Abstract
This work presents a new approach to the analysis of aperiodic pulsatile heteroscedastic time-series data, specifically hormone pulsatility. We have utilized growth hormone (GH) concentration time-series data as an example for the utilization of this new algorithm. While many previously published approaches used for the analysis of GH pulsatility are both subjective and cumbersome to use, AutoDecon is a nonsubjective, standardized, and completely automated algorithm. We have employed computer simulations to evaluate the true-positive, the false-positive, the false-negative, and the sensitivity percentages of several of the routinely employed algorithms when applied to GH concentration time-series data. Based on these simulations, it was concluded that this new algorithm provides a substantial improvement over the previous methods. This novel method has many direct applications in addition to hormone pulsatility, for example, to time-domain fluorescence lifetime measurements, as the mathematical forms that describe these experimental systems are both convolution integrals.
1. Introduction
It has long been recognized that normal function within endocrine systems requires a highly coordinated interplay of events both within and among components of a given hormonal axis. Moreover, it has become increasingly evident that the secretory characteristics of certain endocrine signals are critical determinants of the target organ response. For example, it has been demonstrated that whereas the administration of gonadotropin-releasing hormone (GnRH) in a pulsatile fashion affected the release of luteinizing hormone (LH) and follicle-stimulating hormone (FSH), continuous administration not only failed to stimulate LH and FSH release, but actually inhibited secretion of these hormones (Nakai et al., 1978). More recent studies have further demonstrated that the gene expression of LH and FSH is controlled by the frequency and amplitude characteristics of the pulsatile GnRH signal (Haisenleder et al., 1994). The pulsatile nature of growth hormone (GH) secretion has also received considerable investigative attention and has provided insights into both stimulatory and inhibitory hypothalamic mechanisms that regulate GH secretion (Dimaraki et al., 2001; Hartman, 1991; Maheshwari et al., 2002; Thorner et al., 1997; Van Cauter et al., 1998; Vance et al., 1985; Webb et al., 1984). The secretion of numerous other hormones has also been shown to be pulsatile in nature, including, but not limited to, prolactin, thyroid-stimulating hormone, adrenocorticotropic hormone, parathyroid hormone, cortisol, and insulin.
Given the physiological significance of hormone pulsatility, the ability to separate a pulsatile signal from noise has become crucial with regard to understanding the mechanisms that control the dynamics of this signal. Although earlier attempts to apply computer-based methods to characterize such pulsatile signals provided enhanced objectivity, most methods were not statistically based (Merriam and Wachter, 1982; Santen and Bardin, 1973; Ultra and Cycle) and those that were (Oerter et al., 1986; Veldhuis and Johnson, 1986) only identified perturbations in hormone concentration time series but could not separate pulses into their secretory and clearance components. The application of deconvolution procedures (Evans et al., 1992; Johnson and Veldhuis, 1995; Johnson et al., 2004; Urban et al., 1988; Veldhuis et al., 1987) has been employed effectively to address this limitation, that is, such procedures allow for the “unraveling” of hormone pulses into the secretory event itself and the mechanisms responsible for clearance.
The choice of the model-fitting approach for deconvolution analysis of experimental data is dictated by the properties of the measurement uncertainties that are inherent within such data. Measurement errors within hormone concentration time-series data are typically large, as compared to the size of the secretion events, and are heteroscedastic. In addition, missing values are fairly common and thus it cannot be assumed that the data values are equally spaced in time. Several deconvolution procedures [e.g., those based on the convolution theorem (Jansson, 1984)] may be utilized for this sort of analysis, but most other approaches are incompatible with the properties of data.
Another difficulty with previously published hormone pulsatility analysis approaches (Johnson and Veldhuis, 1995; Johnson et al., 2004; Veldhuis et al., 1987) is that they necessitate a prior knowledge or presumption of the number of secretion events, and initial estimates of the secretion event positions and amplitudes are required to be provided by the user. Finding the number of secretion events and initial parameter estimates is a system identification problem, which is addressed here via a fully automated statistically based approach, AutoDecon, which finds the optimal number of secretion events and the initial parameter estimates while simultaneously performing deconvolution. Moreover, a quintessential data analysis dilemma is in the identification and subsequent characterization of small pulsatile events within a data time series where the amplitudes of these events are comparable to the magnitude of the experimental measurement errors. Event identification is further complicated when the pulsatile events are aperiodic (i.e., they occur at apparently random intervals) and/or when the experimental measurement uncertainties are heteroscedastic (i.e., the measurement uncertainties are variable).
A Monte Carlo technique is utilized to provide information about the operating characteristics of the AutoDecon and other algorithms (e.g., true-positive, false-positive, and false-negative percentages for secretion event identification). This Monte Carlo procedure is an extension of procedures that we (Veldhuis and Johnson, 1988) and others (Guardabasso et al., 1988; Van Cauter, 1988) have employed for this purpose.
The numerical methods outlined in this chapter are generally applicable to many biochemical systems, although this chapter describes this procedure and presents the analysis of GH concentration time-series data as an example of the use of AutoDecon. For example, time-domain fluorescence lifetime measurements (Lakowicz, 1983) are described by the mathematical forms (convolution integrals), which describe pulsatile hormone concentration time series.
2. Methods
AutoDecon implements a rigorous statistical test for the existence of secretion events (Johnson et al., 2008). In addition, the subjective nature defining earlier deconvolution procedures is eliminated by the ability of the program to automatically insert and subsequently test the significance of presumed secretion events. No user intervention is required subsequent to the initialization of the algorithm. This automatic algorithm combines three modules: a parameter-fitting module (analogous to Johnson and Veldhuis, 1995; Veldhuis et al., 1987), a new insertion module that automatically adds presumed secretion events, and a new triage module that automatically removes secretion events, which are deemed to be statistically nonsignificant.
2.1. Overview of deconvolution method
These deconvolution procedures function by developing a mathematical model for the time course of the hormone concentration and then fitting this mathematical model to experimentally observed time-series data with a weighted nonlinear least-squares algorithm. Specifically, this mathematical model (Johnson and Veldhuis, 1995; Veldhuis et al., 1987) is
| (15.1) |
where C(t) is the hormone concentration as a function of time t, S(t) is the secretion into the blood as a function of time, and E(t − z) is elimination from the serum as a function of time. In Eq. (15.1) the limits of integration are from 0 to t and, as a consequence, a second term containing the concentration at time zero, C(0), is included. For the time-domain fluorescence lifetime experimental system, the S(t) function corresponds to the lamp function (i.e., the instrument response function) and the E(t −z) corresponds to the fluorescence decay function.
The hormone concentration elimination function, E(t−z) is assumed to follow a single compartment pharmacokinetic model [Eq. (15.2)]
| (15.2) |
where HL is the one-component elimination half-life, or a two-compartment [Eq. (15.3)] model,
| (15.3) |
where HL1 and HL2 are the elimination half-lives and f2 is the amplitude fraction of the second component.
C(0) in Eq. (15.1) is the concentration of the hormone immediately before the first data point, that is, at time equal to zero. This model is rigorously correct for a single-compartment pharmacokinetic elimination model, Eq.(15.2). However, for the two-compartment elimination model, Eq. (15.3), it is only correct under the assumption that all of the concentration at time t = 0 is the result of an instantaneous secretion event that occurs at this initial moment. This assumption is required as scant information pertaining to the secretion and elimination prior to the first data point is contained within data.
In the original formulation (Johnson and Veldhuis, 1995; Johnson et al., 2004; Veldhuis et al., 1987) the secretion rate is modeled by Eq. (15.4)
| (15.4) |
where the secretion rate is assumed to be the sum of Gaussian-shaped events that occur at different irregularly spaced times, PPk, have differing heights, Hk, and the same width, SecretionSD. Note that the size of the secretion events is expressed as the base 10 logarithm of the height, log Hk, in order to constrain all heights to physiologically relevant positive values. The positive constant So is the basal secretion.
2.2. AutoDecon-Fitting module
The fitting module performs weighted nonlinear least-squares parameter estimations by the Nelder–Mead Simplex algorithm (Nelder and Mead, 1965; Straume et al., 1991). It fits Eq. (15.1) to experimental data by adjusting the parameters of the secretion function, Eq. (15.4), and the elimination function, either Eq. (15.2) or (15.3), so that the parameters have the highest probability of being correct. The module is based on the Amoeba routine (Press et al., 1986), which was modified such that convergence is assumed when both the variance-of-fit and the individual parameter values do not change by more than 2 × 10−5 or when 15,000 iterations have occurred. This is essentially the original Deconv algorithm (Johnson and Veldhuis, 1995; Veldhuis et al., 1987) with the exception that the Nelder–Mead Simplex parameter estimation algorithm (Nelder and Mead, 1965; Straume et al., 1991) is used instead of the damped Gauss–Newton algorithm, which was employed by Deconv. The Nelder–Mead algorithm simplifies the software, as it does not require derivatives.
2.3. AutoDecon insertion module
The insertion module inserts the next presumed secretion event at the location of the maximum of the probable position index, PPI:
| (15.5) |
The parameter Hz in Eq. (15.5) is the amplitude of a presumed secretion event at time z. The index function PPI(t) will have a maximum at the data point position in time where the insertion of a secretion peak will result in the largest negative derivative in the variance of fit versus secretion event size. It is important to note that the partial derivatives of the variance of fit with respect to a secretion event can be evaluated without any additional weighted nonlinear least-squares parameter estimations or even knowing the size of the presumed secretion event, Hz. Using the definition of the variance of fit given in Eq. (15.6), the partial derivative with respect to the addition of a secretion event at time z is shown in Eq. (15.7) where the summation is over all data points,
| (15.6) |
| (15.7) |
where Wi corresponds to the weighting factor for the ith data point and Ri corresponds to the ith residual (see Section 2.8). The inclusion of these weighting factors is the statistically valid method to compensate for the heteroscedastic properties of experimental data.
2.4. AutoDecon triage module
The triage module performs a statistical test to ascertain whether a presumed secretion event should be removed. This test requires two weighted nonlinear least-squares parameter estimations: one with the presumed peak present and one with the presumed peak removed. The ratio of the variance of fit resulting from these two parameter estimations is related to the probability that the presumed secretion event does not exist, P, by an F statistic, as in Eq. (15.8). Typically, a probability level of 0.05 is used:
| (15.8) |
This is the F test for an additional term (Bevington, 1969) where the additional term is the presumed secretion event. The 2′s in Eq. (15.8) are included, as each additional secretion event increases the number of parameters being estimated by 2, specifically the location and the amplitude of the secretion event. The number of degrees of freedom, ndf, is the number of data points minus the total number of parameters being estimated when the secretion event is present. Each cycle of the triage module performs this statistical test for every secretion event in an order determined by size, smallest to largest. If a secretion event is found to not be statistically significant, it is removed and the triage module is restarted from the beginning (i.e., a new cycle starts). Thus, the triage module continues until all nonsignificant secretion events have been removed. Each cycle of the triage module performs m + 1 weighted nonlinear least-squares parameter estimations where m is the current number of secretion events for the current cycle; one where all of the secretion events are present; and one where each of the secretion events has been removed and tested individually.
