Abstract
The association of copper to Riboflavin Binding Protein (RBP) from egg white has been studied by electron paramagnetic resonance (EPR) and X-ray absorption (XAS) spectroscopies. The type II site contains a mix of copper I and II in an oxygen rich environment.
Keywords: Riboflavin binding protein, copper, gallus-gallus
The association of copper to Riboflavin Binding Protein (RBP) from egg white has been studied by electron paramagnetic resonance (EPR) and X-ray absorption (XAS) spectroscopies in order to provide insight into how this essential protein may transport and store copper in avian embryos. Riboflavin Binding Protein, RBP, purified from avian egg white, has been shown to bind copper in a 1:1 molar ratio when dialyzed against copper(II) [1]. While the egg is a unique environment and quite rich in copper, the mechanisms by which this copper is delivered during development and stored for eventual use remain unclear [2]. Since RBP is already identified in the active transport of the cofactor riboflavin to the egg, evidence of its copper binding ability may suggest an additional role for RBP in the transport and storage of copper.
Copper binding to RBP is a recently identified and novel interaction associated with the protein [1]. As purified by the conventional method [3], RBP contains only trace amounts of copper. The loss of copper during purification and subsequent failure to identify a copper binding ability in previous studies may be attributed to the somewhat extreme conditions applied to the protein during the standard purification, specifically to the necessity for pH 4 acetate buffer in several of the elution steps. Aerobic dialysis against buffer containing cupric chloride affords a protein fully loaded with one equivalent of copper, a ratio that has remained fairly constant through various dialysis conditions.
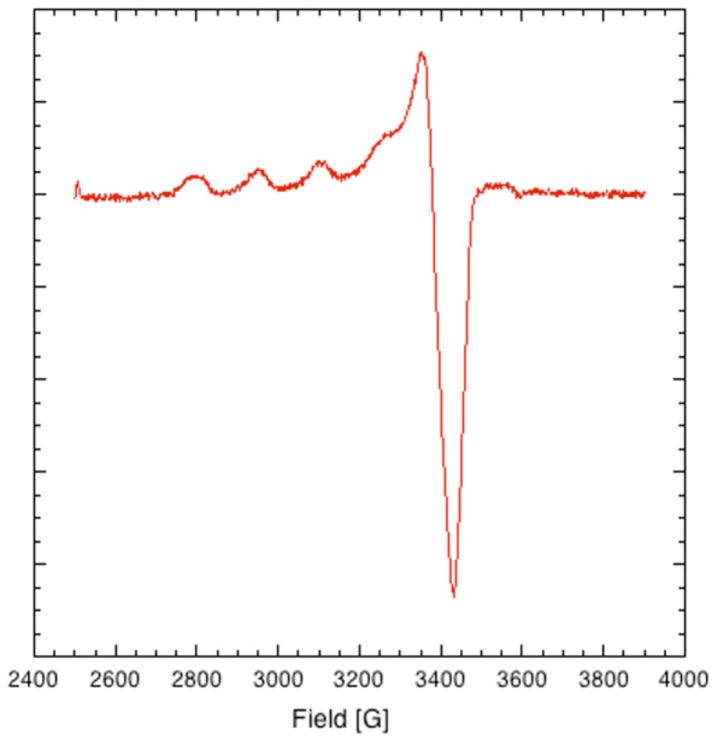
EPR results confirm that the copper binding site is a well-ordered type II site, as shown in Fig. (1). Type II is defined as 4–5 coordinate copper bound to a mixture of N, O and/or S ligands. Spectral parameters unique to this system are: g⊥=2.06 (A⊥=10G) and g||=2.39 (A||=142 G) [1]; these spectral parameters were obtained by simulation, as outlined previously [1] using the XSOPHE software package of Bruker. Simulation results are consistent with a copper binding site containing 3–4 oxygens in the first coordination sphere [4]. The small feature visible in the EPR spectrum at approximately 3600 G is an artifact associated with the cavity.
Fig. 1.
Continuous Wave (CW) EPR of copper loaded Riboflavin Binding Protein (1.65 mM RBP) in 0.10 mM TRIS, pH 7.2. Microwave Frequency: 9.7 GHz. Microwave Power: 2.0 μW, Modulation Frequency: 10 kHz, Modulation Amplitude: 10 G, Temperature: 10K.
