Abstract
Flavin-containing monooxygenase from yeast (yFMO) carries out the O2- and NADPH-dependent oxidation of biological thiols, including oxidizing glutathione to glutathione disulfide. FMO provides a large fraction of the oxidizing necessary for proper folding of disulfide bond-containing proteins; deletion of the enzyme reduces proper folding of endogenous carboxypeptidase Y by about 40%. The enzyme is not essential to cell viability because other enzymes can generate a significant fraction of the oxidizing equivalents required by the cell. However, yFMO is vital to the yeast response to reductive stress. FMO1 deletion mutants grow poorly under reductive stress, and carboxypeptidase Y activity is less than 10% of that in a stressed wild type. The FMO1 gene appears to be under control of an unfolded protein response element and is inducible by factors, such as reductive stress, that elicit the unfolded protein response. Reductive stress can increase yFMO activity at least 6-fold. This increased activity allows the cell to process endogenous disulfide bond-containing proteins and also to allow correct folding of disulfide-bonded proteins expressed from multicopy plasmids. The unfolded protein response is mediated by the Hac1p transcription factor that mediates virtually all of the induction of yFMO triggered by exogenous reducing agents.
Saccharomyces cerevisiae has a single gene, FMO1, coding for the flavin-containing monooxygenase (yFMO) enzyme (1–4). We have cloned this gene, engineered it for easy expression with a His10 tag, purified the protein, and characterized it kinetically (4). We found that yFMO does not oxidize nitrogen-containing compounds, the usual substrates for mammalian FMOs (5–8). Instead, it uses molecular O2 and NADPH to oxidize thiol compounds such as cysteamine, glutathione (γ-glutamylcysteinylglycine, abbreviated as GSH), and cysteine (9). Recently, we constructed a strain of yeast in which the FMO1 gene was deleted (Δfmo) and an FMO1-bearing plasmid that can be used to complement the knockout mutation (pFMO).
It is known that the ratio of GSH/glutathione disulfide (GSSG) is about 100 in eukaryotic cytoplasm and about 1–3 in the endoplasmic reticulum (ER) (10). yFMO, on the cytoplasmic surface of the ER, oxidizes GSH to GSSG, which then is transported into the ER to establish the optimum redox potential necessary to fold disulfide bond-containing proteins. Both wild-type and Δfmo yeast strains were able to express active enzymes from plasmids coding for proteins that lacked disulfide bonds, but Δfmo could not produce active test proteins that contained disulfide bonds when overexpressed (9). The phenotype of Δfmo cells resembles that of cells from which the Ero1 protein has been deleted (11, 12).
It is known that the accumulation of misfolded proteins in the ER, because of improper disulfide bonding or misglycosylation, induces the unfolded protein response (UPR). The UPR triggers up-regulation of various ER resident proteins involved in protein folding (13, 14). In budding yeast, the Hac1p protein, a bZIP transcription factor, is the major signal for such up-regulation when unfolded proteins are detected in the ER (15). Among the proteins known to be up-regulated by the UPR are the chaperonin Kar2p, or BiP (16), the protein disulfide isomerase, Pdi1 (17), the peptidyl-prolyl cis-trans isomerase (18), and Fkb2p (19). Each of these genes contains the UPR element (UPRE) as an upstream activator sequence (20, 21).
We report here that FMO1 is important for optimal cell growth under reductive and oxidative stress and under the stress of protein over expression from a multicopy plasmid. We also show that the FMO1 gene is under the control of the Hac1p-mediated UPR. Situations that lead to an increase in unfolded proteins, such as reductive stress, increase yFMO expression along with other proteins stimulated by the UPR.
Materials and Methods
Materials.
Oligonucleotides for PCR were purchased from Genosys (The Woodlands, TX). All restriction enzymes, T4 DNA ligase, and T4 DNA polymerase were obtained from New England Biolabs. TA Cloning Kit, Zero Blunt PCR Cloning Kit, and pYES2 plasmid were purchased from Invitrogen. Taq DNA polymerase was from Perkin–Elmer, and Pfu DNA polymerase was obtained from CLONTECH.
Media and General Methods.
Complete [yeast extract/peptone/dextrose (YPD)] and synthetic minimal (SC) media are described in ref. 22. All of the yeast strains were grown at 30°C unless otherwise indicated. If required, inositol was added at a final concentration of 50 μg/ml and DTT was added as described in the appropriate figures. To induce GAL1 promoter-dependent gene expression, 2% galactose was added to the mid-log-phase culture (OD600 about 0.6–1.0). Yeast transformation was performed by lithium-acetate procedures (23).
Plasmid and Strain Construction.