2.5. AutoDecon combined modules
The AutoDecon algorithm iteratively adds presumed secretion events, tests the significance of all events, and removes nonsignificant secretion events. The procedure is repeated until no additional secretion events are added. The specific details of how this is accomplished with the insertion, fitting, and triage modules are outlined here.
AutoDecon is generally initialized with the basal secretion, S0, set equal to zero, the concentration at time zero, C(0), set equal to zero, the elimination half-life (HL) set to any physiologically reasonable value, the standard deviation of the secretion events (SecretionSD) set to one-half of the data sampling interval, and zero secretion events. It is possible, but not required, that initial presumed secretion event locations and sizes be included in the initialization. Initializing the program with peak position and amplitude estimates might decrease the amount of computer time required. Note that for the examples presented in this work the initial number of secretion events is set to zero unless specifically noted otherwise.
The next step in the initialization of the AutoDecon algorithm is for the fitting module to estimate only the basal secretion, S0, and the concentration at time zero, C(0). The fitting module will then estimate all of the model parameters except for the elimination half-life and the standard deviation of the secretion events. If any secretion events have been included in the initialization, the second fit will also refine the locations and sizes of these secretion events. Next, the triage module is utilized to remove any nonsig-nificant secretion events. At this point the parameter estimations performed within the triage module will estimate all of the current model parameters, except for the elimination half-life and the standard deviation of the secretion events (SecretionSD).
The AutoDecon algorithm next proceeds with phase 1 by using the insertion module to add a presumed secretion event. This is followed by the triage module to remove any nonsignificant secretion events. Again, the parameter estimations performed within the triage module during phase 1 will estimate all of the current model parameters with exception of the elimination half-life, HL, and the standard deviation of the secretion events, SecretionSD. If during this phase the triage module does not remove any secretion events, phase 1 is repeated to add an additional presumed secretion event. Phase 1 is repeated until no additional secretory events are added in the insertion followed by triage cycle.
Phase 2 repeats the triage module with the fitting module estimating all of the current model parameters, but this time including the elimination half-life, HL, and the standard deviation of the secretion events, SecretionSD.
Phase 3 will repeat phase 1 (i.e., insertion and triage) with the parameter estimations that are performed by the fitting module within the triage module estimating all of the current model parameters, again including the elimination half-life, HL, and the standard deviation of the secretion events, SecretionSD. Phase 3 is repeated until no additional secretion events have been added in the insertion followed by triage cycle.
Phase 4 examines the residuals (i.e., differences between the data points and the fitted curve) for trends that might indicate fitting problems. Specifically, if the first data point is a negative outlier (i.e., the data point is significantly lower than might be expected; see Section 2.7) or the last data point is a positive outlier (i.e., the data point is significantly higher than might be anticipated), then it is possible that the algorithm failed to identify a partial secretion event at either the start or the end of the time series, respectively. If only a portion of the secretion event is present within the data time series, then the AutoDecon algorithm might not resolve this partial event and thus an outlier may be present. When this is observed, then the offending data point is temporarily removed and the entire AutoDecon algorithm is repeated with the current values as the initialization.
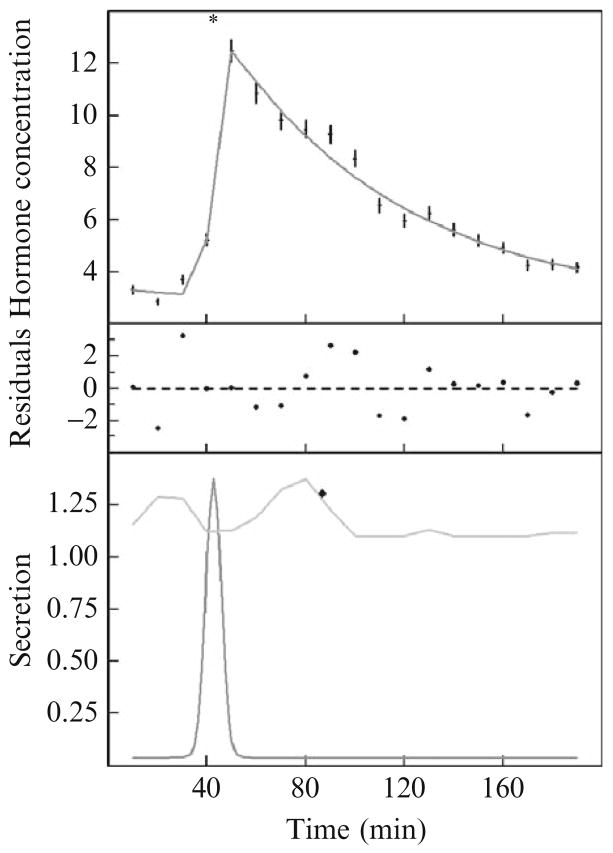
An illustrative example of the results from this procedure is shown in Fig. 15.1. In this example, one secretion event has been located. The upper part of Fig. 15.1 shows the data points and the calculated concentration. The lower part of Fig. 15.1 depicts the observed secretion rate (lower curve) and the probable position index [upper curve calculated per Eq. (15.5)] for addition of the next presumptive secretion event. In this case, the next presumptive secretion event is not statistically significant at a level of 0.05. Thus, the AutoDecon algorithm added this presumptive event, tested it, and subsequently removed it (because it was not significant at a level of 0.05), and then stopped.
Figure 15.1.
An illustrative example of the use of the AutoDecon algorithm. (Top) A set of data points with vertical error bars, with the solid curve corresponding to the predicted concentration, C(t), from Eqs. (15.2), (15.3), and (15.5). The asterisk at the top of the panel marks the location of the single secretion event. (Middle) Weighted differences between the data points and the calculated concentration, with the weighting factors being the Wi in Eq. (15.10). (Bottom) Calculated secretion pattern, S(t), from Eq. (15.5) as a lower curve and the probable position index, PPI(t), as an upper curve. The diamond in this panel marks the location of the next presumed secretory event, which was discarded because it was not statistically significant. Note that the peak of the concentration event in the upper panel occurs significantly later in time than the actual peak of the secretion event. Basal secretion, S0 in Eq. (15.5), is 0.037 per minute. Initial concentration, C0 in Eq. (15.2), is 0.71 min. SecretionSD in Eq. (15.5) is 3.0 min. The elimination half-life, HL in Eq. (15.3), is 50.3 min. The secretion peak position, PP in Eq. (15.5), is 42.4 min.
It is important to note that the peak of the secretion event occurs several minutes before the peak of the corresponding concentration peak. The shape of the secretion event, S(t), dominates the rising portion of the concentration curve, whereas the falling portion of the concentration curve is dominated by the elimination function, E(t). The location of the secretion event (asterisk in the top half of Fig. 15.1) corresponds to a position approximately halfway up the rising portion of the concentration curve.
2.6. Variation on the theme
The objective of phase 1 of the AutoDecon algorithm is to obtain a reasonable initial estimate of the level of basal secretion and find initial estimates of the secretion peak sizes and locations. As outlined, the algorithm works well when the data set being analyzed contains a sufficient amount of information. However, occasionally phase 1 will either overestimate the basal and underestimate the number of the small secretion events or underestimate the basal and overestimate the number of small secretion events. Effectively the algorithm has fallen into the wrong minima of the variance of fit. The later phases of AutoDecon will usually, but not always, correct this situation. This is particularly a problem with limited amounts of data and when thousands of simulated data sets are being analyzed with the totally automated AutoDecon algorithm.
The AutoDecon algorithm is a Systems Identification approach involving multiple weighted nonlinear least-squares parameter estimations. When estimating the parameters of nonlinear equations, by any fitting procedure, the possibility of multiple minima in the variance of fit always exists. The trick to avoiding these incorrect minima is by initializing the fitting algorithm with values that are relatively close to the “correct answers.” Two of the many potential solutions to this multiple minima problem are outlined here; both involve executing the AutoDecon algorithm twice for each data set.
If the AutoDecon algorithm appears to be overestimating the basal secretion and underestimating the number of the small secretion events, then it may be that the P value for the elimination of a secretion event is initially too high. Conversely, if the algorithm underestimates the basal and overestimates the number of small secretion events, then perhaps the initial P value is too small. One potential solution is to execute the AutoDecon algorithm twice. First, execute AutoDecon with a modified P value while keeping the initial estimates of the elimination half-life and SecretionSD constant. AutoDecon will stop after phase 1 since the elimination half-life and SecretionSD are being held constant. Second, execute the AutoDecon algorithm initialized with the results of the first execution, with the desired final P value this time estimating the basal secretion, elimination half-life, and SecretionSD.
For the present GH data it is obvious that the basal secretion is either very low or possibly even zero. In such a case, the AutoDecon algorithm can be executed twice to decrease the possibility of finding multiple minima in the variance of fit. First, execute the AutoDecon algorithm with a very low fixed value for the basal secretion and fixed initial estimates of the elimination half-life and SecretionSD. Again, the program will stop after phase 1 because the elimination half-life and SecretionSD are being held constant. Second, execute the AutoDecon algorithm initialized with the results of the first execution while now estimating the basal secretion, elimination half-life, and SecretionSD.
There are many possible perturbations for the use of AutoDecon. The optimal choice will be dependent on the specific hormone, data collection protocol, and assay characteristics. For the present work we simulated 500 data sets that mimic the results from our experimental results (see data simulation section later) and then tested several perturbations to decide which, if any, was optimal. This second approach was utilized routinely for all of the simulations presented in this chapter. In the current case the secretion rate was initially set to 0.0005 μg/liters/min.
2.7. Outliers
The detection of outliers is a complex statistical issue with a profuse number of divergent criteria depending on the specific application. The mathematical definition of an outlier for this chapter is when the absolute value of the Z score of the particular residual, Zk, is greater than 4. These Z scores are calculated as the particular, kth, residual divided by the root mean squared average value of all of the other residuals, as shown in Eq. (15.9) where N is the total number of residuals:
| (15.9) |
There are a variety of reasons why an outlier may be present in the hormone concentration time series and, as a consequence, in the residuals. An outlier may be the result of a bad data point or may simply be a typographical error that occurred during creation of the data file. An occasional outlier is expected in data due simply to the random measurement uncertainties inherently present within experimental data. Outliers should never merely be discarded, as the apparent existence of such may be caused by some overlooked aspect of data. Thus, in AutoDecon phase 4, the presumed outliers provide useful information about a potential missing secretion event. It is imperative to investigate the cause of an outlier before arbitrarily removing it from the data series.
2.8. Weighting factors
The AutoDecon algorithm is based on a weighted nonlinear least-squares parameter estimation procedure. The required weighting factor corresponds to the inverse of the expected standard error of the mean (SEM) of the particular data point. If every data point were independently sampled and measured more than 15 or 20 times, then the precision of the measured hormone concentrations could be evaluated as the SEM of the replicate samples and measurements. However, with typically only 1 to 3 replicate samples, the SEM of the replicates is far too inaccurate to be used as a realistic estimate of the actual precision of the hormone concentrations. Thus, to estimate the experimental uncertainties in such cases, a variance model was utilized that accounts for the performance characteristics of the clinical laboratory assays. An empirical representation of the variance as a function of the hormone concentration is given in Eq. (15.10):
| (15.10) |
where CV is the % coefficient of variation at the optimal range for the assay, MDC is the minimal detectable concentration, N is the number of replicates, and [Hormone(ti)] is the hormone concentration at time ti. The MDC and CV are measured routinely as part of the quality control measurements performed by the clinical laboratories and thus their values are well known. The minimal detectable concentration is the lowest concentration that can be measured accurately and is measured experimentally as twice the standard deviation (SD) of approximately 15 or 20 samples that contain a hormone concentration of zero.