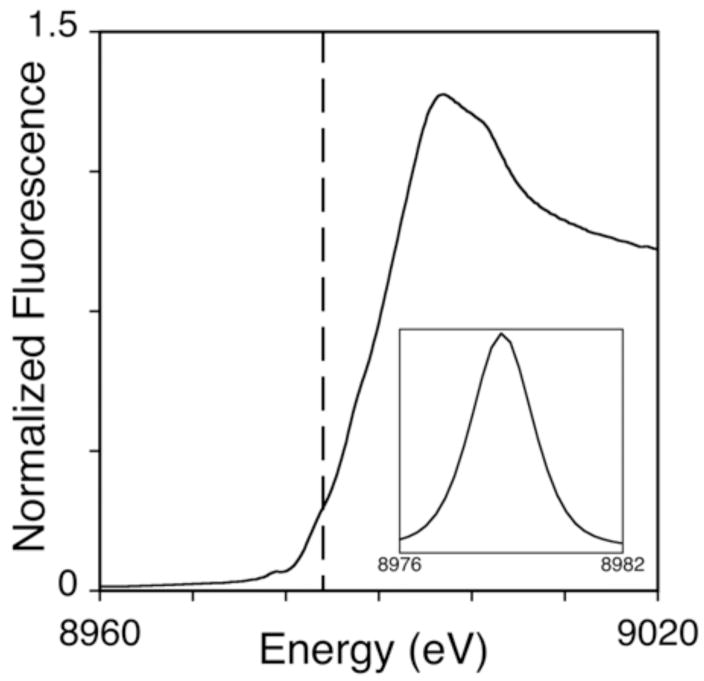
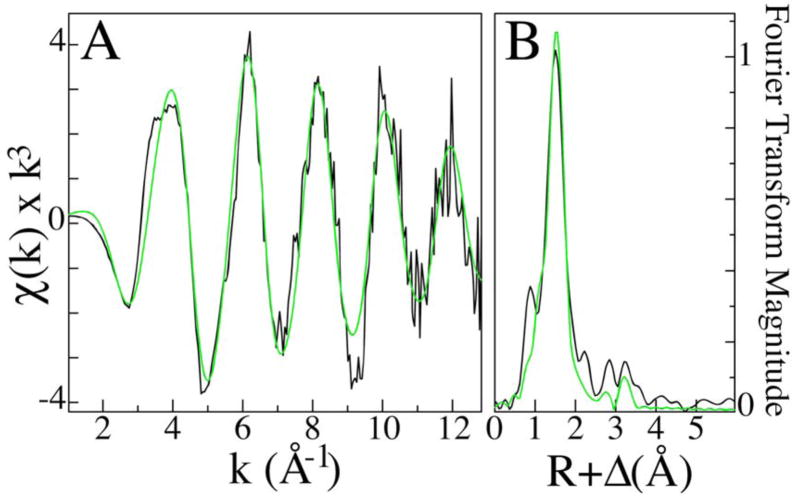
XAS studies, performed on copper loaded RBP, were used to examine the bound metal’s oxidation state and metal-ligand coordination structure. A substantial pre-edge feature at ca. 8978 and the minor feature at 8984 eV, indicative of 1s→3d and 1s→4p electronic transitions typically seen for Cu(II) and Cu(I), respectively, suggest that while the bulk of the copper bound to RBP is Cu(II) a substantial distribution of Cu(I) is also bound to RBP (Fig. 2). EXAFS simulations (Fig. 3A), used to characterize the copper-nearest neighbor ligand structure, indicate an average coordination environment constructed by three oxygen/nitrogen based ligands at an average bond distance of 1.96 Å. These data could not be fit with any copper-sulfur or copper•••copper scattering. The presence of carbon scattering at > 3Å, observed in the Fourier transform of the Cu EXAFS (Fig. 3B), indicates a relative degree of long-range ligand order that is typically seen from carboxylate carbons or imidazole rings [5]. The final EXAFS fits table is provided as supplemental information.
Fig. 2.
XANES of Cu(II) loaded RBP (top). Vertical dashed line indicates the 1s → 4p transition at 8984 eV. Inset shows the expansion of the background-subtracted 1s →3d region of the XANES spectra.
Fig. 3.
EXAFS and Fourier transforms of copper loaded RBP XAS data. Raw EXAFS spectra in black (A) along with the corresponding Fourier transform (B). Simulations of EXAFS and Fourier transform data are shown in green.
