Abstract
The affinity of the poly(ADP-ribose) polymerase-1 (PARP-1) for platinum-damaged DNA was first discovered during photo-crosscroxss-linking experiments using the photoactive compound Pt-BP6 (Zhang, C. X.; Chang, P. V.; Lippard, S. J., J. Am. Chem. Soc. 2004, 126, 6536–6537), an analogue of the anticancer drug cis-diammine-dichloroplatinum(II), cisplatin. Although PARP inhibitors sensitize cancer cells to cisplatin, there are conflicting reports in the literature about their efficacy. In order to improve our understanding of the mechanism by which PARP inhibition might potentiate the cell-killing ability of cisplatin, and to shed light on the source of the discrepancy among different laboratories, we have in the present study probed the influence of three PARP inhibitors in four types of cancer cells, cervical (HeLa), testicular (NTera2), pancreatic (BxPC3) and osteosarcoma (U2OS), on the results of Pt-BP6 photo-cross-linking experiments and cytotoxicity assays. We find that the activity of PARP proteins following exposure to platinum-modified DNA results in the dissociation of DNA-bound proteins. PARP inhibitors were able to sensitize some, but not all, of the cell lines to cisplatin. This cell-line dependence and the potential consequences of PARP-initiated protein removal from platinum-DNA lesions are discussed. Control experiments revealed that NTera2 cells are especially sensitive to PARP inhibition.
INTRODUCTION
The compound cis-diamminedichloroplatinum(II), or cisplatin, is one of the most successful anticancer agents ever discovered. Cisplatin has been used widely to treat a variety of tumor types for more than thirty years.1 Platinum-based therapies remain at the forefront of the fight against cancer, and new strategies are being developed including delivery systems2 and combination therapies.3 Cisplatin kills cells by binding to DNA and blocking template-dependent activities, notably transcription. The most common types of cisplatin-DNA adducts found in patients treated with the drug are 1,2-d(GpG), 1,2-d(ApG), and 1,3-d(GpNpG) intrastrand cross-links, in which one platinum atom binds to the N7 atoms of the two purine bases.1 These platinum-DNA cross-links are recognized by many nuclear proteins, which can signal repair of the adducts, or conversely, lead to cell death.4
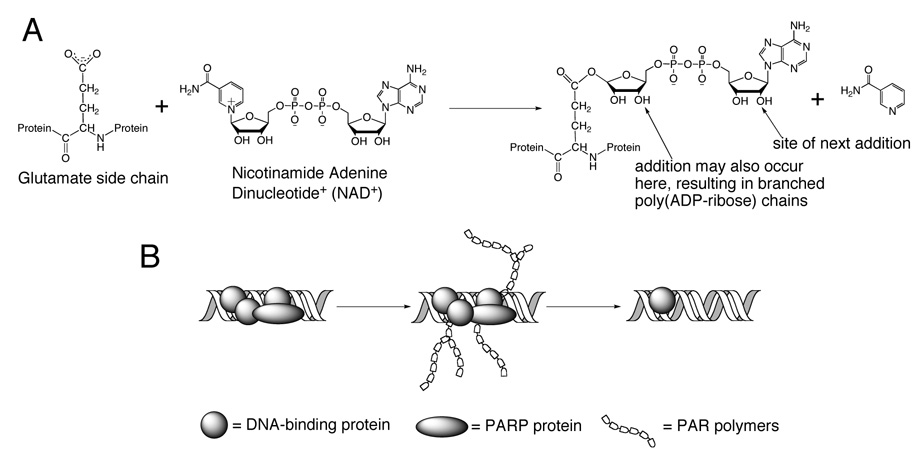
The affinity of poly(ADP-ribose) polymerase-1 (PARP-1) for platinum 1,2-d(GpG) and 1,3-d(GpTpG) intrastrand cross-links on duplex DNA was recently discovered.5,6 The activity of the PARP superfamily of proteins has been implicated in DNA repair, chromatin remodeling, transcriptional control, and inflammation.7 PARP-1 is a 113 kDa protein that contains a DNA-binding domain with two zinc fingers, an automodification domain, and a catalytic domain. The catalytic domain binds nicotinamide adenine dinucleotide (NAD+) and creates long, branched PAR chains in which each unit contains two negatively charged phosphate linkers (Figure 1A). These polymers can be digested by poly(ADP-ribose) glycohydrolase (PARG) to the unmodified form of the protein.7
Figure 1.
A) The catalytic activity of PARP proteins. PARP proteins catalyze the addition of negatively charged poly(ADP-ribose) polymers to acceptor proteins using NAD+ as a substrate. B) This modification is important for DNA repair because the modified proteins will be electrostatically repelled from the DNA. This activity can be eliminated by the addition of PARP inhibitors.
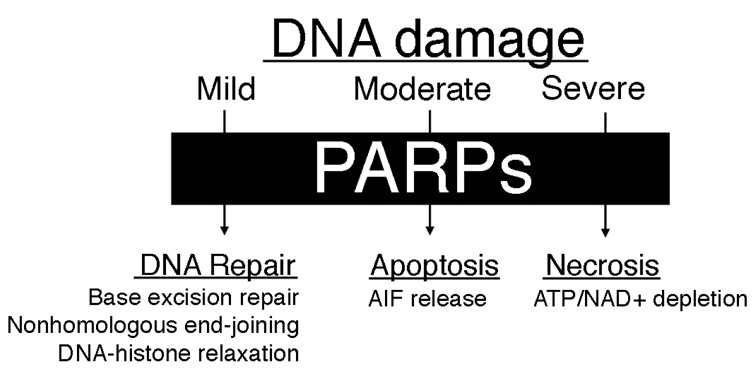
Upon exposure of cells to DNA-damaging agents, PARP proteins are strongly activated. PARP can activate three different cellular pathways (Figure 2).8 Under mild conditions, PARP proteins mediate DNA damage repair. PARP-1 activity has been implicated in base excision repair (BER)9 and in a backup pathway for nonhomologous end-joining (NHEJ) repair of double-strand breaks.10,11 When DNA damage is too severe to be repaired efficiently, the PAR polymers created by PARP proteins signal the release of apoptosis-inducing factor (AIF) from mitochondria, which initiates apoptosis.12 Apoptosis induced by cisplatin is due to both caspase- and AIF-dependent signaling mechanisms.13 Under severe DNA damage conditions, the activity of PARP proteins depletes cellular reservoirs of NAD+, leading to the shutdown of glycolysis. Because cancer cells rely on glycolysis for ATP production, they die by necrosis.14
Figure 2.
Three pathways of PARP activity. The activity of PARP proteins following DNA damage can lead to DNA repair, apoptosis, or necrosis through independent mechanisms.