2.9. AutoDecon performance and comparison with earlier pulse detection algorithms
The present approach for validating pulse detection algorithms involves the creation of synthetic data sets where the number, position, and characteristics of the pulses are “known” a priori (similar to Guardabasso et al., 1988; Van Cauter, 1988; Veldhuis and Johnson, 1988). This chapter modeled these synthetic files upon “real” data obtained from normal control subjects in a clinical study. The pulsatile characteristics observed from these control subjects were incorporated into simulations. AutoDecon and other algorithms were applied to ascertain how well each pulsatile characteristic was recovered by each algorithm.
2.10. Experimental data
Clinical experimental data were obtained from a study approved by the institutional review board of the University of Virginia and the General Clinical Research Center (GCRC). All volunteers gave written informed consent before participating in the study. Sixteen healthy, obese individuals (eight male and eight female) between of the ages of 21 and 55 years of age were recruited by advertisement. The mean (±SD) age (years) of the subjects was 37.1 ± 9.3 [range: 24–55] and the mean (±SD) body mass index (kg/m2) was 34.0 ± 3.0 [range: 30.5–38.4]. Study subjects had maintained a stable weight for the past 6 months (i.e., no change greater than ≈2 kg) and had no intention of losing or gaining weight during the entire study period. Subjects agreed to avoid unusual, unaccustomed, or exceptionally strenuous physical activity from 24 h prior to the start of the study until the conclusion of the study period. Female subjects of reproductive age demonstrated a serum β-human chorionic gonadotropin level consistent with the nongravid state at the prestudy visit. Hormone replacement therapy was allowed if the subject had been on a stable regimen for 3 months or more prior to the study start and the regimen remained stable during the entire study. All subjects had normal renal function and were judged to be in good health on the basis of medical history, physical examination, and routine laboratory tests.
All subjects agreed to avoid the use of any prescription and nonprescription medications and preparations, including any herbal, organic, or nutritional remedy other than a standard approved once-daily multivitamin for the entire study period. No alcoholic beverage consumption occurred for 48 h before or during days when blood samples were drawn. Subjects were permitted to consume two glasses of wine (4 ounces/glass) or two bottles (12 ounces/bottle) of beer or equivalent daily during other times during the study. Grapefruit juice consumption was prohibited for at least 2 weeks prior to the study start and throughout the entire study as grapefruit has been shown to inhibit CYP3A4 activity.
All volunteers were admitted to the GCRC for a 39-h stay. They were encouraged to sleep after 2100 h. Two forearm indwelling venous cannulae were placed in the morning at 0600 for blood sampling and were kept patent with saline and/or heparin flushes. Blood sampling was performed through an indwelling venous cannula every 10 min for 24 h, from 0800 h on day 1 until 088 h on day 2 of admission. Standardized meals were served at 0800, 1300, and 1800 h. Meals were consumed within 30 min with no snacks allowed.
2.11. Growth hormone assay
Serum GH concentrations were measured in duplicate by a fluoroimmunometric assay on an Immulite 2000 analyzer by Diagnostics Products Corporation (DPC). DPC claims an interassay CV of 3.8% at 2.5 μg/liter, 3.5% at 5 μg/liter, and 3.3% at 12.6 μg/liter and an intraassay CV of 3.4% at 2.4 μg/liter, 2.6% at 4.8 μg/liter, and 2.3% at 12 μg/liter and a sensitivity (MDC) of the assay stated to be 0.01 μg/liter. The variability from an analysis of our measured data sets indicates that DPC is conservatively overestimating the actual measurement errors. Our analysis indicates that our instrument has an intraassay CV of approximately 1.64% and a MDC of 0.0066 μg/liter.
2.12. Data simulation
The objective of data simulations is to provide a group of hormone concentration time series where the (1) correct answers (e.g., secretion peak areas and positions) are known a priori and (2) simulations closely mimic the actual experimental observations for a particular hormone within a specific experimental protocol. For this study we simulated 500 data sets for each test condition. These data sets were then analyzed by the AutoDecon algorithm together with several previously published algorithms. A comparison of the apparent results from the AutoDecon software and the correct answers yielded information about the operating characteristics of each of the analysis algorithms (e.g., AutoDecon, Cluster).
The most vital aspect of the data simulation approach is to model actual experimental data in the most exacting way possible. The first step is to analyze a group of actual experimental data with a specific analysis algorithm such as AutoDecon, Cluster, and Pulse in order to create a consensus analysis for each of the actual hormone concentration time series. For the present case, the group of actual experimental data consisted of sixteen 24-h GH concentration time series sampled at 10-min intervals and assayed in duplicate.
Apparent values, based on the analysis of each particular algorithm of the 16 data sets of the basal secretion [S0 in Eq. (15.4)], the SecretionSD [Eq. (15.4)], and the half-life [HL in Eq. (15.2)], are assumed to follow lognormal distributions.
The base 10 logarithms of the individual interpulse intervals are potentially related to log10 of the previous interpulse interval, log10 of the secretion previous event area, log10 of the specific S0, log10 of the specific SecretionSD, log10 of the specific HL, a 24-h cosine wave, and a 24-h sine wave. The relational coefficients were determined by multiple linear regression (MLR) analysis (as in Tables 15.3 and 15.4). The cross-correlations and covariances of the MLR parameters were calculated to verify that this use of the MLR was justified. Sine and cosine functions were included to compensate for the possibility of a 24-h rhythm induced by the sleep–wake cycles of the subjects. Note that the combination of a cosine and a sine wave is a linear model that is mathematically equivalent to a cosine wave with a variable phase. For these simulations, evaluation of the significance of the specific MLR terms is not required. If a term is not significant then it will have negligible amplitude and thus will not contribute to the simulations.
Table 15.3.
Significantly nonzero (p < 0.05) coefficients from multiple linear regressions in Table 15.4a
| Secretion event area | Interpulse interval | |
|---|---|---|
| Log10 previous interval | −1.069 ± 0.123 | −0.245 ± 0.061 |
| Log10 previous event area | 0.264 ± 0.050 | −0.049 ± 0.025 |
| Log10S0 | 1.088 ± 0.228 | |
| Log10SecretionSD | 1.329 ± 0.343 | 0.665 ± 0.158 |
| Cosine (2π TSS/1440) | −0.210 ± 0.051 | 0.056 ± 0.024 |
| Sine (2π TSS/1440) | 0.017 ± 0.049 | 0.117 ± 0.021 |
| R2 | 0.454 | 0.186 |
| Largest cross-correlation | −0.351 | 0.441 |
Backward removal was used to remove nonsignificant coefficients. The sine coefficient for the secretion event area is included because the cosine coefficient is significant.
Table 15.4.
Simulation parameters from a MLR examination of AutoDecon analysis resultsa
| Log10Sec.Arean = N(−0.250,0.326) − 1.073 * [Log10Intervaln−1 − 1.729]+ 0.267 * [Log10Sec.Arean−1 + 0.253 ] + 1.082 * [ Log10S0 + 2.78]+1.324 * [Log10SecretionSD − 0.925] + 0.688 * [Log10HL − 1.151−0.203 * Cos [2πTSS/1440] + 0.026 * Sin [2πTSS/1440] |
| Log10Intervaln = N(1.754,0.063) − 0.054* [Log10Sec.Arean−1 + 0.214] −0.244 * [Log10Intervaln−1 − 1.723 ] + 0.091 * [ Log10S0 + 2.78]+0.674 * [Log10SecretionSD − 0.925] −0.033 * [Log10HL − 1.151]+0.043 * Cos [2πTSS/1440] + 0.108 * Sin [2πTSS/1440] |
| Log10SecretionSD = N(0.935,0.00133) |
| Log10S0 = N(−2.770,0.0.0356) |
| Log10HL = N(1.148,0.00305) |
| MDC = 0.00661 |
| CV = 1.644 |
The n subscript refers to the current interpulse interval and secretion event area, the n − 1 subscript refers to the previous interpulse interval and secretion event area, and TSS is time from 8:00 AM, the start of the hormone sampling period. N (mean, variance) is a pseudo-random variable that follows a normal distribution with the specific mean and variance. Constants were determined by a multiple linear regression based on the experimental measurements. Duplicate samples were simulated unless otherwise noted.
Similarly, the base 10 logarithms of the individual secretion event areas are potentially related to log10 of the previous secretion previous event area, log10 of the interpulse interval, log10 of the specific S0, log10 of the specific SecretionSD, log10 of the specific HL, a 24-h cosine wave, and a 24-h sine wave.
For the actual simulations, the time series is generated to start 24 h prior to the first desired time point and continue for 24 h after the last desired time point. This will normally generate a partial secretion event at both the beginning and the end of the time series. All the secretion events with a position [PPk in Eq. (15.4)] within the desired time are considered to be secretion events, which the various algorithms should be able to locate accurately, including the partial events at the beginning and end of the desired time series.
The simulation is initialized by evaluating unique values for the S0, the SecretionSD, and the HL by generating pseudo-random values based on the corresponding lognormal distribution obtained from experimental data. Next, an interpulse interval and a secretion event area are simulated by generating pseudo-random values that follow the corresponding distributions and dependencies as determined by the multiple linear regressions of results from the analysis of actual experimental data. Initially the time is set to −24 h and the previous secretion event areas and previous interpulse intervals are assumed to be equal to the average values. This process of finding the next interpulse interval and secretion event area is repeated for 24 h past the last desired data point. Gaussian-distributed pseudo-random experimental measurement errors are then added according to Eq. (15.10). Data corresponding to the second of the three simulated days are subsequently utilized as the first simulated hormone concentration time series. This entire process is repeated 500 times to obtain the entire group of 500 simulated hormone concentration time series.
This simulation procedure is substantially different than previous hormone concentration time series simulation algorithms (Guardabasso et al., 1988; Van Cauter, 1988; Veldhuis and Johnson, 1988). The previous algorithms assumed that the interpulse intervals and secretion event sizes each followed independent Gaussian.
2.13. Algorithm operating characteristics
The simulated data sets were analyzed by AutoDecon and several other algorithms for pulse detection. A subsequent comparison between apparent results returned by an algorithm and correct answers from the simulations translates into information about the operating characteristics of each algorithm (e.g., AutoDecon, Cluster). In particular, we used a concordant secretion events criterion (see next section) to evaluate the following.
True positive as the percentage of the apparent secretion events found by the algorithm that closely corresponds in time to actual secretion events
False positive (type I errors) as the percentage of the apparent secretion events found by the algorithm that do not closely correspond in time to actual secretion events
False negative (type II errors) as the percentage of simulated secretion events not located by the algorithm that occur between the first and the last data points
Sensitivity as the percentage of actual simulated secretion events located by the algorithm
Our definition of false negative (type II errors) includes a stipulation that the center of the secretion events must be between the first and the last data points to be considered as a false-negative incident. In approximately 10% of the simulated data sets a secretion event occurred slightly later than the last data point but close enough that the leading edge of the secretion event has a significant contribution to the hormone concentration before the last data point. This stipulation is included so that the failure to find secretion events outside the range of data is not considered as an error. These out-of-range secretion events are, however, included in the calculations of true positives. The analogous partial secretion events that occur before the time of the first data point are incorporated into the C(0) term by the analysis algorithm and are thus not treated as a special case.
