduced nuclear staining in SRF siRNA–treated keratinocytes. Costaining with phalloidin-FITC (green) revealed reduced cell size (see upper panels) and less filopodia (see lower panels) in SRF1 and SRF2 siRNA–treated keratinocytes. Scale bars: 20 μm (upper panels); 5 μm (lower panels). (D) Western blot analysis showing expression of SRF, E-cadherin, γ-catenin (plakoglobin), and β-actin in keratinocytes treated with scrambled or SRF siRNAs. siRNA against an unrelated protein (caspase-5) was used as an additional control. (E) Adhesion efficiency of SRF siRNA–treated keratinocytes is significantly reduced (n = 3). Error bars show mean ± SEM, *P = 0.021 (Bid/SRF1); †P = 0.038 (scrambled/SRF1); **P = 0.005 (Bid/SRF2); ‡P = 0.009 (scrambled/SRF2), ANOVA with Bonferroni’s post hoc test. (F) BrdU incorporation was analyzed in keratinocytes that had been treated for 3 days with SRF siRNAs or with control siRNAs (n = 3 wells per treatment group; 10 microscopic fields per dish were counted). Error bars show mean ± SEM. No significant difference was detected using ANOVA with Bonferroni post hoc test.
To determine whether loss of SRF in keratinocytes directly affected intercellular junctions, we determined the expression of E-cadherin, a major component of adherens junctions and of γ-catenin (plakoglobin), a desmosomal component, in the siRNA-treated cells. As shown in Figure 7D, downregulation of SRF expression did not affect the expression of these proteins, suggesting that these proteins are not directly regulated by SRF in keratinocytes.
Adherence of cells to the culture dish was quantified in a colorimetric assay. This experiment demonstrated that adhesion is significantly reduced in cells treated with siRNA against SRF (Figure 7E).
We finally determined whether SRF knockdown affects proliferation and/or apoptosis of primary human keratinocytes. Cells treated with SRF siRNAs or control siRNAs showed a similar number of apoptotic cells, as determined Western blot analysis of cleaved caspase-3 and by TUNEL staining (Supplemental Figure 5 and data not shown). In addition, the number of proliferating cells was not increased in the cells treated with SRF siRNAs compared with control siRNAs but rather was slightly decreased (Figure 7F). Since SRF regulates the expression of immediate early genes in response to serum (24), we also investigated the effect of FCS on keratinocytes in cultures treated with siRNA against SRF. FCS inhibits proliferation and induces differentiation of primary keratinocytes (25). BrdU incorporation assays in the presence of FCS showed that the growth inhibitory effect of FCS was comparable in all cultures, regardless of the siRNA used (data not shown). Therefore, the hyperproliferation of keratinocytes and the increased apoptosis of these cells seen in Srf mutant skin is likely to result from the enhanced inflammation, whereas the adhesion defect of Srf mutant keratinocytes seems to be a direct consequence of the loss of SRF in these cells.
Discussion
In this study, we show that keratinocytes express the transcription factor SRF and that reduction of SRF expression in these cells severely affects epidermal homeostasis. The underlying mechanism involves disruption of the cytoskeleton and subsequent impairment of cell-cell and cell-matrix adhesion. As a result, an inflammatory response is initiated, which in turn induces keratinocyte hyperproliferation. In addition, we provide what we believe to be the first evidence of a role for epidermal SRF in the pathogenesis of inflammatory skin diseases, including psoriasis.
The deletion of the Srf gene in keratinocytes during development resulted in embryonic lethality. This cannot be fully explained by the skin blistering that we observed in these mice, since other mice with severe blistering phenotypes survived until birth (26–28). Although Cre expression was shown to be restricted to stratified epithelia in the mice that we used for this study (16) and although the investigators who generated these mice as well as ourselves did not detect Cre activity in non-K5–expressing cells at E14.5 using Rosa26 reporter mice (16) (Supplemental Figure 2 and data not shown), we cannot exclude a transient weak expression of Cre in cells of other tissues. Due to the obvious importance of SRF during embryonic development (29–32), recombination in only a few cells may be deleterious, resulting in lethality of Srfflex1/flex1K5Cre embryos.
We detected Srfflex1/flex1K5Cre embryos with severe skin blistering around E15, when epidermal keratinocytes begin to differentiate and hair follicles begin to form (reviewed in ref. 33). Interestingly, the Drosophila SRF homolog was shown to control the differentiation of epithelial-derived intervein cells during wing development, and mutations in this gene have also been associated with a blistering phenotype (34).