Plasmid constructions for the reporter genes and expressions for yeast were described previously, and the construction for FMO1 gene deletion strain also was shown in a previous paper (9). Briefly, the β-galactosidase expression plasmid under control of GAL1 promoter was named as pGAL-gal, one for β-glucuronidase was named as pGAL-glu, one for yeast chitinase was named as pGAL-chit, and one for bacterial alkaline phosphatase was named as pGAL2-bap.
Homologous recombination was used to disrupt the chromosomal yeast HACI gene. A HACI PCR fragment with 5′ and 3′ untranslated regions was amplified by using an upstream primer (5′-ACAAGACCTAGGGCAATATGG-3′) and a downstream primer (5′-GCAGAGCGTGGCTCTTTGAG-3′) together with Taq DNA polymerase. The resulting 1.7-kb fragment was cloned into a TA cloning vector (pCR2.1; Invitrogen). The resulting plasmid (pHAC1) was digested with XmnI, and the 0.8-kb fragment containing the coding sequences of the HAC1 gene was replaced with the URA3 gene fragment obtained from pJR-Ura3 (24) by treating with SmaI, HindIII, and T4 DNA polymerase. The resulting plasmid (pHAC1-Ura3) was restricted with EcoRI, and a linear fragment containing the URA3 marker gene, flanked by segments of the HAC1 gene, was used for transformation of S. cerevisiae DBY1827 (αhis3-Δ200 leu2–3, 112 ura3–52). The URA3 cassette was evicted (9), and the resulting HAC1-disrupted strain was named Δhac1.
To place the N-terminal His-tagged FMO construct (p5′HisFMO) under control of its own promoter, the 802-bp sequence upstream from the start codon of FMO1 was amplified with two primers (the sense primer, 5′-TCGCCTGCCATTAAGATG-3′; and the antisense primer, 5′-CCATGGATGCCTAGTTATTCTTG-3′) together with Pfu DNA polymerase. The 0.8-kb product first was cloned into a Zero Blunt PCR Cloning vector (pCR-Blunt), and the resulting plasmid was named as pCRB-802. pCRB-802 was restricted with EcoRI, filled in with T4 DNA polymerase, and restricted with NcoI to produce the 802-bp upstream sequence. This fragment was cloned into pYX123 (R & D Systems) treated with AgeI/T4 DNA polymerase and NcoI. The resulting plasmid (pYX123–802) was digested with NcoI and XhoI and ligated with the NcoI/XhoI-restricted fragment from pHis-FMO (4) that contained the N-terminal His-tagged FMO1 coding sequences. To make the lacZ reporter gene construct under the control of yeast FMO1 promoter (pFMO-802), the FMO1 promoter sequences (802 bp) was amplified with the sense primer and another antisense primer (5′-GGATCCATGCCTAGTTATTCTTG-3′) containing a BamHI site by using Pfu DNA polymerase. The resulting PCR fragment was cloned into the pCR-Blunt vector. The 0.8-kb fragment restricted using EcoRI and BamHI was placed just before the coding sequences of the lacZ fragment from pSE640 (American Type Culture Collection) digested with EcoRI and BamHI.
Western Blots.
For Western blot analyses of hemagglutinin (HA)-tagged yeast chitinase, cells containing the chitinase expression plasmid (9) were grown in raffinose medium until midlog phase (OD600 between 0.6 and 1.0). Chitinase expression was induced by adding 2% galactose for 6 hr. Cells were washed with water, resuspended with 1× PBS containing 0.1% Triton X-100, and broken with by glass beads. Samples (20-μg proteins from cell extracts per lane) were analyzed by SDS/PAGE; Western blots made use of the 12CA5 antibody conjugated with horseradish peroxidase (Boehringer Mannheim) for HA-tagged proteins. Anti-Hac1p antibody was a gift from Peter Walter (University of California, San Francisco).
Reporter Gene Assay.
The reporter gene assays were determined as described previously (9). To determine the level of UPR, yeast strains were transformed by using a reporter construct (pCF118, kindly provided by Peter Walter) consisting of the lacZ gene under transcriptional control of the KAR2 UPRE (17), grown to midlog phase (OD600 between 0.6 and 1.0), and then subjected to exogenous stresses with varying concentrations of DTT for 2 hr. After stress treatment, the cells were broken and the cell extracts were applied directly to the assay described previously (9).
Carboxypeptidase Y (CPY) Enzymatic Assay.
Yeast cells were grown in YPD at 25°C until midlog phase (0.6 -1.0 OD at 600 nm). DTT was added, to 2 mM, 2 hr before the cells were harvested. Harvested cells were resuspended in 50 mM potassium-phosphate buffer (pH 7.4) containing 50 μM FAD and were ruptured by using a French Pressure cell (SLM–Aminco, Urbana, IL) at 20,000 psi, three times. The membrane and soluble fractions were isolated as described previously (4).