2.14. Concordant secretion events
Determining the operating characteristics of the algorithms requires a comparison of the apparent secretion event positions from an analysis of a simulated time series, with the actual known secretion event positions upon which the simulations were based. This process must consider whether the concordance of the peak positions is statistically significant or whether it is a consequence of a simply random position of the apparent secretion events. For example, when comparing two time series, each having approximately 20 randomly positioned events within a 24-h period, it is unlikely (P < 0.05) that 6 or more of these random events will appear to be coincident within a ±5-min window, that 8 or more will appear to be coincident within a ±10-min window, and that 11 or more will simultaneously occurring within a ±20-min window.
Specifically, the following question could be posed: given two time-series with n and m distinct events, what is the probability that j coincidences (i.e., concordances) will occur based on a random positioning of the distinct events within each of the time series? The resulting probabilities are dependent on the size of the specific time window employed for the definition of coincidence. This question can be resolved easily by a Monte Carlo approach. One hundred thousand pairs of time series are generated with the n and m distinct randomly timed events, respectively. The distribution of the expected number of concordances can then be evaluated by scanning these pairs of random event sequences for coincident peaks where coincidence is defined by any desired time interval.
Obviously, as the coincidence interval increases so will the expected number of coincident events. Thus, the coincidence interval should be kept small. Coincidence windows of ±5, ±10, or maybe ±15 min are reasonable for this study simply because the data sampling is at 10-min intervals. The expected number of coincident events will also increase with the numbers of distinct events, n and m.
3. Results
Growth hormone data chosen to test the AutoDecon algorithm were selected specifically to represent a particularly difficult case. The subjects are neither young nor lean. The size of the observed GH secretion events is expected to be decreased because of both of these factors. In addition, sampling at 10-min intervals makes the resolution of an approximately 14-min elimination half-life somewhat difficult. However, because the hormone concentrations were measured in duplicate with state-of-the-art instrumentation, data from this protocol are typical of the most pristine GH concentration time series data currently available.
3.1. Typical AutoDecon output
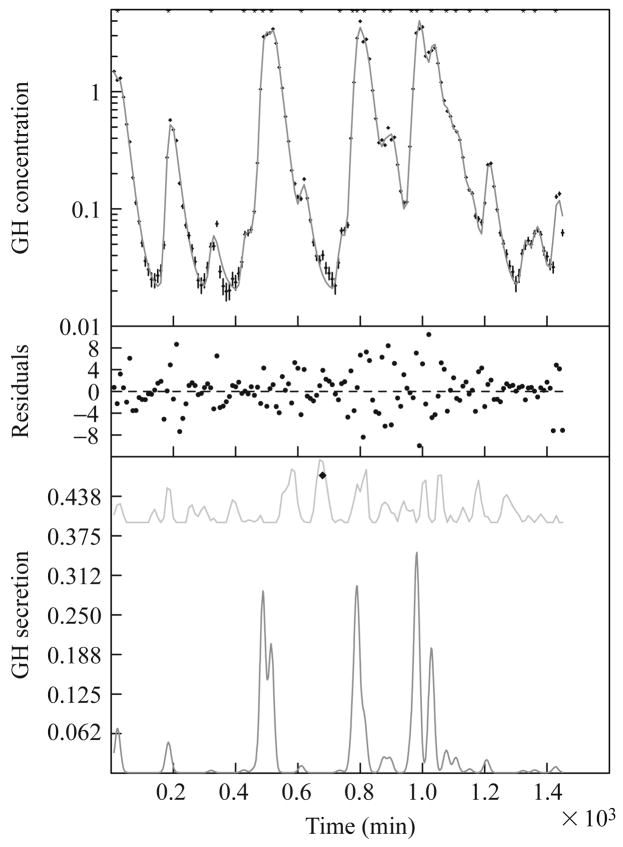
Figure 15.2 presents an example of the analysis of a typical GH time series by the AutoDecon algorithm. The upper part of Fig. 15.2 depicts the GH data points as vertical error bars and best-fit calculated GH concentrations. These are on a logarithmic scale to enhance visualization of the small secretion events. For this specific GH data set, the AutoDecon algorithm identified 24 secretion events with a median interpulse interval of 45.3 min, an elimination half-life of 13.1 min, and a secretion event standard deviation of 8.8 min. The median of the interpulse intervals is given because (as demonstrated later) the distribution of interpulse intervals is not Gaussian (skewness of 0.973 and kurtosis of −0.190), and thus the mean interpulse interval of 56.6 ± 7.9 min is not as informative. The median of 0.522 for the secretion event area (i.e., mass) is also preferable as its distribution is even more non-Gaussian (skewness of 1.594 and kurtosis of 1.281), and thus the mean secretion event area of 1.65 ± 0.46 is even less informative.
Figure 15.2.
An example of the analysis of a typical growth hormone concentration time series by the AutoDecon algorithm. (Top) A set of data points with vertical error bars and a solid curve corresponding to the predicted concentration, C(t), from Eqs. (15.2), (15.3), and (15.5). The asterisk at the top of the panel denotes the location of the single secretion event. (Middle) Weighted differences between the data points and the calculated concentration, with the weighting factors being the Wi in Eq. (15.10). (Bottom) The calculated secretion pattern, S(t), from Eq. (15.5) as a lower curve and the probable position index, PPI(t), as an upper curve. The diamond in this panel marks the location of the next presumed concentration event, which was discarded because it was not statistically significant. Note that the peak of the concentration event in the upper panel occurs significantly later in time than the actual peak of the secretion event.
The probable position index (the upper line in the lower part of Fig. 15.2) indicates that the final presumptive secretion event was located at 666 min into the collection protocol. Visually it appears from the differences between the data points and the calculated curve (upper part of Fig. 15.2) that this might be an actual secretion event. However, this secretion event was not included because of its 0.0505 significance level. Having a significance level over 0.05 does not mean that the secretion event is not present, but simply means that it cannot be demonstrated at the 0.05 significance level. There is another possible secretion event at approximately 200 min into the collection protocol. If a less stringent significance level had been utilized, the number of secretion events would be increased and the apparent basal secretion level would, as a consequence, be lower.
3.2. Experimental data analysis
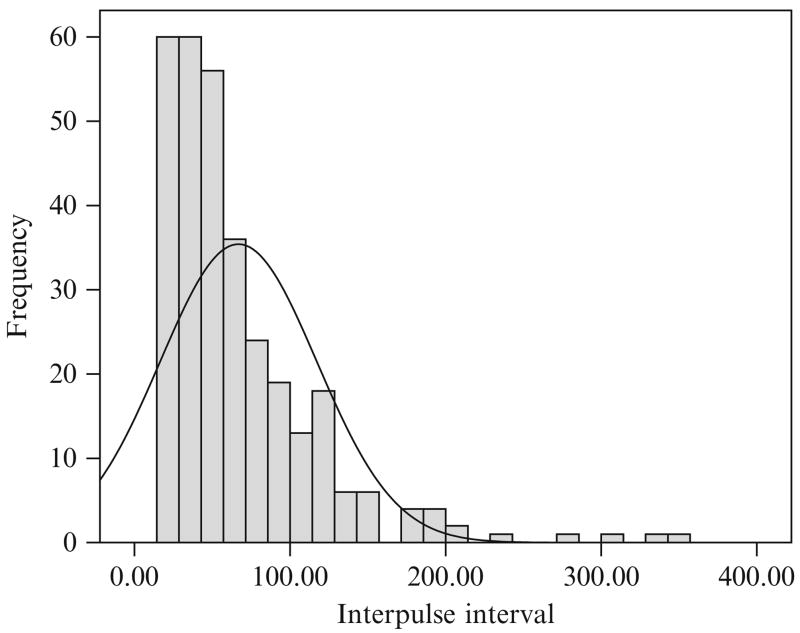
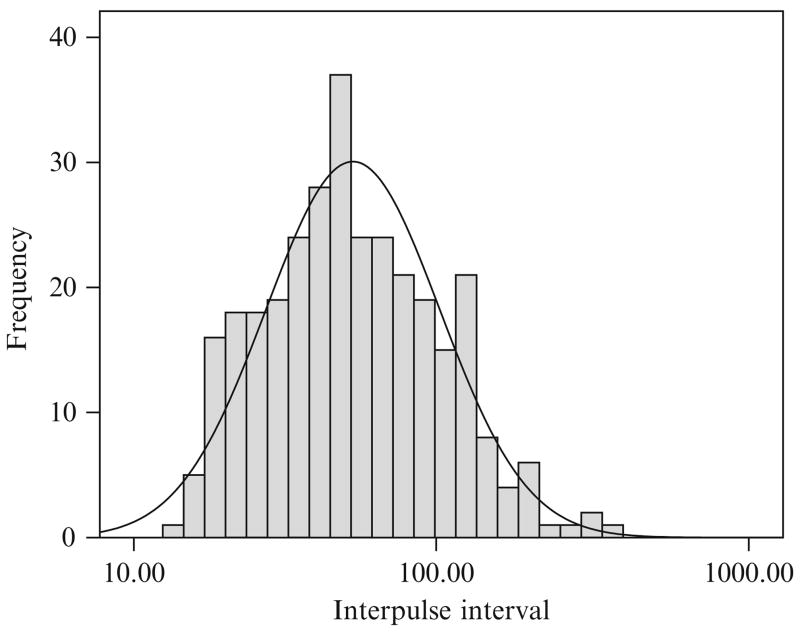
The combined results of the AutoDecon analysis of all 16 of the time series from this protocol are summarized in Table 15.1. In several cases the medians are not close to the means, which implies that the distributions are not Gaussian (i.e., distributed normally). Similarly, the standard deviations of these cases are also sufficiently large that a normal distribution will include a high percentage of values less than zero and thus not physically meaningful. This can clearly be seen in the distribution of interpulse intervals (Fig. 15.3) and in the logarithmic normal distribution of interpulse intervals (Fig. 15.4). The interpulse intervals appear to be distributed approximately lognormally.
Table 15.1.
Summary of secretion properties of the 16 hormone concentration time series analyzed with AutoDecon
| Median | Mean ± SEM | SD | N | Interquartile Range | |
|---|---|---|---|---|---|
| S0 | 0.00138 | 0.00187 ± 0.00024 | 0.00095 | 16 | 0.00138 |
| SecretionSD | 9.02 | 8.89 ± 0.58 | 2.31 | 16 | 2.72 |
| HL | 14.32 | 14.17 ± 0.43 | 1.72 | 16 | 2.08 |
| # events | 18 | 20.56 ± 0.93 | 3.71 | 16 | 7.0 |
| # outliers | 1 | 0.63 ± 0.16 | 0.62 | 16 | 1.0 |
| Interpulse interval | 50.49 | 66.82 ± 2.85 | 51.37 | 313 | 51.54 |
| Event size (area) | 0.51 | 2.14 ± 0.19 | 3.50 | 329 | 2.39 |
| % pulsatile | 93.87 | 93.13 ± 0.68 | 2.74 | 16 | 3.51 |
Figure 15.3.