In Srfflex1/flex1K14CrePR1 mice, we observed recombination of the floxed alleles in the skin at least from E18.5 onwards, which most likely resulted from incomplete suppression of the inducible form of Cre recombinase by heat shock proteins. SRF protein levels were still apparently normal at P2 but markedly reduced at E14, in particular in lesional skin. Thus, it seems that inefficient repression of Cre results in uninduced translocation of this protein into the nucleus. This progresses with time, leading to an increasing number of cells with recombined Srf alleles. Alternatively, clonal expansion of sporadic SRF-deleted basal cells by leaky Cre expression may occur. These cells then repopulate the differentiated layers, resulting in the formation of lesions with SRF-deficient keratinocytes within 1–2 weeks. Mechanical stress or wounding, which further decreases SRF levels, may accelerate this process.
The lesions that appeared in Srf mutant mice were characterized by enhanced keratinocyte proliferation, impaired differentiation of these cells, and inflammation. Since proliferation of cultured keratinocytes was not significantly altered when SRF levels were decreased by SRF siRNA, it seems that the increased proliferation seen in vivo is a secondary effect, probably caused by the secretion of growth factors and cytokines by inflammatory cells. This finding highlights the fact that a primary keratinocyte defect that does not cause increased intrinsic cell proliferation can still lead to hyperproliferation of keratinocytes in vivo. A similar effect was shown in mice lacking IκB kinase 2 (IKK2) in keratinocytes. Loss of IKK2 did not enhance keratinocyte proliferation in vitro but caused a hyperproliferative skin disease in vivo, which was mediated via TNF-α (35).
The enhanced inflammation in SRF-deficient mice may also explain the impaired expression of keratinocyte differentiation markers. In particular, interfollicular expression of K6, K16, and K17 is a hallmark of hyperproliferative epidermis (36) and most likely a consequence of elevated levels of proinflammatory cytokines (37). Alternatively, loss of SRF may directly affect keratin expression, since CArG-like sequences are present in most keratin promoters and since some keratins were upregulated by constitutively active SRF in Srf–/– ES cells (9). However, expression of K5, K15, K6, and K10 was not affected by siRNA-mediated knockdown of SRF in keratinocytes, as shown by real-time RT-PCR for K14 and K6 (data not shown) and by Western blot analysis for K14, K5, and K10 (Supplemental Figure 5). Therefore, the abnormal expression of these keratins in lesional skin is probably not a direct consequence of the loss of SRF.
The most striking abnormalities of Srf mutant epidermis were the severe intercellular gaps. This defect in cell-cell adhesion appears to be a direct consequence of the loss of SRF, since abnormalities in the actin cytoskeleton and reduced cell-cell adhesion were also seen in culture upon siRNA-mediated SRF knockdown. These findings are consistent with the previously described role of SRF in the organization of the actin cytoskeleton and of focal adhesion assembly in ES cells, neurons, smooth muscle cells, and endothelial cells (8, 10, 32, 38). The resulting inflammation may further aggravate this phenotype, since IL-1 was shown to cause actin depolymerization in glial cells (39). In contrast to the direct effect of SRF on the actin cytoskeleton, siRNA-mediated SRF knockdown did not directly affect the levels of E-cadherin or γ-catenin (plakoglobin), which are major components of desmosomes and adherens junctions, respectively. Therefore, it seems most likely that loss of SRF affects cell-cell adhesion through its effect on the actin cytoskeleton.
A second direct effect of the loss of SRF is the reduction in cell-matrix adhesion, which may also explain the blistering phenotype of Srfflex1/flex1K5Cre embryos. The affected adhesion proteins, which are responsible for this phenotype, remain to be determined.
As a result of reduced cell-cell adhesion, impairment of the epidermal barrier may occur. This may allow the penetration of foreign antigens into the skin, which then trigger an inflammatory response (40). The epidermal defect may be aggravated by injury and, together with the observed downregulation of SRF in wounded skin, could explain the preferred occurrence of skin lesions at sites of enhanced mechanical stress. Inflammatory cells release high levels of cytokines that directly stimulate keratinocyte proliferation or induce the expression of keratinocyte mitogens in other stromal cells. Such factors are likely to be responsible for the observed epidermal hyperproliferation in lesional skin of Srf mutant mice. A causative role of a disruption of actin dynamics in the generation of the hyperproliferative phenotype is also supported by results of a recent study in which corneal hyperproliferation combined with abnormal expression of SRF target genes was found in mice with a mutation in the gene encoding destrin, a regulator of actin dynamics (41).