Membrane fractions containing 0.1–0.2 mg of protein were used for the yFMO activity assays as described previously (4). Soluble fractions were used for the yeast CPY activity assays as described (25, 26) by using N-acetyl-dl-phenylalanine p-nitrophenyl ester (Sigma) as substrate.
Metabolic Labeling and Immunoprecipitations.
Metabolic labeling and yeast CPY immunoprecipitation were done as described (27) with minor modifications. Briefly, samples were pulse-labeled for 10 min with 200 μCi of 35S-labeled Met and Cys, in the presence or absence of 2 mM DTT. The samples then were chased with 10 mM each of unlabeled methionine and cysteine and treated with 1 mM cycloheximide to terminate translation. After 30 min, samples were analyzed on nonreducing SDS/PAGE. mAb against CPY (Molecular Probes) and Immunocatcher (CytoSignal, Irvine, CA) were used for the immunoprecipitation.
Results
The Relationship of yFMO to Cellular Growth Under Stress.
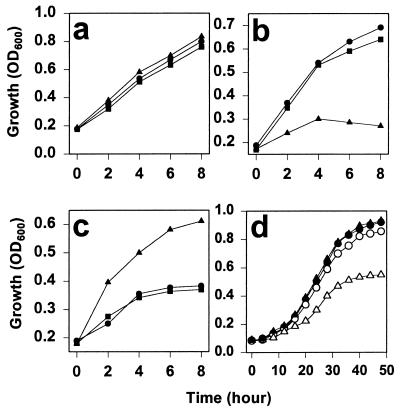
Three different yeast strains were tested for their response to membrane-permeable reductants and oxidants, wild-type, an FMO1 gene-deleted strain (Δfmo), and a deletion strain rescued with a plasmid-borne FMO (pFMO). Without a redox stress, all strains showed normal growth pattern for 8 hr (Fig. 1a). The change in apparent absorbance at 600 nm that we observed can be due to a change in cell density or in cell size; for this work we will subsume both possibilities within the general term “growth.” A reductive stress in the form of 5 mM DTT retarded growth of the Δfmo strain by about 3-fold, and after 4 hr, the cells began to die. In contrast, the strains expressing yFMO grew nearly normally (Fig. 1b).
Figure 1.
Growth of yeast strains under stress. (a) Growth, measured as increasing turbidity, as a function of time for the wild-type strain (●), the Δfmo strain (▴), and the FMO plasmid-rescued strain of the deletion mutant (■). (b) The same strains in the presence of a reductive stress (5 mM DTT). (c) The growth of those strains under the oxidative stress of 2.5 mM diamide. (d) The growth of these strains under the stress of overexpression of chitinase from a plasmid (pGAL-chit). Minimal medium reduces overall growth levels, and chitinase expression was induced by 2% galactose. Symbols are as above except that open figures indicate the expression of chitinase from a plasmid (pGAL-chit).
We next checked the effect of an exogenous oxidative stress on the same strains. The membrane-permeable diazine compound diamide is known to cause formation of disulfide bonds in living cells (28); the test strains were grown in the presence of 2.5 mM diamide. As shown in Fig. 1c, diamide stress retarded growth by the wild-type and pFMO strains by a factor of at least 2 and, after 4 hr, greatly retarded any cell growth. In contrast, the Δfmo strain could grow to nearly normal densities compared with unstressed cells, that is, at least twice the cell density of a wild type and the rescued strain. This result suggests that diamide treatment is complementary to the function of yFMO. The exogenous oxidizing agent can compensate for loss of yFMO function in the knockout mutant, but when both sources of oxidizing equivalents are present the cell is stressed.
It is also likely that ER overloading of disulfide-containing proteins may constitute a stress on the cell. In Fig. 1d, we show the response of yeast strains to the stress of expressing plasmid-borne chitinase with three disulfides. Induction of the plasmid required growth in a minimal medium supplemented with 2% galactose, and so overall growth rates are slower than in Fig. 1 a–c. The figure shows that there is only a slight reduction in growth of wild-type yeast expressing the chitinase load compared with wild-type strains not expressing chitinase. However, the Δfmo strain shows reduced growth under this stress, reaching about one-half the value of the wild-type strains after 40 hr.
yFMO Is Induced as Part of the UPR.