The distribution of interpulse intervals for the entire set of 32 concentration time series found by the AutoDecon algorithm. The solid line corresponds to the best normal (i.e., Gaussian) distribution.
Figure 15.4.
The distribution of interpulse intervals for the entire set of 32 concentration time series found by the AutoDecon algorithm but plotted on a logarithmic X axis. The solid line corresponds to the best lognormal distribution.
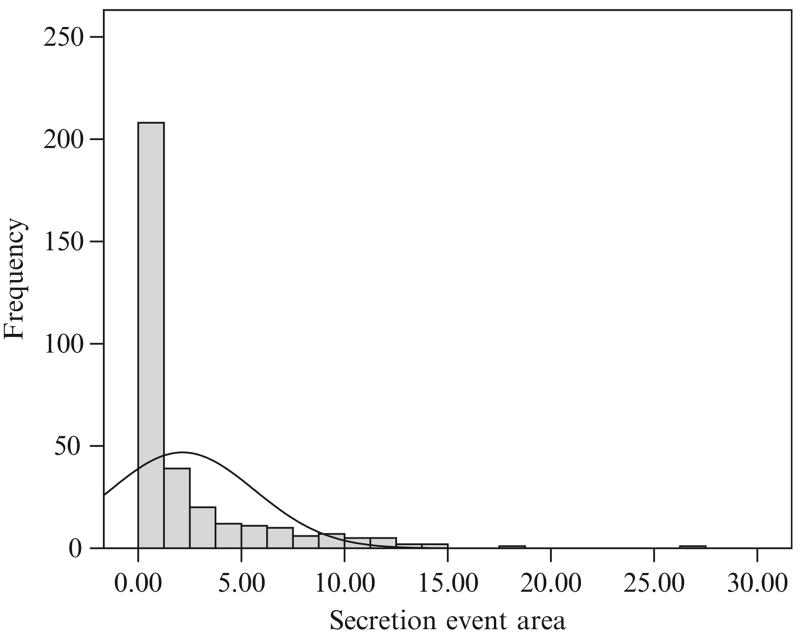
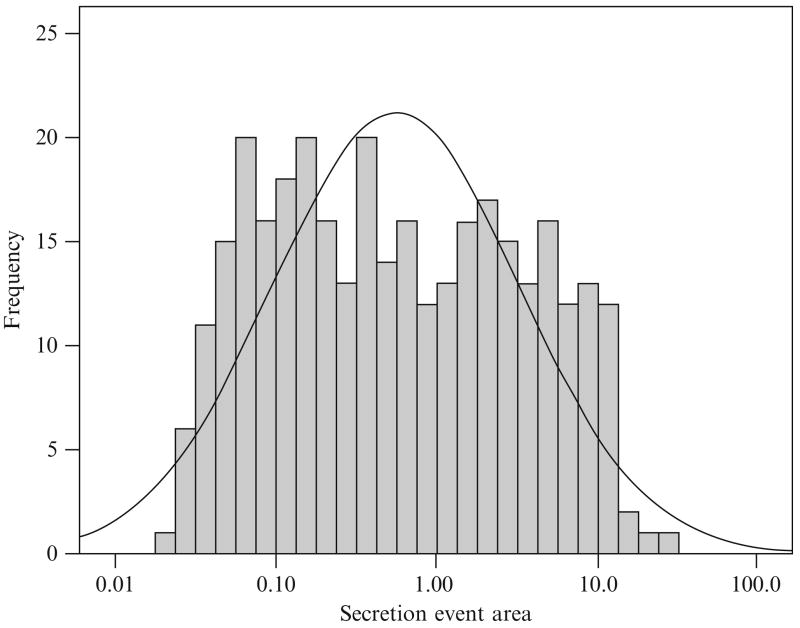
The distributions of the secretion event sizes also cannot be analyzed as a normal distribution (Figs. 15.5 and 15.6). Here the secretion event sizes appear to follow a bimodal (or multimodal) lognormal distribution.
Figure 15.5.
The distribution of secretion event sizes for the entire set of 32 concentration time series found by the AutoDecon algorithm. The solid line corresponds to the best normal (i.e., Gaussian) distribution.
Figure 15.6.
The distribution of secretion event sizes for the entire set of 32 concentration time series found by the AutoDecon algorithm but plotted on a logarithmic X axis. The solid line corresponds to the best lognormal distribution.
The first column of Table 15.2 presents the apparent number of secretion events found by several of the previously published hormone pulse detection algorithms. The Pulse and Pulse2 algorithms have two different entries where the first assumes that the basal secretion is zero and the second assumes that the basal secretion is not zero. Obviously, the operating characteristics of these algorithms need to be determined when applied to GH data so that an appropriate choice of algorithms, and thus results, can be made. The choice of algorithm will be dependent on the specific hormone being investigated, the details of the specific experimental protocol, and the laboratory instrumentation utilized for the hormone concentration measurements.
Table 15.2.
Mean number of secretion events found for the 16 time series by several different algorithms
| Algoritham | Number of secretory events |
Reference | |
|---|---|---|---|
| Original data | Shuffled dataa | ||
| AutoDecon | 20.6 | 0.0 | |
| Clusterb | 12.3 | 22.1 | Veldhuis and Johnson (1986) |
| Pulse (zero basal) | 13.8 | 18.2 | Johnson and Straume (1999) |
| Pulse (variable basal) | 7.2 | 15.6 | Johnson and Straume (1999) |
| Pulse2 (zero basal) | 11.2 | 1.4 | Johnson and Straume (1999) |
| Pulse2 (variable basal) | 12.9 | 0.3 | Johnson and Straume (1999) |
| Pulse4 | 1.0 | 1.0 | Johnson et al. (2004) |
Assuming HL = 14.25 and Secretion SD = 8.95.
Cluster algorithm nadir size and peak size of 2, and both T statistics set to 2.0.
One simple test of the algorithms is to see how they perform when data are shuffled (i.e., randomizing the time sequence of data). This is similar to a previously published procedure for the evaluation of the percentage of false positives (Van Cauter, 1988). The hormone concentration pulsatile events follow a distinct shape, that is, a fast rise and a somewhat slower fall (Johnson and Veldhuis, 1995; Veldhuis et al., 1987). Any operation, such as shuffling the data points, that disrupts the time sequence of the data time series should also disrupt the ability of the hormone detection algorithms to find distinct secretion events within the data sequence. Thus, it is surprising that the Cluster and the Pulse algorithms actually found more apparent events within shuffled (i.e. randomized) data than in original data (Table 15.2, column 2). This is sufficient reason not to use either the Cluster or the Pulse algorithm for GH data.
The Pulse4 algorithm found the fewest peaks of all of the algorithms (Table 15.2), reflecting the conditions of this particular GH protocol, which are well outside the bounds of applicability for the Pulse4 algorithm. Specifically, the Pulse4 algorithm requires more than four data points per elimination half-life while the present protocol samples the data points at a 10-min interval even though GH is known to exhibit an elimination half-life of approximately 14 min, yielding only 1.4 data points per half-life. Thus, Pulse4 clearly is not applicable for this particular GH protocol.
Multiple linear regressions were applied to the interpulse interval and secretion event areas, which yielded some interesting results. The apparent secretion event area is controlled significantly (P < 0.05) by the previous interpulse interval, the previous secretion event area, the basal secretion, and the SecretionSD (see Table 15.3). The interpulse interval is also controlled significantly (P < 0.05) by the previous interpulse interval, the previous secretion event area, and the SecretionSD, but with a much smaller magnitude. Both the intervals and the secretion event areas also appear to be controlled by a 24-h rhythm. These apparent controlling factors may be a reflection of the underlying molecular mechanisms of secretion or they may simply be a limitation of the numerical analysis.
3.3. Comparison between AutoDecon and other algorithms
Table 15.4 presents the simulation parameters utilized to create the first set of 500 simulated hormone concentration time series. This first set was generated to mimic results from the analysis of experimental data by the AutoDecon algorithm. Table 15.4 was created by MLR analysis of the AutoDecon estimated results. In this, and all subsequent simulations, it is assumed that the individual interpulse intervals and secretion event areas could be a function of the previous individual interpulse intervals, secretion event areas, elimination half-life, basal secretion, secretion event standard deviation, and sum of 24-h sine and cosine waves.
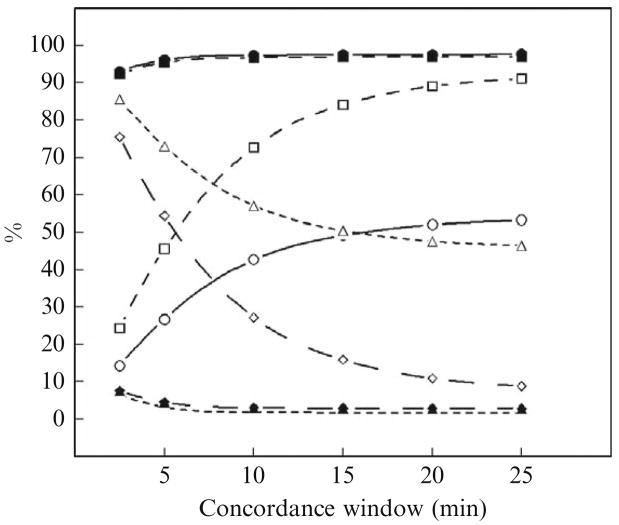
Figure 15.7 presents a comparison of the operating characteristics of the AutoDecon and Cluster (Veldhuis and Johnson, 1986) algorithms when applied to these 500 simulated hormone concentration time series. In Fig. 15.7, true-positive events were determined for a series of different concordance windows. For example, if the algorithm locates a secretion event within a simulated hormone concentration time series to be within ±5 min of the exact location of the secretion event upon which the simulations were based, then it is a true positive with a concordance window of ±5 min or less. AutoDecon performs better by all measures with a ±2.5-min, or longer, concordance window than Cluster with a ±25-min concordance window. For example, with a ±10-min concordance window the sensitivity of AutoDecon is 97.5% and Cluster is 42.7%.
Figure 15.7.
Performance characteristics of the AutoDecon and Cluster algorithms at different concordance windows. These were evaluated by the analysis of 500 simulated data sets generated as described in Table 15.4. Solid symbols are for AutoDecon and open symbols are for Cluster. Solid lines and circles describe the sensitivity %; medium-dashed lines and squares describe the true positive %; long-dashed lines and diamonds describe the false positive %; and short-dashed lines and triangles describe the false negative %.
It is interesting to note that 25% of the false positives identified by AutoDecon corresponded to the first apparent secretion event and that 24% of the false negatives corresponded to the last simulated secretion event in the sequence. These are substantially higher that the ≈5% expected from random chance. It is, however, not unexpected, as the first and last simulated secretion events are contained only partially within the data sequence.