The attachment of PAR polymers to proteins is an important component of several biochemical pathways. Automodified PARP-1 interacts with XRCC1, which may be responsible for the initiation of base excision repair.7 A PAR-binding motif has been identified in several DNA repair proteins, and this type of domain is present in core histones, p53, DNA topoisomerase I, Ku70, XPA, Msh6, DNA Ligase III, and XRCC1.15–17 Negatively charged PAR polymers create an electrostatic repulsion between the proteins they modify and polyanionic DNA (Figure 1B). For example, poly(ADP-ribosyl) ation of histones contributes to chromatin relaxation, making damaged DNA more accessible to repair proteins.7 The C-terminal domain of the chromatin remodeling protein HMGB1 is poly(ADP-ribosyl)ated by PARP-1, causing it to dissociate from chromatin and translocate to the cytosol.18,19
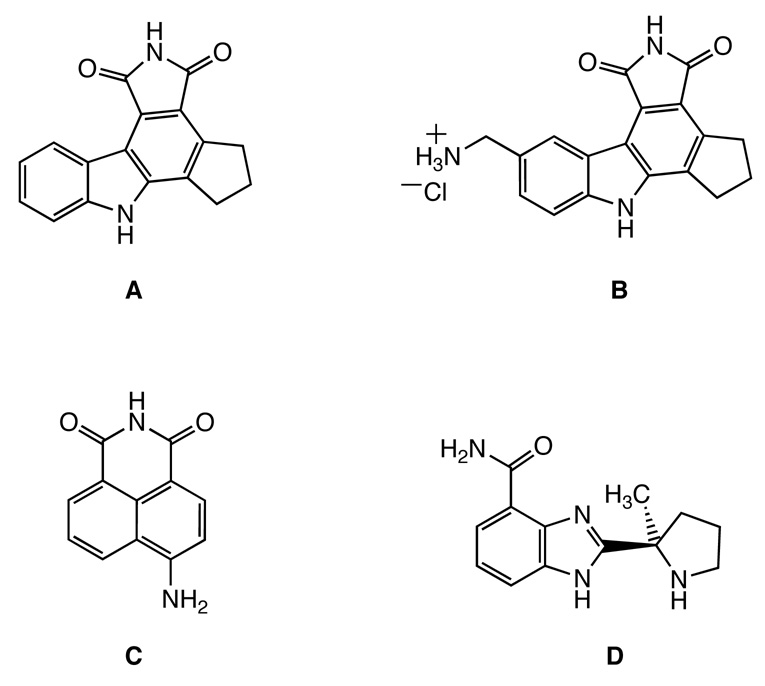
Previous studies to understand PARP activity following DNA damage have focused on DNA methylating agents, such as N-methyl-N-nitro-N-nitrosoguanidine (MNNG).12,20–22 The activity of PARP following cellular exposure cisplatin is less well understood. One study revealed increased levels of PAR polymers upon cisplatin treatment of rat and monkey tumor cells, which was attributed to DNA double-strand break formation during the processing of cisplatin-DNA adducts.23 Another report demonstrated that PARP activity is strongly upregulated following exposure of renal tubular cells to cisplatin, significantly depleting ATP levels within these cells.24 In this work, extremely high concentrations of cisplatin (0.5 mM) were required to induce PARP activity. Recently, a mild increase in PARP activity was reported 24 h after a 4-h treatment of HT29 colon carcinoma cells 10 µM cisplatin.25 Only one cell line was tested using the assay, however, providing little information about the importance of PARP activity in cisplatin sensitivity. HeLa cervical cancer cells were also employed in this study, but PARP activity following cisplatin treatment was not reported.25 Although the activation of PARP upon cisplatin treatment is not well characterized, PARP inhibitors sensitize cells to cisplatin. The compound CEP-6800 (B, Figure 3) sensitizes non-small cell lung carcinoma cells to cisplatin in both culture and xenografts.26 Another newly developed PARP inhibitor, ABT-888 (D, Figure 3), substantially improves tumor response to cisplatin and carboplatin.27 The anticancer potential of PARP inhibitors and platinum drugs in combination therapies is currently being investigated in phase I and II clinical trials.28
Figure 3.
Chemical structures of four PARP inhibitors. CEP-A (A); CEP-6800 (B); 4-amino-1,8-naphthalimide (4-ANI, C); ABT-888 (D).
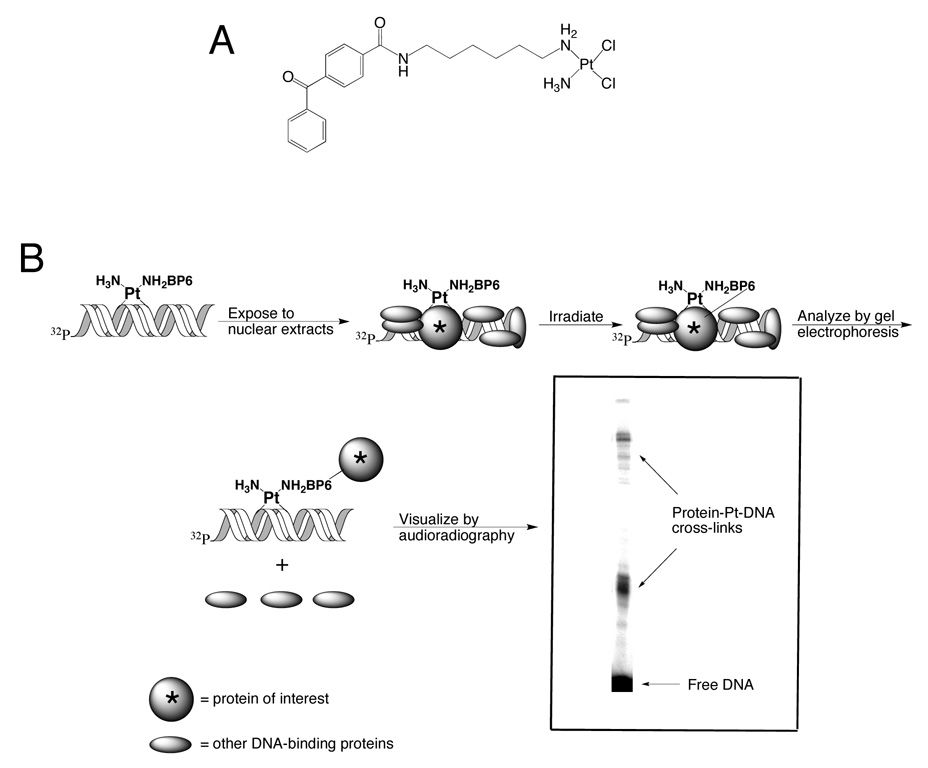
Previously we reported the ability of PARP-1 to bind to platinum-damaged DNA with the use of a novel photo-cross-linking probe.5,6 Photo-cross-linking is achieved by exposing duplex DNA containing a site-specific adduct of a photoactive cisplatin analogue to nuclear extracts from cancer cells. The photoactive compound is cis-[Pt(NH3)(N-(6-aminohexyl)-4-benzophenonamide)Cl2] (Pt-BP6, Figure 4A), which contains a benzophenone moiety tethered to the platinum atom by a hexamethylene linker. Irradiation at 360 nm activates the benzophenone, ultimately leading to covalent bond formation between the platinum-modified DNA and any proteins in the vicinity of the platination site. The products are then resolved by gel electrophoresis, which separates protein-platinum-DNA complexes according to their size. For analytical-scale experiments, radiolabeled DNA allows the protein-Pt-DNA complexes to be visualized by audioradiography (Figure 4B).