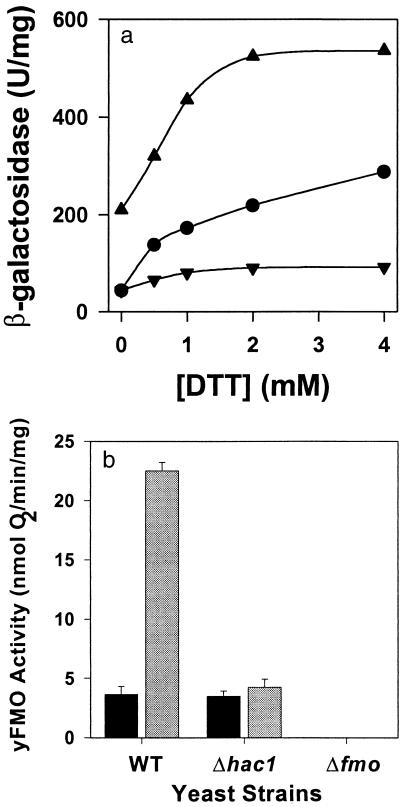
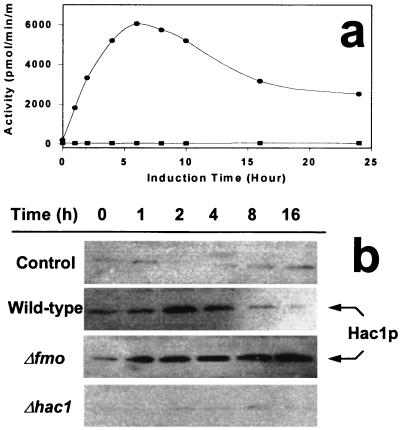
It is clear that yFMO is important to the yeast responses to oxidative and reductive stresses, which probably result in the accumulation of unfolded proteins. Reductive stress, which increases the level of misfolded protein in the ER, is known to induce the UPR through the mediation of Hac1p, a leucine zipper transcription factor (15). We therefore tested the cellular UPR in the presence and absence of the FMO1 gene and the HAC1 gene. The level of the UPR was measured by using a reporter construct consisting of the lacZ gene under transcriptional control of the known KAR2 unfolded protein response element, UPRE (17). The results are shown in Fig. 2a. In wild-type cells the reporter gene has a low level of activity in the absence of exogenous DTT, but increasing concentrations of the reducing agent elicit a modest induction of the reporter gene. The Δfmo strain exhibits a much stronger response. In the absence of exogenous DTT, the Δfmo strain expresses five times the β-galactosidase activity of wild type. The response to exogenous DTT also is more vigorous as manifest by the steeper slope of induction, up to about 2 mM DTT when the reporter gene activity saturates. The HAC1 deletion strain (Δhac1) has wild-type reporter expression in the absence of exogenous DTT. Even 4 mM DTT induces the reporter only 2-fold. yFMO activity itself also was measured in the three strains under normal growth conditions and under reductive stress. As seen in Fig. 2b, 4 mM DTT induces yFMO activity 6-fold in wild-type cells. There is no FMO1 induction over the constitutive levels in the Δhac1 strains, and there is no detectable yFMO activity in the Δfmo strain.
Figure 2.
Induction of UPR genes as a function of reductive stress. (Upper) The β-galactosidase reporter gene was placed downstream from the Kar2 UPR element. The response to increase reductive stress, in the form of exogenous DTT, is observed in wild-type cells (●), the Δfmo strain (▴), and HAC1-deleted (Δhac1) strains (▾). (Lower) Expression of yFMO activity under no stress (solid bars) and in the presence of 2 mM DTT (shaded bars) for the wild-type, HAC1-deleted (Δhac1), and Δfmo strains.
yFMO Activity and the Folding of Disulfide Bond-Containing Proteins.
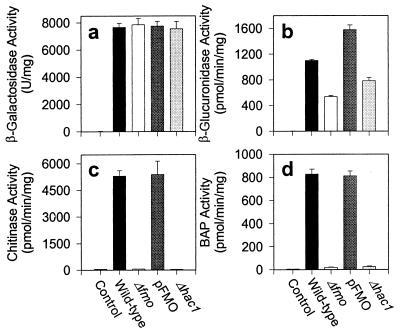
We have shown previously that yFMO is involved in folding disulfide bond-containing proteins expressed from multicopy plasmids by using a variety of test proteins (9). β-Galactosidase has no disulfides and no leader sequence to direct it to the ER; it should be expressed in the cytoplasm. β-Glucuronidase has no disulfides but was engineered with a leader that directs it to the ER. Yeast chitinase has three disulfide bonds and has its own secretion leader, and bacterial alkaline phosphatase, with two disulfides, was placed behind a preproalpha leader to direct it to the ER.