The results presented in this study suggest that downregulation of SRF expression or nuclear exclusion of SRF are generally associated with keratinocyte hyperproliferation. Thus, we found an almost complete loss of nuclear SRF staining in biopsies from patients with the benign hyperproliferative skin disease psoriasis (Figure 1) and a lesser reduction in patients with lichen planus. A mild downregulation was seen in patients showing lichenification as a result of chronic eczematous dermatitis. The appearance of lesions in all these diseases is frequently induced by scratching or wounding (Köbner effect), indicating that injury-induced downregulation of SRF may promote the appearance of the lesions or may even be a prerequisite. Consistent with this hypothesis, we found that wounding of healthy skin induced downregulation of SRF in keratinocytes. During reepithelialization of wounds, keratinocytes reduce their cell-cell contacts, allowing them to migrate and to proliferate (reviewed in ref. 14). Transient downregulation of SRF may thus be a prerequisite for efficient repair of the injured epidermis, whereas constitutive downregulation of SRF is likely to be a major event in the development of hyperproliferative skin disease.
Although a reduction in the levels of nuclear SRF appears to be a relatively unspecific feature of hyperproliferative epidermis, loss of SRF in mice resulted in the development of a phenotype that most closely resembles psoriasis. The importance of actin polymerization and cell adhesion in the pathogenesis of psoriasis is also reflected by the downregulation of LIM kinase 1 in psoriatic epidermis (42). LIM kinases inactivate cofilin and thereby influence actin turnover. This regulatory loop was shown as disrupted in both SRF-deficient neurons (10) and ES cells (D. Hertfelder and A. Nordheim, unpublished observations). Interestingly LIM kinase was also shown to regulate SRF (43–45).
Psoriasis is characterized by upregulation of several keratinocyte mitogens, including TGF-α and amphiregulin, and by enhanced phosphorylation of STAT3 in the epidermis (13). Overexpression of amphiregulin (data not shown) and increased STAT3 phosphorylation were also observed in Srf mutant mice together with upregulation of S100A8 and S100A9, which are also overexpressed in psoriasis (46, 47). Moreover, the inflammatory infiltrate seen in Srf mutant skin is similar to the infiltrate of psoriatic skin (13, 22), and suprabasal integrin expression as well as enhanced angiogenesis were also observed. Although other features of these mice, such as downregulation of K14 and upregulation of K8/K18 expression as well as the presence of a granular layer are not observed in psoriasis and although mouse models cannot fully match the human disease due to important anatomical differences in mouse and human skin, the combination of our mouse and human data strongly suggests a role for SRF in the pathogenesis of psoriasis and possibly other hyperproliferative human skin diseases. Furthermore, our findings further support the concept that epidermal alterations are sufficient to initiate the formation of hyperproliferative skin diseases, including psoriasis (47–49).
Methods
Generation of mice with a keratinocyte-specific deletion within the Srf gene.
Mice homozygous for the conditional Srf allele (Srfflex1) (15) were mated with SrfWT/flex1 mice that were also hemizygous for a transgene encoding Cre recombinase under the control of the bovine K5 promoter (K5Cre) (16) or a transgene encoding a RU486-inducible version of Cre recombinase under the control of the human K14 promoter (K14CrePR1) (17). K5Cre mice were kindly provided by J. Jorcano (Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas [CIEMAT], Madrid, Spain). Recombination of the Srf floxed allele (resulting in Srflx) was verified by PCR as previously described (15). The presence of the Cre recombinase transgene was demonstrated by amplification of a 412-bp fragment using Cre-5′ (5′-TTCGGATCATCAGCTACACC-3′) and Cre-3′ (5′-AACATGCTTCATCGTCGG-3′) primers. All mice were on a C57BL/6J genetic background.
Human skin biopsies.