Figure 4.
A) The structure of the photoactive cisplatin analogue, Pt-BP6. B) Diagrammatic representation of chemical cross-linking of proteins with an affinity for platinum-modified DNA using Pt-BP6 and their identification by gel electrophoresis.
In the present work, the role of PARP proteins in mediating cisplatin cytotoxicity has been investigated in a number of cell lines with varying sensitivities to cisplatin. Included are cervical (HeLa), testicular (NTera2), pancreatic (BxPC3) and osteosarcoma (U2OS) cells. Two previously reported PARP inhibitors were prepared according to literature protocols,29–31 and one step in the synthesis was improved. Photo-cross-linking experiments were preformed in the presence of one of these PARP inhibitors to determine its effect on the binding of nuclear proteins to platinum-damaged DNA. The results indicate that PARP activity in the presence of platinum-damaged DNA induced dissociation of the nuclear proteins from the duplex. In addition, we investigated these two PARP inhibitors for their ability to potentiate cisplatin cytotoxicity in the four cell lines. For comparison purposes, the commercially available and well-characterized PARP inhibitor, 4-ANI, was also evaluated. The commercially available PARP inhibitor, 4-ANI, was also used in these studies to confirm that our synthesized compounds behave similarly to a well-characterized PARP inhibitor. The results indicate that the ability of PARP inhibitors to sensitize cells to cisplatin is cell-line-dependent, an effect that may derive from the nature of PARP activity in the presence of platinum-modified DNA.
MATERIALS AND METHODS
All reactions were performed in oven- or flame-dried round bottom flasks. The flasks were fitted with rubber septa and reactions were conducted under a positive pressure of argon. Stainless steel cannulae or gas-tight syringes were used to transfer air- and moisture-sensitive liquids. Flash column chromatography was performed32 using silica gel (60-Å pore size, 32–63 µm, standard grade). Analytical thin–layer chromatography was carried out by using glass plates pre-coated with 0.25 mm 230–400 mesh silica gel impregnated with a fluorescent indicator (254 nm). Thin layer chromatography plates were visualized by exposure to UV light and an aqueous solution of ceric ammonium molybdate (CAM). Organic solutions were concentrated on rotary evaporators at ~20 Torr (house vacuum) at 25–35 °C. Commercial reagents and solvents were used as received with the following exceptions; dichloromethane, diethyl ether, tetrahydrofuran, and triethylamine were purified as described33 under a positive argon pressure. 1,4-Dioxane (>99.9 % HPLC grade, ≤0.020 % water) and Raney nickel (W.R. Grace and Co. Raney® 3202, slurry, in H2O, active catalyst) were used as received.
Proton nuclear magnetic resonance (1H NMR) spectra were recorded at the MIT Department of Chemistry Instrumentation Facility (DCIF) with an inverse probe 500 MHz spectrometer and are referenced from the residual protium in the NMR solvent peaks (DMSO–d5, δ 2.50). 13C NMR spectra were recorded at 125 MHz and referenced from the carbon resonances of the solvent (DMSO–d6, δ 39.52). High-resolution mass spectra were obtained at the DCIF using a Fourier transform ion cyclotron resonance mass spectrometer with electrospray ionization.
Synthesis of 4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione (A)
To a pale yellow solution of 3a,3b,4,5,6,6a,7,11c-octahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(1H)-dione (E)29 (125 mg, 0.446 mmol, 1 equiv) in 1,4-dioxane (8.1 mL) was added γ-MnO234 (970 mg, 11.2 mmol, 25.0 equiv) and the resulting black suspension was heated to reflux. After 7 h, the suspension was allowed to cool to approximately 60 °C, diluted with THF (4 mL), sonicated for 1 min, and filtered through a plug of celite (diameter 4.0 cm, height 3.5 cm) that was pre-wetted with THF. The reaction flask and plug were rinsed with additional portions of warm tetrahydrofuran (40–50°C, 75 mL total volume), and the clear yellow filtrate was concentrated to give A29 (60.5 mg, 49%) as a bright yellow solid. 1H NMR (DMSO–d6) δ ppm: 11.91 (s, 1H), 10.93 (s, 1H), 8.80 (d, J = 7.5 Hz, 1H), 7.56 (d, J = 8.0 Hz, 1H), 7.51 (t, J = 7.7 Hz, 1H), 7.27 (t, J = 7.7 H, 1H), 3.23 (t, J = 7.5 Hz, 2H), 3.15 (t, J = 7.5 Hz, 2H), 2.27 (quintet, J = 7.5 Hz, 2H). 13C NMR (DMSO–d6) δ ppm: 171.8, 171.8, 142.7, 141.4, 139.8, 133.2, 128.7, 126.5, 125.7, 121.8, 121.1, 120.8, 118.6, 112.7, 31.9, 30.8, 26.3.
Synthesis of 10-(aminomethyl)-4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione (B)
This compound was prepared as described in the literature.29 A suspension of 2-(1,3-dioxo-2,3,4,5,6,7-hexahydro-1H-cyclopenta-[a]pyrrolo[3,4-c]carbazol-10-yl)acetonitrile29 (47.0 mg, 0.156 mmol, 1 equiv) and Raney nickel (W.R. Grace and Co. Raney® 3202, slurry, in H2O, active catalyst, 250 mg, 532 wt. %) in dimethylformamide (5.0 mL) was saturated with ammonia by passage of a stream of ammonia gas (1 atm) for 10 min. The reaction vessel was placed in a hydrogenation apparatus and the apparatus was purged three times with dihydrogen (60 psi), then maintained under dihydrogen (60 psi) with vigorous stirring of the reaction mixture. After 48 h, the hydrogenation apparatus was opened and an additional portion of Raney nickel (250 mg, 532 wt. %) was added, the suspension was purged with ammonia gas (1 atm) for 10 min, and the vessel was purged with H2 (3 × 60 psi) then maintained under H2 (60 psi). After an additional 48 h another portion of Raney Nickel (250 mg, 532 wt. %) was added in the same fashion, and the reaction mixture was maintained under H2 (60 psi) for 96 h. The reaction mixture was gently vacuum-filtered through a plug of celite (diameter 2.5 cm, height 2.5 cm) that was pre-wetted with dimethylformamide, and the reaction flask and celite were rinsed with additional portions of dimethylformamide (35 mL total). The bright yellow filtrate was concentrated to a yellow residue, which was dissolved in aqueous HCl (0.021 N, 30 mL). The aqueous solution was washed with ethyl acetate (2 × 15 mL) prior to lyophilization to give B29 (5.5 mg, 12%) as a bright yellow solid. 1H NMR (DMSO–d6) δ ppm: 12.17 (s, 1H), 11.00 (s, 1H), 8.82 (s, 1H), 7.66 (d, J = 8.5 Hz, 1H), 7.61 (d, J = 8.5 Hz, 1H), 4.16 (m, 2H), 3.23 (t, J = 7.2 Hz, 2H), 3.16 (t, J = 7.2 Hz, 2H), 2.27 (quintet, J = 7.5 Hz, 2H). HRMS–ESI (m/z): calcd for C18H15N3O2Na [M + Na]+: 328.1056, found: 328.1050.