Since photoreduction of copper is common in XAS, EPR spectroscopy was used to quantitate the relative amounts of copper(I) and copper(II) bound to the protein by comparison to copper sulfate standard samples. Spin quantitation shows that the protein, as loaded aerobically with copper II in the absence of external reducing agents contains between 30 and 50% copper I in various samples tested, suggesting that while some of the signal for copper I seen in the XANES data might be attributed to photoreduction, some reduction of the copper II is occuring as a result of the binding interaction.
The reduction of copper upon binding to RBP is an interesting result. Dialyses are performed on holo-RBP (RBP bound to its riboflavin cofactor) under aerobic conditions in the absence of any external reducing agents. Furthermore, a stable copper I site featuring no sulfur or histidyl ligands would be most unusual. One possible source of reducing equivalents from the protein would be the bound riboflavin moiety. Experiments are ongoing to identify the specific ligands to copper in the binding site and to identify the reducing agent, in addition to a modification of the purification protocol so that we might detect natively bound copper in the protein as purified.
Since RBP is known to be involved in the active transport of riboflavin in egg, and since the delivery system for copper into the egg is not fully understood, the implication of the current work is that RBP could participate in the active transport of copper into the egg. Furthermore, since RBP has been shown to be capable of reducing the copper, the protein may play some role in the stabilization of the copper redox state for bound metal. The possibility of a new copper transport protein that also modulates the redox properties of its substrate are of extreme interest, as copper metal is utilized ubiquitously at different locations in the egg during all stages of embryonic development.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Diabetes and Digenstive and Kidney Diseases R01DK068139 (TLS). Special thanks to Professor John McCracken and the Michigan Center for Structural Biology for use of the EPR facilities.
ABBREVIATIONS
- RBP
Riboflavin Binding Protein
- EPR
Electron paramagnetic resonance
- XAS
X-ray absorption spectroscopy
- XANES
X-ray absorption near edge spectroscopy
- EXAFS
Extended X-ray absorption fine structure
Footnotes
General Chemicals
TRIS, and CuCl2 were purchased from Sigma (St. Louis, MO).
Protein Purification
Purification and concentration quantification RBP from egg whites followed published protocols [3].
Dialysis for Copper Loading
Lyophilized Protein was re-suspended in 0.1 M pH 7.2 Tris and dialyzed overnight aerobically against Tris buffer containing 10 mM CuCl2. This dialysis was followed with dialysis against pure Tris buffer to remove any excess copper. Total copper was quantitated using graphite furnace atomic absorption spectroscopy as previously reported [1]. Protein is digested in a nitric acid solution, followed by the addition of 3% Hydrogen Peroxide. Copper is measured by atomic absorption spectroscopy by comparison a standard curve of analytical aqueous copper solutions.
EPR Spectroscopy
Continuous wave electron paramagnetic resonance studies were performed at 10K on a Bruker E680X spectrometer equipped with an Oxford CF935 cryostat and either a 5mm dielectric resonator or a 4 mM dielectric resonator equipped with an ENDOR accessory.
XAS Experiments
XAS studies were carried out at both the Stanford Synchrotron Radiation Laboratory (SSRL), on beamline 9-3, and at the National Synchrotron Light Source (NSLS), on beam-line X9b. SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program. NSLS, located at Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886. Spectra were collected on multiple reproducible samples. Data collection and analysis protocols have been previously reported [6].
SUPPORTIVE/SUPPLEMENTARY MATERIAL
A table containing the final EXAFS fits is included as supplemental information.
References
- 1.Smith SR, Pala I, Benore-Parsons M. J Inorg Biochem. 2006;100:1730. doi: 10.1016/j.jinorgbio.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 2.Richards MP. Poultry Sci. 1997;76:152. doi: 10.1093/ps/76.1.152. [DOI] [PubMed] [Google Scholar]
- 3.Miller MS, White HB., III Meth Enzymol. 1986:227. doi: 10.1016/0076-6879(86)22174-6. [DOI] [PubMed] [Google Scholar]
- 4.Peisach J, Blumberg WE. Arch Biochem Biophys. 1974;165:691. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- 5.Hakemian AE, Tinberg CE, Kondapalli KC, Tesler J, Hoffman BM, Stemmler TL, Rosenzweig AC. J Am Chem Soc. 2005;127:17142. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC, Stemmler TL. Inorg Chem. 2006;45:8372. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.