Biopsies were obtained anonymously from patients with stable psoriatic skin lesions located in the upper and lower extremities. Normal skin was obtained from skin transplants of patients seen at the Dermatology Department, University of Cologne, for surgery. All samples included the dermis and the epidermis. The biopsies were immediately fixed in 4% paraformaldehyde and embedded in paraffin. Alternatively, they were frozen in liquid nitrogen and used for the preparation of protein lysates. None of the patients had received oral or topical treatment for the last 8 weeks before biopsy. All patients provided informed consent for the Department of Dermatology, University of Cologne, and the use of biopsies for scientific purposes was approved by the Institutional Commission of Ethics (Az. 9645/96). Biopsies from patients with lichen planus or lichenification resulting from chronic eczematous dermatitis were obtained anonymously. Informed consent for research was obtained prior to routine diagnostic services at the University Hospital of Lausanne. The biopsies were immediately fixed in 4% paraformaldehyde and embedded in paraffin.
Wounding and preparation of wound tissue.
BALB/c mice were anesthetized by i.p. injection of ketamine (75 mg/kg) and xylazine (5 mg/kg). Four full-thickness excisional wounds of 5-mm diameter were generated on the backs of mice by excising the skin and the s.c. muscle panniculus carnosus (50). Wounds were left uncovered and harvested at different time points after injury. All animal experiments were approved by the local veterinary authorities of Zurich, Switzerland (Kantonales Veterinäramt Zürich).
Primary human keratinocyte cell culture, siRNA transfection, and cell culture assays.
Primary keratinocytes were isolated from human foreskin and cultured up to passage 5 in K-SFM (Invitrogen) supplemented with epidermal growth factor and bovine pituitary extract. Cells were transfected with siRNA using INTERFERin (Polyplus-transfection). Experiments were performed 48 hours or 72 hours after transfection. Proliferation was determined by BrdU incorporation. In brief, cells were incubated for 2 hours with BrdU (100 μM), fixed in 4% PFA/PBS, and stained with an FITC-coupled anti-BrdU mAb (Roche) according to standard procedures. Cell adhesion was analyzed in a colorimetric assay, based on p-nitrophenyl N-acetyl-β-d-glucosaminide (Sigma-Aldrich) (51). The following siRNAs were used: SRF1, 5′-AUGUUUGCCAUGAGUAUUAdTdT-3′; SRF2, 5′-GAAUGAGUGCCACUGGCUUdTdT-3′; Bid, 5′-CCGUGAUGUUUUCACACAdTdT-3′; caspase-5, 5′-UGGAUAACUUCGUGAUAAAdTdT-3′; and scrambled, 5′-UUCUCCGAACGUGUCACGUdTdT-3′.
Production of SRF-specific mAbs.
Murine Srf cDNA (IMAGE clone 6404515; Deutsches Ressourcenzentrum für Genomforschung) was cloned in frame of 6 downstream histidine codons into the expression vector pET20b. Purification of bacterially overexpressed SRF-6xHis fusion protein was performed by fast liquid protein chromatography (FPLC) with a HisTrap HP column (GE Healthcare). Approximately 50 μg of SRF fusion protein was injected both i.p. and s.c. into Lou/C rats using CPG2006 (TIB MOLBIOL) and incomplete Freund’s adjuvant. After an 8-week interval, a final boost was given i.p. and s.c. 3 days before fusion. Fusion of the myeloma cell line P3X63-Ag8.653 with the rat immune spleen cells was performed using standard procedures. Hybridoma supernatants were tested in a solid-phase immunoassay. A mouse mAb specific for the histidine tail of the fusion protein (His 3D5; Invitrogen) was coated overnight at a concentration of 3 μg/ml. After blocking with nonfat milk, the SRF fusion protein or an irrelevant His-tagged fusion protein were incubated, followed by the hybridoma supernatants. Bound rat mAbs were detected with a cocktail of biotinylated mouse mAbs against the rat IgG heavy chains (α-IgG1, α-IgG2a, α-IgG2b [ATCC], α-IgG2c [Ascenion]), thus avoiding IgM mAbs. The biotinylated mAbs were visualized with peroxidase-labeled Avidin (Alexis) and o-phenylenediamine as chromogen in the peroxidase reaction. Supernatants were then tested in Western blots with lysates from mouse brain. Those supernatants able to detect endogenous SRF were further tested by immunohistochemistry on vibratome and paraffin sections from WT and SRF-deficient mouse brains. Finally, the 3 best clones, designated 2C5, 6F1 and 7D9 (all of the IgG2a subclass), were stably subcloned.