Cell culture
HeLa, NTera2, BxPC3, and U2OS cells were grown in DMEM with 10% FBS at 37 °C in an atmosphere of 5% CO2. HeLa YS cells were prepared as previously described5 and grown in DMEM with 10% FBS supplemented with 100 µg/mL zeocin selection reagent (Invitrogen). Nuclear extracts were prepared as previously described.5,6
Photo-cross-linking in the presence of PARP inhibitors
Photo-cross-linking experiments were carried out as previously described.5,6 A 25-bp DNA duplex containing a site-specific 1,2-d(GpG) or 1,3-d(GpTpG) intrastrand cross-link of Pt-BP6 was exposed to HeLa nuclear extracts in the presence of 0, 0.01, 0.05, 0.1, 0.3, or 1.0 µM CEP-A prior to photo-cross-linking. The inhibitor was dissolved in DMF and diluted to the desired concentration with the final solution containing 0.02% DMF. Photo-cross-linking was also performed without DMF as a control. Photo-cross-linking experiments were then repeated using nuclear extracts from NTera2, BxPC3, U2OS, and HeLa YS cell lines, with or without 1.0 µM CEP-A, for both types of Pt-BP6 cross-link. The audioradiographs were quantitatedquantified using ImageQuant data analysis software.
Cytotoxicity assays
HeLa, NTera2, BxPC3 and U2OS cells were plated at 500–1000 cells/well in a 96-well plate. The following day, the cells were treated with varying concentrations of PARP inhibitors CEP-A (A), CEP-6800 (B), and 4-amino-1,8-naphthalimide (4-ANI, C) to determine the maximum tolerated dose of inhibitor in each cell line. After 96 h, the viability of the cells was assed by the MTT assay. To each well was added 5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and the plates were incubated at 37 °C for four h. The media was revomed from each well by vacuum, and replaced with 100 µL of DMSO. The number of viable cells was determined by measuring the absorbance of each well at 562 nm. The cytotoxicity assays were then repeated with the maximum tolerated dose of PARP inhibitor plus varying concentrations of cisplatin.
RESULTS
Overview
The effect of PARP inhibition on the ability of nuclear proteins to bind platinum-modified DNA was assessed using photo-cross-linking experiments5,6 in which a radiolabeled 25-bp duplex DNA containing a site-specific adduct of a photoactive analogue of cisplatin is incubated with nuclear extracts from cancer cells and then irradiated at 360 nm. Such irradiation causes a covalent bond to be formed between the platinum-modified DNA and a nearby bound protein (Figure 4). Previous work of this kind identified several proteins that bind to platinum-modified DNA, including PARP-1.5,6 In the present study, the addition of a PARP inhibitor CEP-A (A, Figure 3) to the photo-cross-linking reaction increased the total photo-cross-linking yield. The extent of this effect varied between cell lines and platinum cross-links tested. The ability of PARP inhibitors to sensitize the cell lines to cisplatin was also assessed.
Synthesis of CEP-A (A) and CEP-6800 (B)
Pyrrolocarbazole compounds A and B were prepared according to the concise approach in the literature29–31 (Scheme S.1, Supporting Information). In this sequence, in situ N-carboxylation of indole followed by directed lithiation at C2 and trapping with cyclopentanone provided the corresponding tertiary alcohol,35 which underwent dehydration upon treatment with hydrochloric acid. Subsequent [4+2] cycloaddition with maleimide upon heating a finely dispersed solid mixture afforded the adduct E.
Double dehydrogenation of E using 2,3-dichloro-5,6-dicyano-p-benzoquinone (DDQ) as an oxidant provided the pyrrolocarbazole product containing varying amounts of inseparable DDQ derived byproducts. Alternatively, in a procedure optimized during this work, heating a mixture of E and freshly prepared γ-MnO234 in refluxing 1,4-dioxane cleanly provided pyrrolocarbazole A as a bright yellow solid after simple filtration of the hot reaction mixture. Subsequent introduction of the methylamino group was performed as previously described.29 Regioselective bromination, coupling of the bromide with copper cyanide, and hydrogenation using Raney nickel in the presence of ammonia provided the primary amine B. Isolation of B as a hydrochloric acid salt was achieved by extraction into aqueous solution and lyophilization. Spectral properties of A and B were identical to those reported previously.29
Photo-cross-linking in the presence of the PARP inhibitor CEP-A (A)
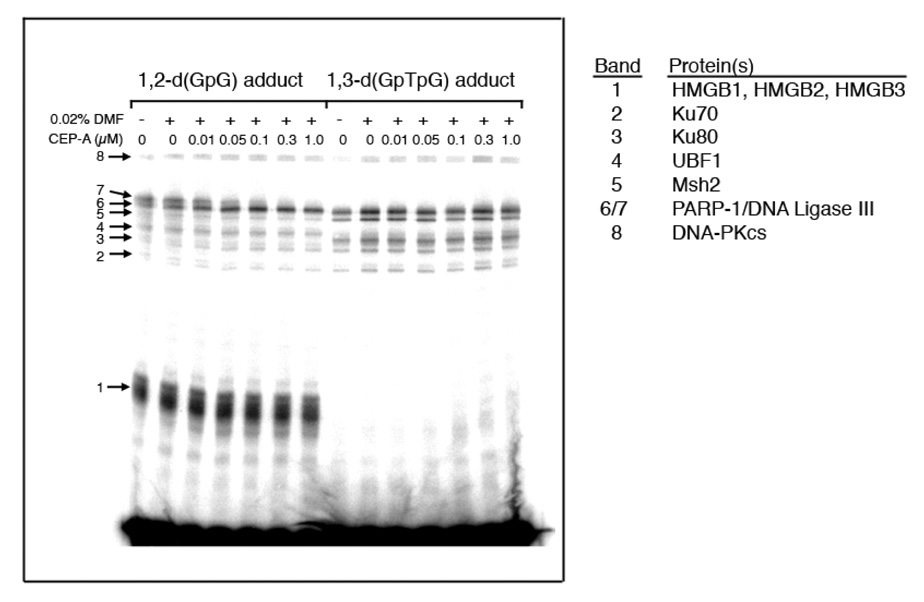
Photo-cross-linking experiments were not affected by the presence of 0.02% DMF, necessary to dissolve the PARP inhibitor CEP-A (Figure 5). For a 25-bp DNA duplex containing a 1,2-d(GpG) adduct of Pt-BP6 in HeLa nuclear extracts, increasing amounts of CEP-A (A) in the photo-cross-linking experiment resulted in a decrease in intensity of high-molecular weight band 7, and band 6 directly below increased in intensity (Figure 5). The proteins present in these bands, identified and discussed in previous work,5,6 are labeled on the figure. For the 25-bp DNA containing a Pt-BP6 1,3-d(GpTpG) cross-link, PARP inhibition caused no significant effect on any of the bands (Figure 5).