These constructs were expressed in wild type (DBY1827), FMO1 gene-deleted strain (Δfmo), the FMO plasmid-rescued strain of the FMO1 deletion (pFMO), and, finally, in the HAC1 gene-deleted strain (Δhac1). To check the proper folding of expressed proteins from these constructs in the cell, enzyme activities were measured. The enzyme activities for these constructs are shown in Fig. 3. The level of β-galactosidase activity was identical in the cytoplasm of each strain (Fig. 3a). Neither yFMO nor Hac1p activity influences the cytoplasmic expression of this protein, which lacks disulfide bonds. The ER expression of a protein lacking disulfides, β-glucuronidase, also is not affected significantly by yFMO or by Hac1p activity (Fig. 3b). The FMO1 and HAC1 deletion strains each produce about 50–60% of the activity of wild type or the yFMO-rescued strain. Fig. 3 c and d show the ER expression of proteins containing disulfide bonds, chitinase and alkaline phosphatase. In both cases the wild type and yFMO-rescued strain show equal activity. In strong contrast, the FMO1 and HAC1 deletion strain shows essentially no activity of either test protein.
Figure 3.
Expression of test protein constructs in the wild-type, FMO1-deleted (Δfmo), FMO-rescued (pFMO), and HAC1-deleted (Δhac1) strains of yeast. The wild-type strain (DBY1827), transformed with the parent vector (pYES2), was used as the control. Expression of test proteins was induced for 6 hr at midlog phase (OD600 between 0.6 and 1.0) by adding 2% galactose, except bacterial alkaline phosphatase-expressing cells, which were automatically induced at late-log phase.
Fig. 3 shows that essentially no active chitinase is produced by either the yFMO or the HAC1 deletion strains. We used Western blots to test whether inactive chitinase protein was produced under these conditions, as we had done previously (9). The control strain did not express any chitinase, the wild-type strain expressed high levels of chitinase, and the Δfmo and Δhac1 strains showed only about 20–40% of the expression level of wild type (data not shown). Chitinase protein is expressed, but it is not active and presumably is misfolded in these two deletion strains.
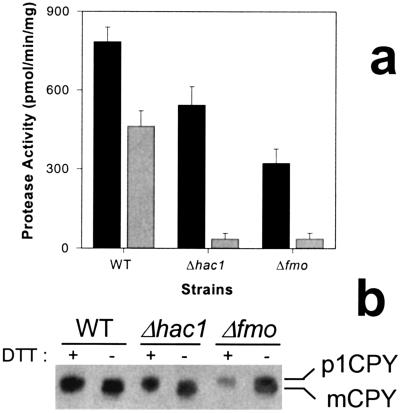
In addition to monitoring the expression of plasmid-encoded proteins, we examined the expression and activity of an endogenous disulfide bond-containing protease, CPY. CPY is synthesized as an inactive 69-kDa precursor; its five disulfide bonds are formed in the ER and are required for routing to vacuoles for activation by proteolytic cleavage to the mature, 61-kDa form (29). Fig. 4a shows proteolytic activity in wild-type, Δfmo, and Δhac1 yeast strains under unstressed conditions and in the presence of 2 mM DTT. Fig. 4b is a nonreducing SDS/PAGE autoradiogram showing the concentrations of unprocessed CPY and of mature CPY from these various strains and conditions. Exogenous DTT reduces protease activity by about 40%; the autoradiogram suggests, although the bands are not cleanly resolved, that the total CPY protein expression remains roughly constant but that more of the proenzyme remains unprocessed and inactive. The Δhac1 mutant expresses roughly 70% of wild-type activity under unstressed conditions, but exogenous DTT reduces this activity 10-fold. The autoradiogram clearly suggests the cause is a failure to process and activate the proCPY. A similar pattern is seen in the Δfmo strain. Deletion of the yFMO enzyme allows expression of about 40% of wild-type enzyme activity, and the autoradiogram shows that this results from a failure to produce the mature CPY form. Exogenous DTT stress reduces protease activity 7-fold, and the gel shows almost no detectable mature CPY.
Figure 4.
Expression of CPY in several strains of yeast. (a) Protease activity as measured by hydrolysis of using N-acetyl-dl-phenylalanine p-nitrophenyl ester, a synthetic substrate for CPY. The solid bars represent protease activity under nonstress conditions, and the gray bars represent activity in the presence of 2 mM DTT. (b) Western blots of CPY expression. The unprocessed and inactive, 69-kDa proenzyme band is labeled p1CPY, and the active, mature, 61-kDa band is labeled mCPY.
yFMO Deletion Results in a Prolonged UPR Under Stress.