Electron microscopy.
Mouse skin samples were excised and immediately transferred to 4% paraformaldehyde in PBS, fixed overnight, rinsed, and stored in PBS. Before embedding, samples were treated with 2% OsO4 for 2 hours. After washing, they were stained in 1% uranyl acetate, dehydrated through a series of graded ethanols, and embedded in Araldite resin. Ultrathin sections (30–60 nm) were processed with a diamond knife and placed on copper grids. Transmission electron microscopy was performed using a 902A electron microscope (Zeiss). EM902A with Megaview III digital image acquisition system with iTEM software, version 5.0.
(Olympus-SIS) was used for morphometric analysis of the number of desmosomes along the cell membrane at the intercellular space between keratinocytes of at least 10 randomized selected epidermal areas per mouse sample at a magnification of ×20,000. The number is calculated from the number of desmosomes/μm of measured cell membrane length. The individual length of the desmosomes was measured directly on the computer screen using the acquisition software.
Statistics.
Statistical analysis was performed using SPSS software (SPSS). Multiple treatment data were analyzed using 1-way ANOVA, and individual groups were compared using a Bonferroni’s post hoc test, whereas 2-tailed Student’s t test was used for experiments examining differences among groups.
Supplementary Material
Acknowledgments
We thank Nicole Hallschmid and Anja Müller for excellent technical assistance, José Jorcano for kindly providing K5-Cre transgenic mice, and Pierre Coulombe for kindly providing the K17 antibody. This work was supported by grants from the Swiss National Science Foundation (3100A9-109340/1 to S. Werner), the European Union (Ulcer Therapy, LSHB-CT-2005-512102 to S. Werner), and the Deutsche Forschungsgemeinschaft (SFB 446, B7 to A. Nordheim and SFB 829 to C. Mauch and W. Bloch).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: K, keratin; SRF, serum response factor.
Citation for this article: J. Clin. Invest. 119:899–910 (2009). doi:10.1172/JCI37771
References
- 1.Fuchs E., Raghavan S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 2.Shore P., Sharrocks A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 3.Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 4.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 5.Sun Q., et al. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miano J.M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/S0022-2828(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 7.Posern G., Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Schratt G., et al. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philippar U., et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 10.Alberti S., et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan-Albuquerque N., Van Putten V., Weiser-Evans M.C., Nemenoff R.A. Depletion of serum response factor by RNA interference mimics the mitogenic effects of platelet derived growth factor-BB in vascular smooth muscle cells. Circ. Res. 2005;97:427–433. doi: 10.1161/01.RES.0000179776.40216.a9. [DOI] [PubMed] [Google Scholar]
- 12.Poser S., Impey S., Trinh K., Xia Z., Storm D.R. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–4966. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowes M.A., Bowcock A.M., Krueger J.G. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 14.Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 15.Wiebel F.F., Rennekampff V., Vintersten K., Nordheim A. Generation of mice carrying conditional knockout alleles for the transcription factor SRF. Genesis. 2002;32:124–126. doi: 10.1002/gene.10049. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez A., et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 17.Berton T.R., et al. Characterization of an inducible, epidermal-specific knockout system: differential expression of lacZ in different Cre reporter mouse strains. Genesis. 2000;26:160–161. doi: 10.1002/(SICI)1526-968X(200002)26:2<160::AID-GENE20>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 18.Kellendonk C., et al. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 1996;24:1404–1411. doi: 10.1093/nar/24.8.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koster M.I., Roop D.R. The role of p63 in development and differentiation of the epidermis. J. Dermatol. Sci. 2004;34:3–9. doi: 10.1016/j.jdermsci.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/S0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 21.Carroll J.M., Romero M.R., Watt F.M. Suprabasal integrin expression in the epidermis of transgenic mice results in developmental defects and a phenotype resembling psoriasis. Cell. 1995;83:957–968. doi: 10.1016/0092-8674(95)90211-2. [DOI] [PubMed] [Google Scholar]