Figure 5.
Results of photo-cross-linking of Pt-BP6-modified 25-bp duplexes in HeLa nuclear extracts as a function of increasing concentrations of the PARP inhibitor, CEP-A. For the 1,2-d(GpG) adduct, the intensity of band 6 increases, while that of band 7 decreases with increasing concentrations of CEP-A.
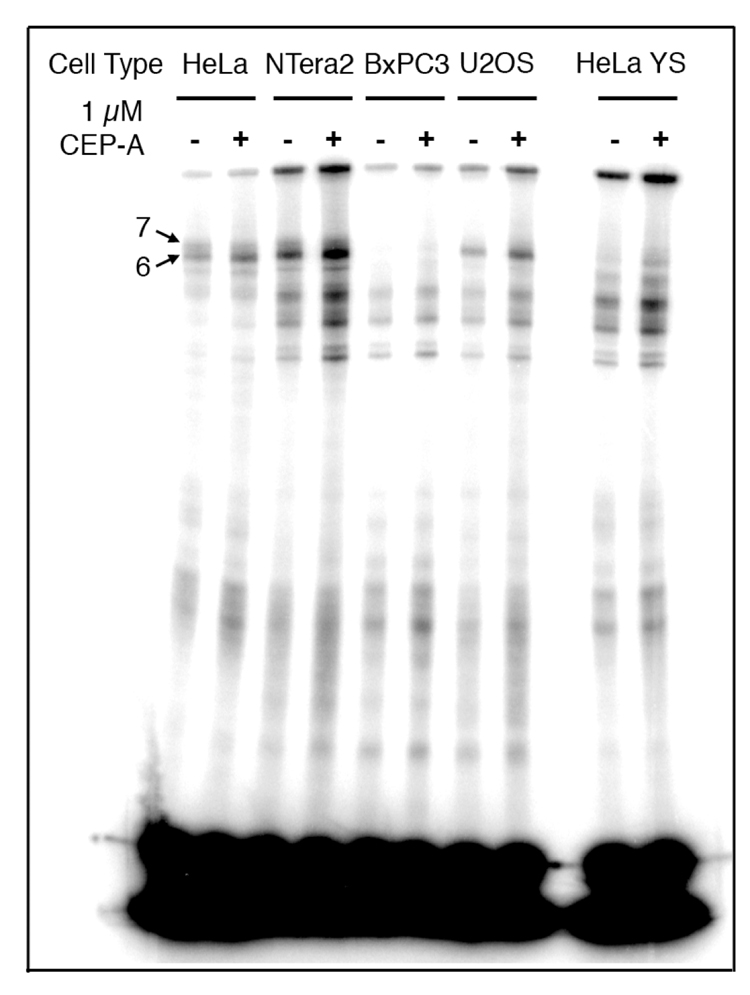
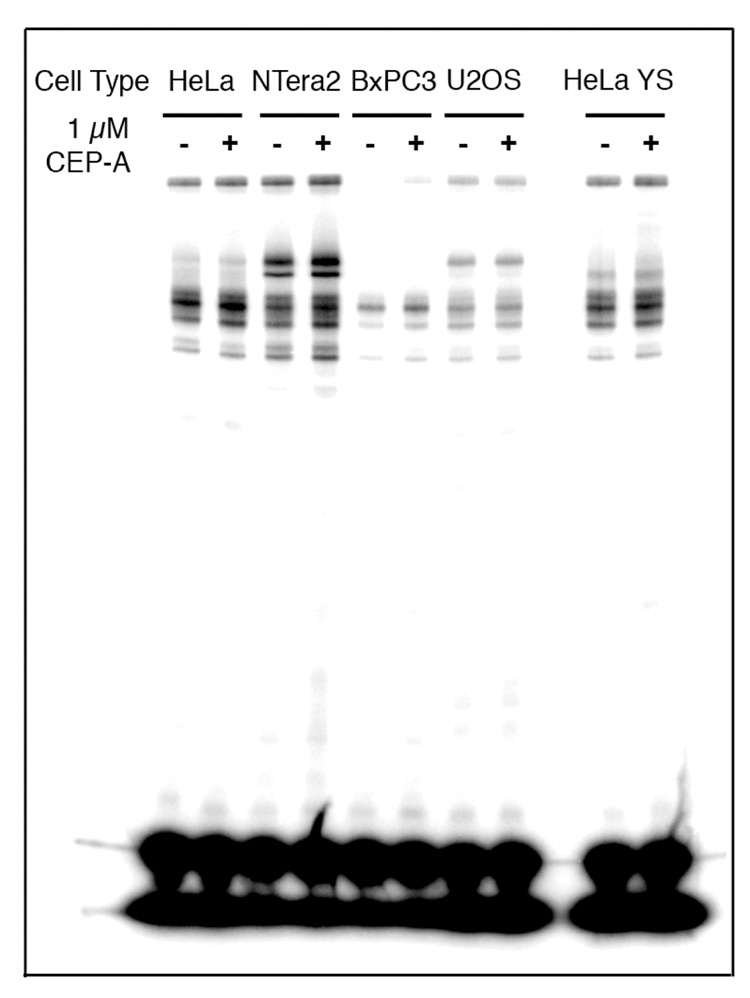
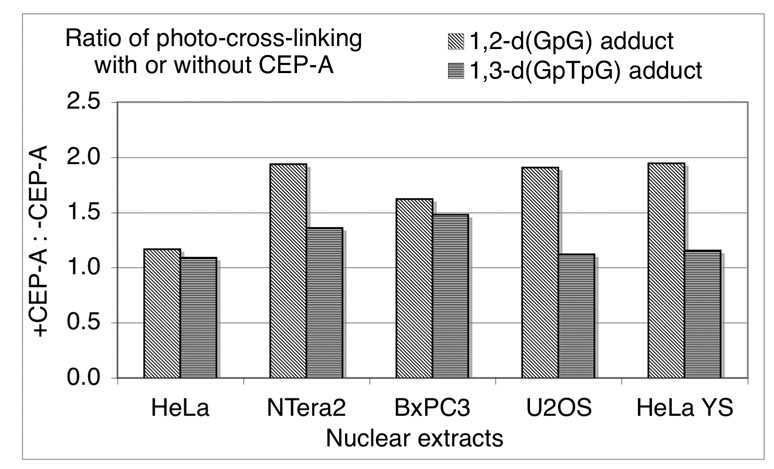
These experiments were repeated with 1 µM CEP-A (A) using nuclear extracts from HeLa, NTera2, BxPC3, U2OS, as well as HeLa cells in which PARP-1 has been silenced using RNAi (HeLa YS, Figure 6 and Figure 7). The behavior of the high-molecular weight bands for the duplex containing a 1,2-d(GpG) intrastrand cross-link with HeLa nuclear extracts was consistent with the foregoing results, but for nuclear extracts from the other cell lines, the behavior was different (Figure 6). The total amount of photo-cross-linking for this probe increased upon addition of the PARP inhibitor for all cell lines tested by 20–100% of the original intensity (Figure 6 and Figure 8). Consistent with the experiment using HeLa nuclear extracts presented in Figure 6, the addition of CEP-A (A) did not significantly affect the photo-cross-linking of the duplex containing a 1,3-d(GpTpG) intrastrand cross-link in nuclear extracts from any of the cell lines tested (Figure 7). The total amount of photo-cross-linking by the 1,3-d(GpTpG) probe increased upon addition of the PARP inhibitor, but to a lesser degree than for the 1,2-d(GpG) probe. These results are presented graphically in Figure 8.