We showed above that DTT elicited an exaggerated induction of the UPR in Δfmo yeast. We also wanted to observe the effect of stress represented by an accumulation of unfolded protein. To do this, we transformed various yeast strains with a multicopy chitinase expression plasmid and monitored the expressed chitinase activity with time. The data are shown in Fig. 5. The control is wild-type yeast transformed with the parent plasmid lacking chitinase. It shows no detectable chitinase activity with time (Fig. 5a). The plasmid-expressed chitinase reached maximum at 6 hr and then declined and leveled off at about one-half maximum values. This protein synthesis burden constitutes a form of stress on the cells, and we used antibodies against Hac1p (15) to monitor the time dependence of Hac1p expression in the yeast strains (Fig. 5b). The control strain shows no significant increase in Hac1p production over time, presumably because the cell is not under stress. Not surprisingly, HAC1-deleted strains did not show significant expression of Hac1p either. Wild-type yeast that express chitinase from a multicopy plasmid also increase the concentration of Hac1p with time. Hac1p reaches a maximum at around 2 hr and then declines to base levels by 16 hr. That is, the UPR signal initially rises and then decays as the stress of overloading is dealt with by the cell. In the FMO1-deleted strain, however, the stress is sensed, as indicated by the increase in Hac1p production, but it rises continuously out to 16 hr.
Figure 5.
yFMO affects the duration of the Hac1p-mediated UPR. (a) The expression of chitinase as a function of time after induction in wild-type control cells with pYES2 (■) and those transformed with a chitinase expression plasmid (pGAL-chit) (●). (b) Western blot showing the time course of Hac1p expression in control cells with the parental vector, pYES2, and in wild-type, Δfmo, and Δhac1 strains bearing the expression plasmid (pGAL-chit). Chitinase expression was induced by 2% galactose; 20 μg of protein from cell extracts was loaded into the wells.
Discussion
Cells originally evolved in a reducing atmosphere, and the cytoplasm of modern cells retains this heritage, maintaining a reducing environment. This is clearly reflected in the ratio of GSH/GSSG of about 100 (10). As photosynthesis generated O2, the earth's environment became oxidizing, and modern cells evolved to accommodate this change. The cell must counteract the effect of nonspecific oxidation reactions like those involving oxygen radicals and peroxides. Specific oxidizing chemistry generally is compartmentalized to separate them from the cytoplasm. The ER is the site of a great deal of the processing of newly synthesized proteins, and, in particular, it is the site of the oxidative formation of protein disulfide bonds. Proteins are extruded into this specialized vesicle, where they can be operated on by a variety of enzymes that catalyze the formation and rearrangement of these bonds. The specialized environment is more oxidizing than the cytoplasm and exhibits a GSH/GSSG ratio of about 1. The glutathione redox buffer is the major one in eukaryotic cells, but it is not the only one. Cells unable to synthesize the glutathione tripeptide are still viable and can oxidize protein thiols in the ER by using other oxidants (11).
As the yeast cell carries out the continuous process of disulfide bond formation, the ER requires a continuous influx of oxidizing equivalents. yFMO is a major, and perhaps the major, source of these chemicals. It uses O2 and NADPH to generate GSSG and can also generate cysteamine and cysteine on the cytoplasmic surface of the ER. The oxidizing units presumably are transported into the ER, and we have shown that GSSG, generated by the enzyme, concentrates in the ER (9). In addition to yFMO, yeast contain other enzymes that can generate GSSG. The best known example is glutathione peroxidase (30), a cytoplasmic and mitochondrial enzyme that uses H2O2 to oxidize GSH.
In this paper we are concerned with the role and regulation of yFMO. It is clear from Fig. 1 that yFMO is not required for cell viability. In the absence of stress, yeast cells grow about as well with the FMO1 gene deleted as when it is present. However, as Fig. 4 shows, deletion of yFMO activity interferes with the proper processing of those cellular proteins that contain disulfide bonds. In the absence of stress, the Δfmo strain fails to activate over half of its CPY protein, a process dependent on proper disulfide formation. The fact that about 40% of the protease activity is expressed suggests that redundant systems, perhaps glutathione peroxidase, are still operating in the absence of yFMO.
The role of yFMO comes into sharper focus when the yeast cells are placed under stress. Exposure to exogenous reducing agents, like DTT, constitute a stress on the cell, presumably because the DTT can raise the reduction potential of the ER and retard proper processing of disulfide bond-containing proteins. yFMO activity can partially counteract this by generating additional oxidizing equivalents. It is not surprising, therefore, that the gene is inducible by such stress. The role of yFMO in the cellular stress response is emphasized by noting that loss of yFMO activity greatly retards the growth of cells exposed to exogenous DTT (Fig. 1b). This retardation in growth is likely coupled to the decreased ability of the stressed cells to fold disulfide bond-containing proteins as clearly seen in Fig. 4, where FMO1 deletion retards the folding and processing of CPY. Under DTT stress, the Δfmo yeast express only about 7% of the protease activity seen in wild type yeast under the same stress and an autoradiogram shows essentially all CPY protein in the unprocessed form indicative of improper folding in the ER.