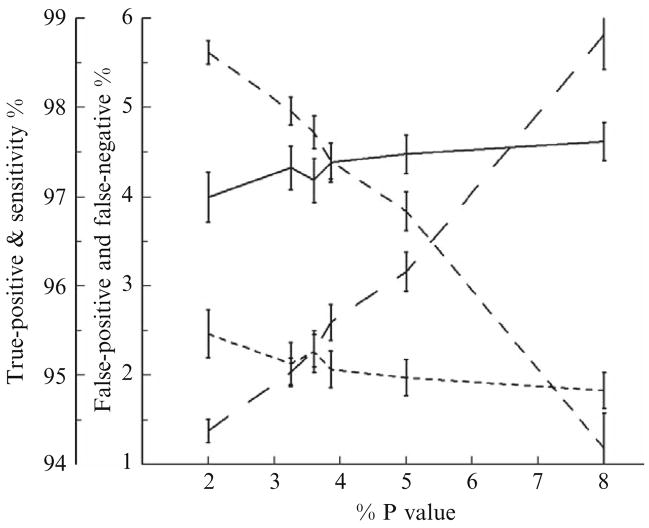
Figure 15.8 presents the corresponding mean true positive %, false positive %, false negative %, and sensitivity of the AutoDecon algorithm when different P values are used by the triage module. These were evaluated with the same 500 simulated hormone concentration time series used in Fig. 15.7. Clearly, the AutoDecon algorithm outperforms the Cluster algorithm for all triage P values between 0.02 and 0.08. The false positive % and false negative % are both approximately 2.1% when the triage P value is set to 0.0325. When the triage P value is set to 0.039, both the true positive % and the sensitivity are approximately 97.4%. Please note that these values only apply to GH data collected with this specific protocol. The actual choice triage P value is obviously dependent on which of these operating characteristics is deemed the most important. For the remainder of this chapter, the triage P value will be set to 0.0325, which represents a reasonable value to jointly optimize the false positive % and false negative %.
Figure 15.8.
Performance characteristics of AutoDecon as a function of the P value used by the algorithm’s triage module for the removal of nonsignificant secretion events. Solid lines describe the sensitivity %; medium-dashed lines describe the true positive %; long-dashed lines describe the false positive %; and short-dashed lines describe the false negative %.
Table 15.5 presents a comparison of the operating characteristics of five of the analysis algorithms when applied to these 500 simulated hormone concentration time series. It is clear from Table 15.5 that the AutoDecon algorithm performs far better than the other algorithms when simulated data are based on results obtained by the AutoDecon algorithm. It is important to note that the operating characteristics shown in Table 15.5. apply only to the analysis of GH sampled every 10 min with the experimental measurement errors shown in Table 15.4. These operating characteristics will undoubtedly vary with different hormones and possibly even with different experimental protocols for the measurement of GH.
Table 15.5.
Comparison of performance of analysis algorithms evaluated from simulated data based on AutoDecon results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon (P value = 0.0325)a | |||
| %true positive | 100.00 | 97.96 ± 0.16 | 4.76 |
| %false positive | 0.00 | 2.04 ± 0.16 | 4.76 |
| %false negative | 0.00 | 2.13 ± 0.24 | 0.00 |
| %sensitivity | 100.00 | 97.32 ± 0.25 | 5.26 |
| Clusterb | |||
| %true positive | 90.00 | 89.13 ± 0.50 | 18.18 |
| %false positive | 10.00 | 10.87 ± 0.50 | 18.18 |
| %false negative | 47.37 | 47.64 ± 0.49 | 13.82 |
| %sensitivity | 52.00 | 52.08 ± 0.49 | 13.61 |
| Pulse with no basal secretion and half-life = 14.25 mina | |||
| %true positive | 75.00 | 74.91 ± 0.54 | 16.67 |
| %false positive | 25.00 | 25.09 ± 0.54 | 16.67 |
| %false negative | 37.50 | 37.41 ± 0.47 | 13.11 |
| %sensitivity | 61.91 | 62.25 ± 0.47 | 13.19 |
| Pulsea with basal secretion and half-life = 14.25 mina | |||
| %true positive | 100.00 | 99.00 ± 0.19 | 0.00 |
| %false positive | 0.00 | 1.00 ± 0.19 | 0.00 |
| %false negative | 55.28 | 53.97 ± 0.60 | 18.18 |
| %sensitivity | 44.44 | 45.78 ± 0.60 | 18.13 |
| Pulse2 with no basal secretiona | |||
| %true positive | 76.92 | 76.73 ± 0.59 | 19.85 |
| %false positive | 23.08 | 23.27 ± 0.59 | 19.84 |
| %false negative | 23.91 | 25.51 ± 0.63 | 16.23 |
| %sensitivity | 75.00 | 74.09 ± 0.64 | 16.67 |
| Pulse2 with basal secretiona | |||
| %true positive | 90.00 | 86.78 ± 0.57 | 21.43 |
| %false positive | 10.00 | 13.22 ± 0.57 | 21.43 |
| %false negative | 25.00 | 25.98 ± 0.58 | 17.19 |
| %sensitivity | 75.00 | 74.61 ± 0.58 | 21.43 |
| Pulse4 with variable half-lifea | |||
| %true positive | 0.00 | 41.15 ± 2.19 | 100.00 |
| %false positive | 100.00 | 58.85 ± 2.19 | 100.00 |
| %false negative | 100.00 | 94.14 ± 0.64 | 5.89 |
| %sensitivity | 0.00 | 5.84 ± 0.64 | 5.89 |
| Pulse4 with half-life = 14.25 mina | |||
| %true positive | 100.00 | 66.33 ± 2.10 | 100.00 |
| %false positive | 0.00 | 33.67 ± 2.10 | 100.00 |
| %false negative | 94.59 | 94.23 ± 0.44 | 6.67 |
| %sensitivity | 5.26 | 5.74 ± 0.44 | 6.25 |
Requiring alignment of secretion event positions to be within ±10 min.
Requiring alignment of secretion event positions to be within ±20 min, evaluated with Cluster parameters of 2 × 2 and T statistics = 2.0.
Table 15.6 presents an analogous analysis based on 500 simulated hormone concentration time series generated to mimic the results of the analysis of experimental data by the Cluster algorithm. Clearly, the AutoDecon algorithm significantly outperforms the Cluster algorithm, even when data are simulated to mimic the Cluster algorithm results.
Table 15.6.
Comparison of performance of AutoDecon and Cluster algorithms evaluated from simulated data based on Cluster results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon P (value = 0.0325)a | |||
| %true positive | 100.00 | 96.20 ± 0.25 | 8.33 |
| %false positive | 0.00 | 3.81 ± 0.25 | 8.33 |
| %false negative | 6.91 | 6.29 ± 0.31 | 9.09 |
| %sensitivity | 92.86 | 93.33 ± 0.31 | 10.00 |
| Clusterb | |||
| %true positive | 87.50 | 85.15 ± 0.51 | 13.89 |
| %false positiv | 12.50 | 14.85 ± 0.51 | 13.89 |
| %false negative | 28.57 | 29.15 ± 0.51 | 14.54 |
| %sensitivity | 71.43 | 70.56 ± 0.51 | 14.14 |
Peak alignment within 10 min.
Peak alignment within 20 min.
In both Tables 15.5 and 15.6 the window to accept a match between the simulated secretion event positions and the apparent event locations found was taken to be 20 min for the Cluster algorithm and 10 min for the AutoDecon algorithm. Thus, the AutoDecon algorithm outperformed the Cluster algorithm even with a significant test bias that favors the Cluster algorithm.
A similar analysis was performed for the Pulse and Pulse2 algorithms with and without basal secretion and the results presented in Tables 15.7–15.10. In every case the AutoDecon algorithm performed significantly better than either the Pulse or the Pulse2 algorithm.
Table 15.7.
Comparison of performance of AutoDecon and Pulse algorithms evaluated from simulated data based on Pulse with no basal secretion results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon (P value = 0.0325)a | |||
| %true positive | 100.00 | 97.85 ± 0.17 | 5.26 |
| %false positive | 0.00 | 2.16 ± 0.17 | 5.26 |
| %false negative | 0.00 | 0.67 ± 0.09 | 0.00 |
| %sensitivity | 100.00 | 98.96 ± 0.11 | 0.00 |
| Pulse with no basal secretion and half-life = 14.25 mina | |||
| %true positive | 93.54 | 93.19 ± 0.35 | 10.00 |
| %false positive | 6.46 | 6.81 ± 0.35 | 10.00 |
| %false negative | 23.08 | 21.36 ± 0.63 | 22.03 |
| %sensitivity | 76.47 | 78.35 ± 0.63 | 10.00 |
Peak alignment within 10 min.
Table 15.10.
Comparison of performance of AutoDecon and Pulse2 algorithms evaluated from simulated data based on Pulse2 with basal secretion results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon (P value = 0.01)a | |||
| %true positive | 100.00 | 98.91 ± 0.12 | 0.00 |
| %false positive | 0.00 | 1.09 ± 0.12 | 0.00 |
| %false negative | 0.00 | 0.28 ± 0.15 | 0.00 |
| %sensitivity | 100.00 | 99.44 ± 0.16 | 0.00 |
| AutoDecon (P value = 0.02)a | |||
| %true positive | 100.00 | 98.06 ± 0.17 | 0.00 |
| %false positive | 0.00 | 1.94 ± 0.17 | 0.00 |
| %false negative | 0.00 | 0.28 ± 0.15 | 0.00 |
| %sensitivity | 100.00 | 99.44 ± 0.16 | 0.00 |
| AutoDecon (P value = 0.0325)a | |||
| %true positive | 100.00 | 96.98 ± 0.20 | 6.67 |
| %false positive | 0.00 | 3.02 ± 0.20 | 6.67 |
| %false negative | 0.00 | 0.24 ± 0.15 | 0.00 |
| %sensitivity | 100.00 | 99.47 ± 0.16 | 0.00 |
| Pulse2 with basal secretiona | |||
| %true positive | 100.00 | 97.50 ± 0.21 | 5.56 |
| %false positive | 0.00 | 2.50 ± 0.21 | 5.56 |
| %false negative | 0.00 | 5.64 ± 0.35 | 8.71 |
| %sensitivity | 94.44 | 94.08 ± 0.35 | 9.09 |
Peak alignments within 10 min.
The optimal P value for the removal of nonsignificant secretion events was determined (see Fig. 15.8) to be approximately 0.035 for simulated data based on the AutoDecon analysis of experimental data. However, for simulated data based on the Pulse2 analysis of experimental data (see Table 15.10), the optimal P value is substantially lower, on the order of 0.01 or less. The origin of this difference is that the AutoDecon algorithm is identifying more, and smaller, secretion events not identified by the Pulse2 algorithm. Thus, AutoDecon will have fewer small secretion events to identify when it is applied to simulated data based on the Pulse2 analysis. Hence, AutoDecon will have a lower false-negative percentage with these data. Consequently, the optimal P value is dependent on characteristics of the particular experimental data being analyzed.
3.4. Optimal sampling paradigms for AutoDecon
The general idea of examining a large number of simulated hormone concentration time series may also be employed to address questions such as: What if the samples were assayed in singlicate or triplicate instead of duplicate? Would 5-min sampled singlicates be better than 10-min sampled duplicates or 15-min sampled triplicates? What would happen if an older instrument with a larger measurement uncertainty (i.e., MDC and CV) was used for the assays? The easiest way to answer these questions is to simulate the appropriately corresponding hormone concentration time series and then determine how the particular analysis algorithm will perform with those series of data.