Figure 6.
Results of photo-cross-linking of 25-bp duplex containing a 1,2-d(GpG) adduct of Pt-BP6 in various cancer cell extracts in the presence of the PARP inhibitor, CEP-A. The inhibitor increases the intensity of photo-cross-linking in each cell line. This effect is quantitated in Figure 8.
Figure 7.
Results of photo-cross-linking of 25-bp duplex containing a 1,3-d(GpTpG) adduct of Pt-BP6 in various cancer cell extracts in the presence of the PARP inhibitor, CEP-A. The inhibitor does not have a significant effect in any of the cell lines. This effect is quantitated in Figure 8.
Figure 8.
Intensity of total photo-cross-linking for site-specifically platinated DNA, with or without addition of the PARP inhibitor CEP-A. The effect of PARP inhibitor CEP-A in photo-cross-linking experiments reveals an increase in total intensity of photo-cross-linking. The effect is greater for the 1,2-d(GpG) than the 1,3-d(GpTpG) intrastrand cross-link. The effect is also cell-line dependent, with HeLa cells exhibiting the smallest increase.
Cytotoxicity assays
The toxicity of PARP inhibitors was investigated for each cell line to determine the maximum tolerated dose (Figure S.1 and Table S.1, Supporting Information. These values were calculated by evaluating the highest concentration at which at least 90% of the cells survive. The compound CEP-A was toxic even at 0.1 µM in all cell lines (Figure S.1). Despite this toxicity, cells co-treated with 0.1 µM CEP-A and cisplatin behaved in an identical manner to those co-treated with nontoxic doses of the other inhibitors, as discussed below (Figure 9).
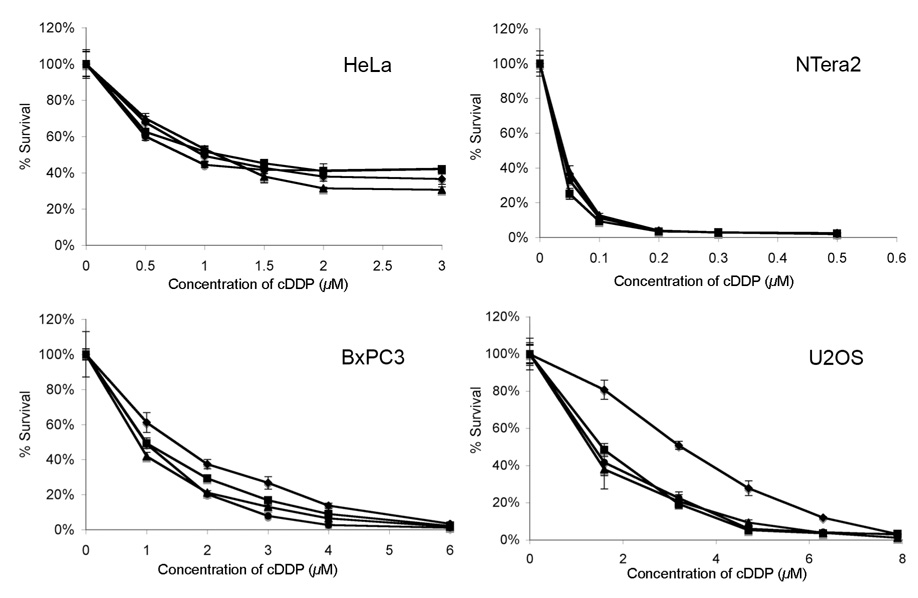
Figure 9.
Results of cytotoxicity assays of cisplatin co-treated with PARP inhibitors in HeLa, NTera2, BxPC3, and U2OS cells. Cells were treated with cisplatin alone (◆) or co-treated with maximum tolerated dose of 4-ANI (■), CEP-A (▴) or CEP-6800 (●). The IC50 values are summarized in Table 1.
Cells were treated with the maximum tolerated dose of inhibitor and varying concentrations of cisplatin. The results of the MTT assays are presented in Figure 9 and summarized in Table 1. The sensitivity of HeLa and NTera2 cells to cisplatin was unchanged by addition of PARP inhibitors, but BxPC3 and U2OS cells were sensitized to cisplatin by factors of 1.6 and 3.3, respectively.
Table 1.