- 22.Stratis A., et al. Pathogenic role for skin macrophages in a mouse model of keratinocyte-induced psoriasis-like skin inflammation. J. Clin. Invest. 2006;116:2094–2104. doi: 10.1172/JCI27179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhardt C., Nemeth J., Angel P., Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006;72:1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillai S., Bikle D.D., Hincenbergs M., Elias P.M. Biochemical and morphological characterization of growth and differentiation of normal human neonatal keratinocytes in a serum-free medium. J. Cell. Physiol. 1988;134:229–237. doi: 10.1002/jcp.1041340208. [DOI] [PubMed] [Google Scholar]
- 26.Georges-Labouesse E., et al. Absence of integrin alpha 6 leads to epidermolysis bullosa and neonatal death in mice. Nat. Genet. 1996;13:370–373. doi: 10.1038/ng0796-370. [DOI] [PubMed] [Google Scholar]
- 27.van der Neut R., et al. Epithelial detachment due to absence of hemidesmosomes in integrin beta 4 null mice. Nat. Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- 28.Dowling J., Yu Q.C., Fuchs E. Beta4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arsenian S., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miano J.M., et al. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parlakian A., et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco C.A., et al. Serum response factor is required for sprouting angiogenesis and vascular integrity. Dev. Cell. 2008;15:448–461. doi: 10.1016/j.devcel.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Mack J.A., Anand S., Maytin E.V. Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res. C Embryo Today. 2005;75:314–329. doi: 10.1002/bdrc.20055. [DOI] [PubMed] [Google Scholar]
- 34.Montagne J., et al. The Drosophila Serum Response Factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development. 1996;122:2589–2597. doi: 10.1242/dev.122.9.2589. [DOI] [PubMed] [Google Scholar]
- 35.Pasparakis M., et al. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417:861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
- 36.McGowan K.M., Coulombe P.A. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei L., Debets R., Hegmans J.J., Benner R., Prens E.P. IL-1 beta and IFN-gamma induce the regenerative epidermal phenotype of psoriasis in the transwell skin organ culture system. IFN-gamma up-regulates the expression of keratin 17 and keratinocyte transglutaminase via endogenous IL-1 production. J. Pathol. 1999;187:358–364. doi: 10.1002/(SICI)1096-9896(199902)187:3<358::AID-PATH253>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Angstenberger M., et al. Severe intestinal obstruction on induced smooth muscle-specific ablation of the transcription factor SRF in adult mice. Gastroenterology. 2007;133:1948–1959. doi: 10.1053/j.gastro.2007.08.078. [DOI] [PubMed] [Google Scholar]
- 39.Namekata K., Harada C., Kohyama K., Matsumoto Y., Harada T. Interleukin-1 stimulates glutamate uptake in glial cells by accelerating membrane trafficking of Na+/K+-ATPase via actin depolymerization. Mol. Cell. Biol. 2008;28:3273–3280. doi: 10.1128/MCB.02159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segre J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdoni A.M., Aoyama N., Ikeda A., Ikeda S. Effect of destrin mutations on the gene expression profile in vivo. Physiol. Genomics. 2008;34:9–21. doi: 10.1152/physiolgenomics.00285.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honma M., Benitah S.A., Watt F.M. Role of LIM kinases in normal and psoriatic human epidermis. Mol. Biol. Cell. 2006;17:1888–1896. doi: 10.1091/mbc.E05-12-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geneste O., Copeland J.W., Treisman R. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 2002;157:831–838. doi: 10.1083/jcb.200203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arber S., et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 45.Yang N., et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 46.Broome A.M., Ryan D., Eckert R.L. S100 protein subcellular localization during epidermal differentiation and psoriasis. J. Histochem. Cytochem. 2003;51:675–685. doi: 10.1177/002215540305100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenz R., et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 2005;437:369–375. doi: 10.1038/nature03963. [DOI] [PubMed] [Google Scholar]
- 48.Sano S., et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- 49.Cook P.W., Pittelkow M.R., Piepkorn M. Overexpression of amphiregulin in the epidermis of transgenic mice induces a psoriasis-like cutaneous phenotype. J. Invest. Dermatol. 1999;113:860. doi: 10.1046/j.1523-1747.1999.00753.x. [DOI] [PubMed] [Google Scholar]
- 50.Werner S., et al. The function of KGF in morphogenesis of epithelium and reepithelialization of wounds. Science. 1994;266:819–822. doi: 10.1126/science.7973639. [DOI] [PubMed] [Google Scholar]
- 51.Montanez E., et al. Analysis of integrin functions in peri-implantation embryos, hematopoietic system, and skin. Methods Enzymol. 2007;426:239–289. doi: 10.1016/S0076-6879(07)26012-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.