Another stress on the oxidizing capacity of the cell is a chemical directive to express an unusually heavy load of disulfide bonded proteins. This is seen in Fig. 3, where multicopy plasmids are used to express proteins that do, or do not, contain disulfide bonds. Proper folding is measured by the enzyme activity of these proteins. β-Galactosidase, a protein lacking disulfide bonds, shows the same expressed activity in wild-type and Δfmo strains. In sharp contrast, chitinase activity is reduced several hundredfold in the Δfmo strain. Western blots show that this disulfide bond-containing protein is expressed but it is inactive, presumably because of improper formation of its three disulfide bonds. [It should be noted that as seen in Fig. 3b, and as addressed earlier (9), β-glucuronidase activity is reduced about 50% in the deletion strains. Presumably this reflects an unspecified change in the ER environment, caused by loss of yFMO activity, and is deleterious to optimum folding even of proteins that lack disulfide bonds.] It is also clear that redundant systems, like glutathione peroxidase, are unable to compensate for the loss of oxidizing equivalents normally generated by yFMO. As a consequence, the effect of FMO1 deletion on chitinase expression from a multicopy plasmid is even more dramatic than on expression of an endogenous protein like CPY.
Our work also shows that the FMO1 gene is under the control of an inducible promoter. As seen in Fig. 2 Lower, yFMO is expressed at a significant background level under normal, unstressed growth conditions. The enzyme uses O2 to generate a large share of the oxidizing equivalents required to generate protein disulfide bonds. UPR-controlled enzymes are induced by reductive stress, as shown in Fig. 2, and may reach full induction at 2 mM exogenous DTT. Under such a reductive stress, yFMO activity can increase at least 6-fold. The additional yFMO activity helps counteract the reducing stress; this is shown in Fig. 3a, where wild-type cells under reductive stress express 11 times more endogenous CPY protease activity than do Δfmo mutants.
Unfolded proteins in the ER trigger the unfolded response, UPR, which is mediated largely by the transcription factor Hac1p (15). Stresses, like exogenous reducing agents, that cause unfolded proteins to accumulate in the ER indirectly elicit the UPR. The UPR involves the mobilization of many proteins involved in protein folding (31, 32), including the chaperonin BiP (16, 21, 33), Pdi (17), proline isomerase (18), and Ero1, the initial oxidizing protein in the ER lumen (11). It is now clear that the FMO1 promoter is induced by the UPR and must be added to the above list. The importance of Hac1p to the induction of the FMO1 gene is shown in Fig. 2b, where deletion of the HAC1 gene eliminates the induction of yFMO activity by exogenous stress.
The effect of HAC1 deletion on the expression of disulfide bond-containing proteins is more complicated to interpret, presumably because Hac1p affects the expression of many proteins involved in protein folding. Nevertheless, the effects are consistent with the important role of Hac1p in controlling yFMO induction and its activity, in turn, facilitating folding of disulfide bond-containing proteins. Fig. 4 shows that the Δhac1 strain is far less robust than wild type in its ability to fold CPY. Under reductive stress, the wild-type strain generates about 70% of protease activity seen in an unstressed cell. With yFMO and other proteins elicited by the UPR intact, the cell can fold disulfide bonded proteins even under stress. However, the HAC1 gene deletion strain can generate only about 10% of the protease activity seen in the unstressed cell. This poor performance probably reflects the fact that all of the UPR proteins, including yFMO, cannot be strongly induced. However, the importance of yFMO to this total response is attested to by the observation that under stress, (CPY) protease activity in the Δhac1 strain is nearly identical to that in the Δfmo strain. That is, the loss of yFMO expression alone is as deleterious to CPY folding as the failure to induce the entire spectrum of UPR proteins. A similar impact is seen on the expression of disulfide bond-containing proteins from plasmids (Fig. 3). Whereas loss of the Hac1p signal has no effect on the expression of β-galactosidase activity, it essentially eliminates expression of chitinase activity, producing roughly the same expression of activity as the Δfmo strain. yFMO can be strongly induced under stress, and it appears that the base levels of yFMO activity seen in Δhac1 strains are essentially ineffective in dealing with the oxidation requirements of folding plasmid-expressed chitinase. The loss of yFMO must cause unfolded chitinase to accumulate, and this triggers a prolonged UPR as seen in Fig. 5.
It has been shown that Hac1p binds to an upstream control element called the UPRE (16). This element contains a pseudo-palindrome separated by a single base, usually C, and can function in either orientation (20, 21). We have shown that the 802-bp upstream region of FMO1 contains information that allows the Hac1p-mediated induction of yFMO or of a reporter gene. In this study we have not carried out any analysis of the chemical nature of this control region, but it is very likely that the UPRE lies between −145 and −124 bp upstream of the yFMO start codon. As shown in Fig. 6, that sequence strongly resembles known UPREs compared previously by Mori et al. (21) and is probably the recognition site for Hac1p.
Figure 6.