IC50 values for four cell lines co-treated with cisplatin at their maximum tolerated dose of three PARP inhibitors.
| Cell line | cDDP alone (µM) | + 4-ANI (µM) | + CEP-A (µM) | + CEP-6800 (µM) | Sensitization |
|---|---|---|---|---|---|
| HeLa | 1.0 | 1.1 | 0.8 | 1.1 | none |
| NTera2 | 0.04 | 0.04 | 0.04 | 0.04 | none |
| BxPC3 | 1.5 | 0.9 | 1.0 | 1.0 | 1.6-fold |
| U2OS | 3.3 | 1.4 | 1.4 | 1.6 | 2.3-fold |
IV. DISCUSSION
PARP-1 activity dissociates proteins from platinum-modified DNA in nuclear extracts
The activity of poly(ADP-ribose) polymerase (PARP) proteins in the presence of DNA damage can lead to repair or, conversely, signal cell death (Figure 2). It was recently discovered that PARP-1 binds to platinum-modified DNA.5,6 PARP-1 and the PARP family catalyze the addition of poly(ADP-ribose) (PAR) polymers onto acceptor proteins in a reaction that consumes NAD+ (Figure 1A).15 Each unit of the polymer contains two negatively charged phosphate moieties, which can electrostatically repel DNA molecules from PAR-modified proteins (Figure 1B).7 PARP-1 automodification leads to dissociation of the enzyme from DNA, and the protein can also catalyze the modification other proteins, including histones, which relaxes histone-DNA interactions.15 In the present work, we studied the consequences of PARP activity upon exposure of nuclear proteins to platinum-modified DNA using photo-cross-linking experiments.
The method utilizes DNA containing a site-specific adduct of a benzophenone-modified cisplatin analogue Pt-BP6. Photo-cross-linking with such probes enables the study of nuclear proteins that bind to platinum-modified DNA (Figure 4). Several platinum-modified DNA-binding proteins have been identified in this manner, as discussed elsewhere.5,6 Here we performed photo-cross-linking experiments in the presence of the PARP inhibitor CEP-A (A). The addition of CEP-A (A) to nuclear extracts prior to photo-cross-linking generally increased the amount of proteins photo-cross-linked to Pt-BP6-modified DNA (Figure 6–Figure 8). This result is consistent with a model in which PARP activity stimulated by platinum-DNA cross-links results in the PAR-modification of DNA-binding proteins, causing them to dissociate from the duplex (Figure 1B).7 Inhibition of PARP activity by CEP-A (A) eliminates this effect, resulting in more stable protein-DNA interactions and, consequently, increased amounts of photo-cross-linking.
Our experiments indicate that the addition of PARP inhibitor significantly increases the photo-cross-linking of proteins to the platinum-modified DNA containing a 1,2-d(GpG) intrastrand adduct of Pt-BP6 in each type of nuclear extract examined except for HeLa (Figure 6 and Figure 8). Nuclear extracts from HeLa cells exhibited only a modest increase in photo-cross-linking following addition of the PARP inhibitor (Figure 5, Figure 6 and Figure 8). In these nuclear extracts exclusively, a high molecular weight band decreases in intensity with the addition of PARP inhibitor (Figure 5, band 7). This result indicates that PARP-1 activity in HeLa extracts following exposure to platinum-damaged DNA is unique.
Photo-cross-linking was more significantly affected for the 1,2-d(GpG) than the 1,3-d(GpTpG) intrastrand cross-link (Figure 6–Figure 8). This effect was consistent across all cell lines tested, although to a lesser degree for BxPC3 extracts, indicating that the 1,2-d(GpG) intrastrand cross-link more efficiently activates the protein. Experiments using extracts from HeLa cells in which PARP-1 has been silenced with RNAi (HeLa YS) reveal an increase in photo-cross-linking, similar to the behavior of NTera2, BxPC3 and U2OS cellular extracts. This result most likely indicates that, in the PARP-1-silenced cell line, other PARP isoforms are present having the same activity as PARP-1.
NTera2 cells are sensitive to PARP inhibition
The toxicities of three PARP inhibitors (Figure S.1, Supporting Information) were first determined for the cell lines tested to obtain the maximum tolerated dose that could be used to potentiate the cell-killing ability of cisplatin. NTera2 cells are extremely sensitive to PARP inhibitors, behavior that hampers our ability to assess their capacity to enhance cisplatin sensitivity. This finding is perplexing given that NTera2 cells express high levels of PARP-1.5 PARP-1 is commonly mutated in germ cells, specific variants being Val762Ala and Lys940Arg, two residues in the catalytic domain of the protein.36 Compromised activity of the enzymeprotein by these mutations may render it especially sensitive to PARP inhibitors. It is also possible that NTera2 cells are deficient in certain DNA repair pathways that could strongly sensitize themlead to a strong sensitivity to PARP inhibitors, as for similar to BRCA-mutated cancers.37 The reliance of NTera2 cells on PARP activity, even without the addition of DNA-damaging agents, warrants further investigation.
The potentiation of cisplatin sensitivity by PARP inhibitors is cell line-dependent
Reports in the literature demonstrate that certain cell lines are unaffected by the presence of PARP inhibitors, whereas others are sensitized to cisplatin. For example, PARP inhibitors were unable to sensitize human ovarian tumor cell lines SK-OV-3, OAW-42, and the rat ovarian tumor cell line O-342 to cisplatin,38 but could sensitize B16F10 murine melanoma, 9L rat glioma, HCT-116 human colon carcinoma, DOHH-2 human B-cell lymphoma, MX-1 human breast carcinoma, and Calu-6 human non-small cell lung carcinoma cells to the drug.26,27 The use of new PARP inhibitors CEP-6800 (B, Figure 3) and ABT-888 (D, Figure 3) for experiments involving the B16F10, 9L, HCT-116, DOHH-2, MX-1, and Calu-6 cell lines is one reason for this discrepancy, because these compounds are more water soluble and are able to enter cells and more efficiently inhibit PARP proteins.26,27 The present work demonstrates that there is a cell line dependence to this effect. Testicular (NTera2) and cervical (HeLa) cancer cells were unaffected, but pancreatic (BxPC3) and osteosarcoma (U2OS) cancer cells are sensitized to cisplatin by PARP inhibition by factors of 3.3 and 1.6, respectively (Figure 9, Table 1). These results were consistently obtained for both the newly developed PARP inhibitors CEP-A (A) and CEP-6800 (B) as well as a commercially available compound 4-ANI (C).
A model for the cell line-dependence of sensitization to cisplatin by PARP inhibitors
The sensitization of certain cell lines to cisplatin by PARP inhibitors may be caused by differences in the processing of platinum-DNA adducts in the absence of PARP activity. This possibility was investigated by performing photo-cross-linking studies in the presence of the PARP inhibitor CEP-A, as described above. Experiments using extracts from HeLa cells show the smallest increase in photo-cross-linking compared to the other types of extracts tested (Figure 6). Although the total amount of photo-cross-linking does not increase significantly, one band appears to shift upon addition of PARP inhibitor to the reaction (Figure 5, band 7). This band might be due to poly(ADP-ribosyl)ated PARP-1, which would migrate slightly more slowly owing to an increase in molecular weight than the unmodified protein. Alternatively, it might be due to the recruitment of another DNA-binding protein, such as DNA Ligase III. In either case, the data indicate that PARP-1 in NTera2, BxPC3, and U2OS nuclear extracts modifies other proteins to a greater degree, causing them to dissociate from DNA, an effect not reproduced with HeLa nuclear extracts.