Table 15.11 addresses the question of singlicate vs duplicate vs triplicate assays. The data series samples were simulated as in Table 15.4 with the exception that three groups of 500 hormone concentration time series were simulated with singlicate, duplicate, and triplicate assays, respectively. It is, of course, expected that the results will improve as the number of replicates is increased. Based on the analysis of these simulated time series, it appears that the major effect is that the false-negative percentage improves as the number of replicates increases. Virtually every secretion event identified by the AutoDecon algorithm is an actual secretion event, and thus there is little room for improvement in the false-positive percentages.
Table 15.11.
Comparison of performance of the AutoDecona algorithm when samples are simulated in singlicate, duplicate, and triplicate
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| Singlicate samples | |||
| %true positive | 100.00 | 97.76 ± 0.18 | 5.26 |
| %false positive | 0.00 | 2.24 ± 0.18 | 5.26 |
| %false negative | 0.00 | 3.27 ± 0.23 | 5.88 |
| %sensitivity | 00.00 | 96.06 ± 0.24 | 6.67 |
| Duplicate samples | |||
| %true positive | 100.00 | 97.96 ± 0.16 | 4.76 |
| %false positive | 0.00 | 2.04 ± 0.16 | 4.76 |
| %false negative | 0.00 | 2.13 ± 0.24 | 0.00 |
| %sensitivity | 100.00 | 97.32 ± 0.25 | 5.26 |
| Triplicate samples | |||
| %true positive | 100.00 | 97.56 ± 0.19 | 5.26 |
| %false positive | 0.00 | 2.44 ± 0.19 | 5.26 |
| %false negative | 0.00 | 2.06 ± 0.23 | 0.00 |
| %sensitivity | 100.00 | 97.47 ± 0.24 | 5.00 |
Peak alignment within 10 min, P value = 0.0325.
Table 15.12 addresses the question of 5-min sampled singlicates vs 10-min sampled duplicates vs 15-min sampled triplicates. In this case, each of the groups incurs the same expense in terms of assay samples and thus addresses the question of the effects of the sampling interval at a constant cost for the assays. Again, the samples were simulated as in Table 15.4 with the exception that the groups of 500 hormone concentration time series were simulated with 5-min sampled singlicates, 10-min sampled duplicates, etc. The expectation was that the results would improve as the interval between the data points decreases, as this provides a better temporal resolution for the secretion events. Based on the analysis of these simulated time series, it is apparent that the false-negative and true-positive rates both improve as the sampling interval decreases.
Table 15.12.
Performance comparison of the AutoDecon algorithm when samples are simulated in 5-min sampled singlicates, 10-min sampled duplicates, etc.
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| 5-min singlicate samplesa | |||
| %true positive | 100.00 | 97.44 ± 0.31 | 4.76 |
| %false positive | 0.00 | 2.36 ± 0.24 | 4.76 |
| %false negative | 0.00 | 2.84 ± 0.30 | 5.26 |
| %sensitivity | 100.00 | 96.89 ± 0.30 | 5.56 |
| 10-min duplicate samplesa | |||
| %true positive | 100.00 | 97.96 ± 0.16 | 4.76 |
| %false positive | 0.00 | 2.04 ± 0.16 | 4.76 |
| %false negative | 0.00 | 2.13 ± 0.24 | 0.00 |
| %sensitivity | 100.00 | 97.32 ± 0.25 | 5.26 |
| 15-min triplicate samplesa | |||
| %true positive | 100.00 | 97.26 ± 0.19 | 5.88 |
| %false positive | 0.00 | 2.74 ± 0.19 | 5.88 |
| %false negative | 0.00 | 5.36 ± 0.40 | 6.67 |
| %sensitivity | 97.92 | 93.94 ± 0.41 | 9.52 |
| 20-min quadruplicate samplesa | |||
| %true positive | 94.12 | 94.24 ± 0.30 | 8.33 |
| %false positive | 5.88 | 5.76 ± 0.30 | 8.33 |
| %false negative | 7.70 | 12.12 ± 0.57 | 17.65 |
| %sensitivity | 90.69 | 87.20 ± 0.57 | 13.99 |
Peak alignment within 10 min, P value = 0.0325.
Table 15.13 presents an analogous simulation except that the experimental measurement errors are simulated at three different levels: (1) the level observed in our experiments, (2) the level that the manufacturer claims for the Immulite 2000 instrumentation, and (3) the level that the manufacturer claims for the Immulite 1000 instrument. These measurement uncertainty levels represent the levels typical of the currently available instrumentation. Results from these simulations demonstrate that the true positive (%) and false positive (%) do not degrade rapidly as the measurement uncertainty increases. However, the percent false negative and percent sensitivity do diminish significantly with increasing measurement uncertainties.
Table 15.13.
Comparison of performance of the AutoDecona algorithm when samples are simulated with different measurement uncertainties
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| Our observed noise level | |||
| MDC = 0.00661 and CV = 1.644% | |||
| %true positive | 100.00 | 97.96 ± 0.16 | 4.76 |
| %false positive | 0.00 | 2.04 ± 0.16 | 4.76 |
| %false negative | 0.00 | 2.13 ± 0.24 | 0.00 |
| %sensitivity | 100.00 | 97.32 ± 0.25 | 5.26 |
| Immulite 2000 reported noise level | |||
| MDC = 0.01 and CV = 3% | |||
| %true positive | 100.00 | 97.80 ± 0.17 | 5.26 |
| %false positive | 0.00 | 2.20 ± 0.17 | 5.26 |
| %false negative | 0.00 | 4.37 ± 0.29 | 6.67 |
| %sensitivity | 100.00 | 95.24 ± 0.30 | 7.14 |
| Immulite 1000 reported noise level | |||
| MDC = 0.01 and CV = 6% | |||
| %true positive | 100.00 | 97.25 ± 0.21 | 5.56 |
| %false positive | 0.00 | 2.76 ± 0.21 | 5.56 |
| %false negative | 5.88 | 7.03 ± 0.24 | 11.11 |
| %sensitivity | 93.93 | 92.55 ± 0.39 | 11.88 |
Peak alignment within 10 min, P value = 0.0325.
4. Discussion
The process of validating AutoDecon or any other data analysis software involves analyzing a large number of data sets where the expected results are known a priori. Given data sets with known results, it is simple to calculate how well the software and algorithms perform with the specific data sets. There are basically two methods of obtaining hormone concentration time series data where the answers are known: an experimental approach and a mathematical approach. The experimental approach involves manipulation of the biological system such that the hormone pulses are secreted at specific known times and with specific known amounts. This manipulation can theoretically be achieved by administering drugs or hormones at specific times that will induce and/or inhibit the secretion or elimination of the hormone of interest. Obviously, this experimental approach is extremely difficult, if not impossible, to control and achieve while also being time-consuming and expensive. The alternative mathematical approach is to simulate a large number of synthetic data sets with assumed (i.e., known) values of the model parameters and realistic amounts of random experimental measurement errors (Guardabasso et al., 1988; Van Cauter, 1988; Veldhuis and Johnson, 1988). The more realistic the simulations, the more realistic the conclusions about the operating characteristics of the software and algorithms can be.
Within this context, a particularly difficult set of GH data was chosen to test the AutoDecon algorithm. The size of the observed GH secretion events is small because the subjects are neither young nor lean. In addition, resolution of an approximately 14-min elimination half-life from data sampled at 10-min intervals is challenging. However, because the hormone concentrations were measured in duplicate with state-of-the-art instrumentation, data from this protocol are typical of the best GH concentration time series data currently available.
The best method for validation of the software is to try all possibilities for all of the variables involved in the simulation. Thus, the variables required for these simulations include the number of replicate assays, the sampling interval, the MDC, the CV, the mean interpulse interval, the standard deviation of the mean interpulse interval, the mean secretion event amplitude and its standard deviation, the SecretionSD, and the HL. However, it is clearly impossible to test all possible combinations and correlations for all of these variables. Indeed, generalizing these operating characteristics to all possible data sets cannot be achieved except in the simplest of cases simply due to the large number of potential variables involved. The more realistic approach is to create synthetic data that mimic the actual hormone concentration time series obtained by a specific experimental protocol and group of subjects. The resulting operating characteristics can then be applied to that specific hormone, experimental protocol, and its subjects.
Key to the simulation approach is the modeling of actual experimental data in the most rigorously precise way possible. Figures 15.3–15.6 clearly demonstrate that neither the interpulse intervals nor the secretion event areas can be described by Gaussian distributions. However, the interpulse intervals and the secretion event sizes are commonly modeled and reported in the literature as a Gaussian distribution (i.e., simply as a mean and standard deviation). If modeled this way, the actual number of secretion events will be underestimated significantly (because the distribution of intervals between secretion events will be overestimated) and the distribution of sizes of secretion events will be overestimated. Simulations with too few excessively large secretion peaks will be much easier to analyze and will, in all likelihood, falsely inflate the operating characteristics of the algorithm. This will, as a consequence, lead to an overestimation of the significance of experimental results.
This chapter modeled the lognormal distributions of interpulse intervals and secretion event areas by multiple linear regressions in order to generate data sets with known answers that closely resemble the original data set. The multiple linear regression approach is excellent for this purpose because it emulates the distribution and sizes of the measurable secretion events and simultaneously allows for the coupling of these to each other and other variables such as basal secretion, half-life, and a 24-h rhythm. This multiple linear regression model is arbitrary and thus provides only limited information about the underlying molecular mechanisms of GH secretion. It does, however, provide an excellent basis to generate simulated hormone concentration time series data (with known answers) that closely mimic actual experimental data.
Clearly, the AutoDecon algorithm outlined in this work performs significantly better than several of the commonly used algorithms when applied to the analysis of simulated GH concentration time series data (see Tables 15.2 and 15.5–15.10). Use of the AutoDecon algorithm is not subjective, as it is a totally automated algorithm. The user need only specify approximate values for the SecretionSD and the one- or two-component half-live(s). The algorithm then adjusts these parameters automatically and also simultaneously finds the locations and amplitudes of the secretion events that have the highest probability of being correct. Moreover, AutoDecon is not overly sensitive to the magnitude of the experimental measurement errors (see Table 15.13), and, as a consequence, to the required number of replicate measurements (see Table 15.12). Thus, within reasonable bounds, the choice of the frequency of data collection and required number of replicate assays is a cost-value judgment for the investigator.
This analysis reveals some interesting aspects about the mechanisms controlling the secretion of GH. The analysis presented here suggests that many more secretion events are present within the data series than previously thought and that these secretion events have a wide variation in size. An argument can be made that the numerous small secretion events are not clinically relevant and thus should be ignored. However, if the goal is to understand the molecular and physiological mechanisms that lead to the secretion and elimination of GH, then the accurate quantification of these small secretion events is critically important.
These results indicate that approximately 6% of the total GH secreted occurs in a basal, or constant, nonpulsatile mode. However, Fig. 15.2 shows small secretion events that are not considered as actual secretion events because they are not statistically significant at a 0.05 level. In all likelihood there are many of these small events that are not identified as distinct secretion events by the AutoDecon, or any other algorithm, which may in fact be secretion events that are simply too small to rise to the level of statistical significance. Under such circumstances, these small events are lumped into the apparent basal secretion. Thus, the 6% basal secretion should be considered as an upper limit for the basal secretion, which might in fact be substantially lower. Accompanying this, the estimate of approximately 18 secretion events per 24-h period should be considered as a lower bound for the actual number of secretion events.