Comparison of the upstream region of the FMO1 gene with known UPR elements. The sequences above the line are known UPREs (21), and a sequence upstream of FMO1 is compared with them. The arrows mark the characteristic pseudo-palindrome separated by a C base. The light numbers flanking the 22 base elements indicate their positions upstream of the gene; either orientation of the element is permitted.
In summary, the FMO1 gene is expressed at a low, but significant level under normal cell conditions and contributes substantially to the pool of oxidizing equivalents required to process endogenous disulfide bond-containing proteins. Stresses that trigger the UPR, such as exogenous reducing agents, induce the gene strongly, and the increased yFMO activity is crucial to maintaining an environment capable of folding disulfide bond-containing proteins. The induction is mediated by Hac1p, which binds to a consensus UPRE, and accounts for virtually all of the induction of FMO1 by exogenous reducing agents.
Acknowledgments
This work was supported by Grant GM 30048 from the National Institutes of Health, Grant MCB-9601096 from the National Science Foundation, and by grants from the Foundation for Research and the Welch Foundation.
Abbreviations
- FMO
flavin-containing monooxygenase
- yFMO
FMO from yeast
- GSH
glutathione
- GSSG
glutathione disulfide
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- UPRE
UPR element
- CPY
carboxypeptidase Y
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Johnston M, Andrews S, Brinkman R, Cooper J, Ding H, Dover J, Du Z, Favello A, Fulton L, Gattung S, et al. Science. 1994;265:2077–2082. doi: 10.1126/science.8091229. [DOI] [PubMed] [Google Scholar]
- 2.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 3.Mewes H W, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver S G, et al. Nature (London) 1997;387,(Suppl.):7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 4.Suh J K, Poulsen L L, Ziegler D M, Robertus J D. Arch Biochem Biophys. 1996;336:268–274. doi: 10.1006/abbi.1996.0557. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler D M. Annu Rev Pharmacol Toxicol. 1993;33:1979–1999. doi: 10.1146/annurev.pa.33.040193.001143. [DOI] [PubMed] [Google Scholar]
- 6.Cholerton S, Smith R L. In: N-oxidation of Drugs: Biochemistry, Pharmacology, Toxicology. Hlavic P, Damani L A, editors. London: Chapman & Hall; 1991. pp. 107–131. [Google Scholar]
- 7.Ziegler D M. In: N-oxidation of Drugs: Biochemistry, Pharmacology, Toxicology. Hlavic P, Damani L A, editors. London: Chapman & Hall; 1991. pp. 50–70. [Google Scholar]
- 8.Ziegler D M, Poulsen L L. In: Drug Metabolism: Toward the Next Millennium. Gooderham N J, editor. Amsterdam: IOS Press; 1998. pp. 30–38. [Google Scholar]
- 9.Suh J K, Poulsen L L, Ziegler D M, Robertus J D. Proc Natl Acad Sci USA. 1999;96:2687–2691. doi: 10.1073/pnas.96.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang C, Sinskey A J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 11.Frand A R, Kaiser C A. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 12.Pollard M G, Travers K J, Weissman J S. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 13.Sidrauski C, Cox J S, Walter P. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 14.Sidrauski C, Chapman R, Walter P. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 15.Cox J S, Walter P. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 16.Mori K, Sant A, Kohno K, Normington K, Gething M J, Sambrook J F. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J S, Shamu C E, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 18.Lodish H F, Kong N. J Biol Chem. 1991;266:14835–14838. [PubMed] [Google Scholar]
- 19.Partaledis J A, Berlin V. Proc Natl Acad Sci USA. 1993;90:5450–5454. doi: 10.1073/pnas.90.12.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 21.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. J Biol Chem. 1998;273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- 22.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 23.Ito H, Fukuda Y, Murata K, Kmura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roca J, Gartenberg M R, Oshima Y, Wang J C. Nucleic Acids Res. 1992;20:4671–4672. doi: 10.1093/nar/20.17.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aibara S, Hayashi R, Hata T. Agric Biol Chem. 1971;35:658–666. [Google Scholar]
- 26.Bai Y, Hayashi R, Hata T. J Biochem. 1975;78:617–626. doi: 10.1093/oxfordjournals.jbchem.a130948. [DOI] [PubMed] [Google Scholar]
- 27.Simons J F, Ferro-Novick S, Rose M D, Helenius A. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosower N S, Kosower E M. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 29.Jamsa E, Simonen M, Makarow M. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- 30.Galiazzo F, Schiesser A, Rotilio G. Biochem Biophys Res Commun. 1987;147:1200–1205. doi: 10.1016/s0006-291x(87)80197-3. [DOI] [PubMed] [Google Scholar]
- 31.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 32.Chapman R, Sidrauski C, Walter P. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- 33.Nikawa J, Akiyoshi M, Hirata S, Fukuda T. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]