One possible model to tie together the in vitro and in vivo results is that PARP-1 activity in BxPC3 and U2OS cells dissociates proteins from damaged DNA, allowing the repair apparatus to access the site. Chemical inhibition of PARP-1 would eliminate this effect, inhibiting repair and leading to sensitization of the cells to cisplatin. HeLa cells do not experience this sensitization because PARP-1 activity in HeLa does not significantly affect other platinum damage-binding proteins. Our photo-cross-linking results in NTera2 nuclear extracts cannot be explained by this model, but these cells may be too sensitive to PARP inhibitors to allow an accurate measure of cisplatin sensitization, as already discussed.
V. CONCLUSIONS
Photo-cross-linking studies in the presence of a PARP inhibitor indicate that the activity of PARP proteins bound to platinum-damaged DNA leads to dissociation of PARP-1 itself, as well as other proteins, from the damaged duplex. We also discovered that PARPs are better activated in nuclear extracts by a 1,2-d(GpG) than a 1,3-d(GpTpG) Pt-BP6 intrastrand cross-link. Several studies in the literature report varying degrees of sensitization of cancer cells to cisplatin by PARP inhibitors. It has thus far been difficult to determine whether these inconsistencies are due to the cell lines or the inhibitors used, since both are varied. We present here the finding that PARP inhibitors sensitize cells to cisplatin in a manner that is cell line-dependent. In our work, PARP inhibition resulted in the greatest increase in cisplatin sensitivity for U2OS osteosarcoma cells. NTera2 testicular carcinoma cells do not show this effect, but are very sensitive to PARP inhibitors themselves. This sensitivity may be due to PARP-1 mutations, which are common in germ cells. We present a model in which PARP inhibitors are able to sensitize cells to cisplatin if PARP activity in that cell line causes the dissociation of nuclear proteins from platinum-damaged DNA.
Supplementary Material
Abbreviations
- AIF
apoptosis-inducing factor
- BER
base excision repair
- CAM
ceric ammonium molybdate
- NAD+
nicotinamide adenine dinucleotide
- MNNG
N-methyl-N-nitro-N-nitrosoguanidine
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NHEJ
nonhomologous end-joining
- PAR
poly(ADP-ribose)
- PARG
poly(ADP-ribose) glycohydrolase
- PARP
poly(ADP-ribose) polymerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 2.Feazell RP, Nakayama-Ratchford N, Dai H, Lippard SJ. J. Am. Chem. Soc. 2007;129:8438–8439. doi: 10.1021/ja073231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Lippard SJ. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 4.Jung Y, Lippard SJ. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 5.Guggenheim ER, Xu D, Zhang CX, Chang PV, Lippard SJ. 2008 submitted. [Google Scholar]
- 6.Zhang CX, Chang PV, Lippard SJ. J. Am. Chem. Soc. 2004;126:6536–6537. doi: 10.1021/ja049533o. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber V, Dantzer F, Amé J-C, de Murcia G. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 8.Nguewa PA, Fuertes MA, Alonso C, Perez JM. Mol. Pharmacol. 2003;64:1007–1014. doi: 10.1124/mol.64.5.1007. [DOI] [PubMed] [Google Scholar]
- 9.Bouchard VJ, Rouleau M, Poirier GG. Exp. Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 10.Audebert M, Salles B, Calsou P. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S-W, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Zhang C, Narayani N, Du C, Balaji KC. Cancer Lett. 2007;255:127–134. doi: 10.1016/j.canlet.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Amaravadi RK, Thompson CB. Clin. Cancer Res. 2007;13:7271–7279. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 15.Kim MY, Zhang T, Kraus WL. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 16.Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 18.Ditsworth D, Zong W-X, Thompson CB. J. Biol. Chem. 2007;282:17845–17854. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanuma S, Johnson GS. J. Biol. Chem. 1983;258:4067–4070. [PubMed] [Google Scholar]
- 20.Keil C, Gröbe T, Oei SL. J. Biol. Chem. 2006;281:34394–34405. doi: 10.1074/jbc.M606470200. [DOI] [PubMed] [Google Scholar]
- 21.Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. J. Biol. Chem. 2005;280:15773–15785. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- 22.Zong W-X, Ditsworth D, Bauer DE, Wang Z-Q, Thompson CB. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bürkle A, Chen G, Küpper JH, Grube K, Zeller WJ. Carcinogenesis. 1993;14:559–561. doi: 10.1093/carcin/14.4.559. [DOI] [PubMed] [Google Scholar]
- 24.Shino Y, Itoh Y, Kubota T, Yano T, Sendo T, Oishi R. Free Radic. Biol. Med. 2003;35:966–977. doi: 10.1016/s0891-5849(03)00470-2. [DOI] [PubMed] [Google Scholar]
- 25.Gambi N, Tramontano F, Quesada P. Biochem. Pharmacol. 2008;75:2356–2363. doi: 10.1016/j.bcp.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, Klein-Szanto A, Dionne C, Ruggeri B. Mol. Cancer. Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 27.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu G-D, Rosenberg SH, Giranda VL, Frost DJ. Clin. Cancer Res. 2007;13:2728–2739. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 28.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 29.Chatterjee S, Diebold JL, Dunn D, Hudkins RL, Dandu R, Wells GJ, Zulli AL. Appl. No. 11/455,356. U.S. Patent. 2006 (Cephalon Inc.)
- 30.Hudkins RL, Park CH. J. Heterocyclic Chem. 2003;40:135–142. [Google Scholar]
- 31.Tao M, Park CH, Bihovsky R, Wells GJ, Husten J, Ator MA, Hudkins RL. Bioorg. Med. Chem. Lett. 2006;16:938–942. doi: 10.1016/j.bmcl.2005.10.099. [DOI] [PubMed] [Google Scholar]
- 32.Still WC, Kahn M, Mitra A. J. Org. Chem. 1978:2923–2925. [Google Scholar]
- 33.Pangborn AB, Giardello MA, Grubbs RH, Rosen RK, Timmers FJ. Organometallics. 1996;15:1518–1520. [Google Scholar]
- 34.Fatiadi AJ. Synthesis. 1976;2:65–104. [Google Scholar]
- 35.Katritzky AR, Akutagawa K. Tetrahedron Lett. 1985;26:5935–5938. [Google Scholar]
- 36.Shiokawa M, Masutani M, Fujihara H, Ueki K, Nishikawa R, Sugimura T, Kubo H, Nakagama H. Jpn. J. Clin. Oncol. 2005;35:97–102. doi: 10.1093/jjco/hyi028. [DOI] [PubMed] [Google Scholar]
- 37.De Soto JA, Deng C-X. Int. J. Med. Sci. 2006;3:117–123. doi: 10.7150/ijms.3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernges F, Zeller WJ. J. Cancer Res. Clin. Oncol. 1996;122:665–670. doi: 10.1007/BF01209029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.