It is tempting to conclude that the asymmetry of the lognormal distribution of interpulse intervals, as depicted in Fig. 15.4, is a consequence of the inability of the algorithm to detect small secretion events. These missed events would yield overestimates of the interval and, when averaged over the entire data set, may be the cause of the skewed distribution. This could change the shape of the distribution to an unknown extent. It is clear that with data sampled at a 10-min interval it will become increasingly difficult to resolve interpulse intervals as the interpulse intervals decrease to this 10-min limit. Thus, it is expected that the observed distribution of inter-pulse intervals will be underestimated for short interpulse intervals and similarly artificially high for the longer interpulse intervals. The bimodal (or multimodal) nature of the secretion event areas (Fig. 15.6) will also be affected by the probable existence of multiple small secretion events that are below the limit of detectability.
In summary, AutoDecon is a novel, user-friendly deconvolution method that provides both an objective approach to initial secretory burst selection and a statistically based verification of candidate secretory bursts. When applied to synthetic GH concentration time series, AutoDecon performs substantially better than a number of alternative pulse detection algorithms in terms of optimizing the detection of true-positive secretory events while at the same time minimizing the detection of false-positive and false-negative events. Although AutoDecon will require validation with regard to application to other hormonal systems, it would seem to hold substantial promise as a biomathematical tool with which to identify and characterize a variety of pulsatile hormonal signals.
The Concordance and AutoDecon algorithms are part of our hormone pulsatility analysis suite. They can be downloaded from http://www.mljohnson.pharm,virginia.edu/pulse_xp/.
Table 15.8.
Comparison of performance of AutoDecon and Pulse algorithms evaluated from simulated data based on Pulse with basal secretion results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon (P value = 0.0325)a | |||
| %true positive | 100.00 | 94.51 ± 0.36 | 10.00 |
| %false positive | 0.00 | 5.49 ± 0.36 | 10.00 |
| %false negative | 0.00 | 1.46 ± 0.17 | 0.00 |
| %sensitivity | 100.00 | 98.13 ± 0.19 | 0.00 |
| Pulse with basal secretion and half-life = 14.25 mina | |||
| %true positive | 80.00 | 76.33 ± 0.96 | 41.89 |
| %false positive | 20.00 | 23.67 ± 0.96 | 41.89 |
| %false negative | 14.29 | 16.49 ± 0.52 | 15.00 |
| %sensitivity | 86.62 | 83.17 ± 0.53 | 15.00 |
Peak alignment within 10 min.
Table 15.9.
Comparison of performance of AutoDecon and Pulse2 algorithms evaluated from simulated data based on Pulse2 with no basal secretion results
| Median | Mean ± SEM | Interquartile range | |
|---|---|---|---|
| AutoDecon P (value = 0.0325)a | |||
| %true positive | 100.00 | 97.40 ± 0.21 | 6.25 |
| %false positive | 0.00 | 2.60 ± 0.21 | 6.25 |
| %false negative | 0.00 | 0.08 ± 0.03 | 0.00 |
| %sensitivity | 00.00 | 99.56 ± 0.08 | 0.00 |
| Pulse2 with no basal secretiona | |||
| %true positive | 100.00 | 96.54 ± 0.32 | 7.14 |
| %false positive | 0.00 | 3.46 ± 0.32 | 7.14 |
| %false negative | 5.41 | 6.65 ± 0.45 | 10.00 |
| %sensitivity | 93.33 | 93.02 ± 0.45 | 10.00 |
Peak alignment within 10 min.
Acknowledgments
This work was supported in part by NIH Grants RR-00847 to the General Clinical Research Center at the University of Virginia, U54 HD28934, R01 RR019991, R25 DK064122, R21 DK072095, P30 DK063609, R01 DK076037, and R01 DK51562.
References
- Bevington PR. Data Reduction and Error Analysis for the Physical Sciences. McGraw Hill; New York: 1969. [Google Scholar]
- Dimaraki EV, Jaffe CA, Demott-Friberg R, Russell-Aulet M, Bowers CY, Marbach P, Barkan AL. Generation of growth hormone pulsatility in women: Evidence against somatostatin withdrawal as pulse initiator. Am J Physiol Endocrinol Metab. 2001;280:E489–E495. doi: 10.1152/ajpendo.2001.280.3.E489. [DOI] [PubMed] [Google Scholar]
- Evans WS, Sollenberger MJ, Booth RA, Rogol AD, Urban RJ, Carlsen EC, Johnson ML, Veldhuis JD. Contemporary aspects of discrete peak-detection algorithms: II. The paradigm of the luteinizing hormone pulse signal in women. Endocrine Rev. 1992;13:81–104. doi: 10.1210/edrv-13-1-81. [DOI] [PubMed] [Google Scholar]
- Guardabasso V, De Nicolao G, Rocchetti M, Rodbard D. Evaluation of pulse-detection algorithms by computer simulation of hormone secretion. Am J Physiol. 1988;255:E775–E783. doi: 10.1152/ajpendo.1988.255.6.E775. [DOI] [PubMed] [Google Scholar]
- Haisenleder D, Dalkin A, Marshall J. Regulation of gonadotropin gene expression. Phys Reprod Edition. 1994;2:1793. [Google Scholar]
- Hartman ML, Faria AC, Vance ML, Johnson ML, Thorner MO, Veldhuis JD. Temporal structure of in vivo growth hormone secretory events in humans. Am J Physiol. 1991;260:E101–E110. doi: 10.1152/ajpendo.1991.260.1.E101. [DOI] [PubMed] [Google Scholar]
- Jansson PA. Deconvolution with Applications in Spectroscopy. Academic Press; New York: 1984. p. 22. [Google Scholar]
- Johnson ML, Straume M. Innovative quantitative neuroendocrine techniques, in (sex-steroid interactions with growth hormone) Serona Symposia. 1999:318–326. [Google Scholar]
- Johnson ML, Veldhuis JD. Evolution of deconvolution analysis as a hormone pulse detection algorithm. Methods Neurosci. 1995;28:1–24. doi: 10.1016/S0076-6879(04)84004-7. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Virostko A, Veldhuis JD, Evans WS. Deconvolution analysis as a hormone pulse-detection algorithm. Methods Enzymol. 2004;384:40–54. doi: 10.1016/S0076-6879(04)84004-7. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Pipes L, Veldhuis PP, Farhi LS, Boyd D, Evans WS. Validation of AutoDecon, a deconvolution algorithm for identification and characterization of luteinizing hormone. Anal Biochem. 2008;381:8–17. doi: 10.1016/j.ab.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. Plenum Press; New York: 1983. [Google Scholar]
- Maheshwari HG, Pezzoli SS, Rahim A, Shalet SM, Thorner MO, Baumann G. Pulsatile growth hormone secretion persists in genetic growth hormone-releasing hormone resistance. Am J Physiol Endocrinol Metab. 2002;282:E943–E951. doi: 10.1152/ajpendo.00537.2001. [DOI] [PubMed] [Google Scholar]
- Merriam GR, Wachter KW. Algorithms for the study of episodic hormone secretion. Am J Physiol. 1982;243:E310–E318. doi: 10.1152/ajpendo.1982.243.4.E310. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Plant TM, Hess DL, Keogh EJ, Knobil E. On the sites of negative and positive feedback actions of estradiol in the control of gonadotropin secretion in the rhesus monkey (Macaca mulatto) Endocrinology. 1978;102:1015–1018. doi: 10.1210/endo-102-4-1008. [DOI] [PubMed] [Google Scholar]
- Nelder JA, Mead R. A Simplex method for function minimization. Comput J. 1965;7:308–313. [Google Scholar]
- Press WH, Flannery BP, Teukolsky SA, Vettering WT. Numerical Recipes: The Art of Scientific Computing. Cambridge Univ. Press; Cambridge: 1986. [Google Scholar]
- Oerter KE, Guardabasso V, Rodbard D. Detection and characterization of peaks and estimation of instantaneous secretory rate for episodic pulsatile hormone secretion. Comp Biomed Res. 1986;19:170–191. doi: 10.1016/0010-4809(86)90014-5. [DOI] [PubMed] [Google Scholar]
- Santen RJ, Bardin CW. Episodic luteinizing hormone secretion in man: Pulse analysis, clinical interpretation, physiologic mechanisms. J Clin Invest. 1973;52:2617–2628. doi: 10.1172/JCI107454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume M, Frasier-Cadoret SG, Johnson ML. Least-squares analysis of fluorescence data. Top Fluorescence Spectrosc. 1991;2:171–240. [Google Scholar]
- Thorner MO, Chapman IM, Gaylinn BD, Pezzoli SS, Hartman ML. Growth hormone-releasing hormone and growth hormone-releasing peptide as therapeutic agents to enhance growth hormone secretion in disease and aging. Recent Prog Horm Res. 1997;52:215–244. [PubMed] [Google Scholar]
- Urban RJ, Evans WS, Rogol AD, Kaiser DL, Johnson ML, Veldhuis JD. Contemporary aspects of discrete peak detection algorithms: I. The paradigm of the luteinizing hormone pulse signal in men. Endocrine Rev. 1988;9:3–37. doi: 10.1210/edrv-9-1-3. [DOI] [PubMed] [Google Scholar]
- Van Cauter E. Estimating false-positive and false-negative errors in analysis of hormonal pulsatility. Am J Physiol. 1988;254:E786–E794. doi: 10.1152/ajpendo.1988.254.6.E786. [DOI] [PubMed] [Google Scholar]
- Van Cauter E, Plat L, Copinschi G. Interrelations between sleep and the somatotropic axis. Sleep. 1998;21:553–566. [PubMed] [Google Scholar]
- Vance ML, Kaiser DL, Evans WS, Furlanetto R, Vale W, Rivier J, Thorner MO. Pulsatile growth hormone secretion in normal man during a continuous 24-hour infusion of human growth hormone releasing factor (1–40): Evidence for intermittent somatostatin secretion. J Clin Invest. 1985;75:1584–1590. doi: 10.1172/JCI111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis JD, Carlson ML, Johnson ML. The pituitary gland secretes in bursts: Appraising the nature of glandular secretory impulses by simultaneous multiple-parameter deconvolution of plasma hormone concentrations. Proc Natl Acad Sci USA. 1987;84:7686–7690. doi: 10.1073/pnas.84.21.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis JD, Johnson ML. Cluster analysis: A simple, versatile, and robust algorithm for endocrine pulse detection. Am J Physiol. 1986;250:E486–E493. doi: 10.1152/ajpendo.1986.250.4.E486. [DOI] [PubMed] [Google Scholar]
- Veldhuis J, Johnson ML. A novel general biophysical model for simulating episodic endocrine gland signaling. Am J Physiol. 1988;255:E749–E759. doi: 10.1152/ajpendo.1988.255.6.E749. [DOI] [PubMed] [Google Scholar]
- Webb CB, Vance ML, Thorner MO, Perisutti G, Thominet J, Rivier J, Vale W, Frohman LA. Plasma growth hormone responses to constant infusions of human pancreatic growth hormone releasing factor: Intermittent secretion or response attenuation. J Clin Invest. 1984;74:96–103. doi: 10.1172/JCI111424. [DOI] [PMC free article] [PubMed] [Google Scholar]