Abstract
Notch signaling regulates smooth muscle cell phenotype and is critical for vascular development. One Notch target is smooth muscle α-actin (SMA), a differentiated smooth muscle cell marker. The Notch intracellular domain (NotchICD) forms a complex with CBF-1 (C-promoter– binding factor-1) and directly induces SMA expression. Using primary human smooth muscle cells, we show that expression of the constitutive active ICD of human Notch1, Notch2, or Notch4 receptors increase SMA levels. NotchICD also induce expression of the transcriptional repressors HRT1 (Hairy-related transcription factor 1) and HRT2, in a CBF-1–dependent manner. However, unlike the activating effects of NotchICD, HRT1 or HRT2 represses basal SMA expression, and both are strong antagonists of NotchICD-induced SMA upregulation. This antagonism does not depend on histone deacetylase activity and occurs at the transcriptional level. Competitive coimmunoprecipitation experiments demonstrate that HRT does not disrupt the association of NotchICD and CBF-1, which form a complex in the presence or absence of HRTs. However, HRT suppresses NotchICD/CBF-1 binding to the SMA promoter, as measured by chromatin immunoprecipitation, and transactivation of an SMA promoter reporter spanning sequences −124/+32. SMA expression was regulated similarly following endogenous Notch activation in smooth muscle cells by coculture with endothelial cells, and this effect was also sensitive to HRT inhibition. Temporally defined HRT activity may constitute a negative feedback mechanism of Notch signaling. Our study presents a novel mechanism by which a balance between Notch signaling and HRT activity determines the expression of smooth muscle differentiation markers including SMA.
Keywords: vascular smooth muscle cells, smooth muscle α-actin, signaling pathways, endothelial cells
Notch signaling is important in vascular development and the pathogenesis of vascular diseases.1,2 Following Notch activation by Jagged/Delta ligands, Notch intracellular domain (ICD) is proteolytically liberated, enters the nucleus, and participates in a multiprotein complex including CBF-1 (C-promoter– binding factor-1) that transcriptionally activates genes, including the Notch effector genes Hes, HRT1 (Hairy-related transcription factor 1) (also known as Hey1, Herp2, Hesr1), and HRT2 (Hey2, CHF1, Herp1, Hesr2).3–5 In smooth muscle cells (SMCs), Notch/HRT regulates cell proliferation, survival, and migration.6–11
Smooth muscle α-actin (SMA) is a marker of the differentiated SMCs with well-characterized transcriptional regulation.12–14 NotchICD/CBF-1 activity induces SMA expression in multiple cell types.15 This is consistent with studies showing that Jagged1 activation of Notch signaling supports SMC differentiation10 and that Notch signaling is required in vivo for proper SMC differentiation from neural crest progenitors.16 Interestingly, other studies indicate that Notch1 or Notch3 activation or HRT activity suppresses SMA expression and cell differentiation6 and that Notch1ICD or HRT2 expression inhibits myocardin-activated SMC marker gene transcription.17 We focused on SMA regulation in human primary aortic SMCs (HASMCs) and found that Notch1, Notch2, and Notch4 activation increases SMA expression in a CBF-1–dependent manner. We characterized a novel negative-feedback pathway by which expression of HRT1 or HRT2, alone or in combination with Notch activation, suppresses SMA transcript and protein levels. This is the first report showing that SMA regulation by Notch signaling depends on a balance between Notch activity and HRT activity, providing a mechanism for the activation and suppression of SMA expression. We propose a model by which HRT interacts with SMA promoter elements to consequently inhibit NotchICD/CBF-1 activation of SMA transcription.
Materials and Methods
Cell Culture
HASMCs, human aortic endothelial cells (HAECs), and human umbilical vein endothelial cells (HUVECs) were purchased from Cambrex (Walkersville, Md). HASMCs were maintained in SmGM2 medium and used between passages 4 and 7. Endothelial cells were maintained in EGM2 medium and used between passages 3 and 7. For coculture, HAECs or HUVECs were cultured in 6-well plates until confluent and overlaid with HASMCs (control or expressing CBF-1 luciferase or HRTs). Endothelial cells were retrovirally transduced with green fluorescent protein (GFP) or soluble Jagged1.18 γ-Secretase inhibitor IX (Calbiochem) was used at 10 µmol/L. Cell lysates were collected for luciferase assays or immunoblot.
Constructs and Gene Expression
Adenoviral constructs in pAdlox were generated as described.19 The Notch1ICD, Notch2ICD, Notch4ICD, HRT1, and HRT2 constructs contain C-terminal epitope tags.8 pCMX-CBF-1 for wild-type CBF-1 expression and pEF-BOSneo-R218H for dominant-negative (DN)-CBF-1 have been characterized by T. Honjo and colleagues,20 and pEF-BOSneo-R218H was provided by I. Prudovsky (Maine Medical Center Research Institute, Scarborough). The mouse HRT2 mutant constructs, mHRT2 B(−) and HRT2 C(−) were provided by E. Olson (University of Texas Southwestern Medical Center, Dallas).5 Cells were transduced 12 hours after plating for 24 hours with 100 TCID50 (tissue culture 50% infective dose) virus particles per cell, optimized empirically by immunohistochemistry to achieve >90% transduction with minimal cytotoxicity. Endothelial cells were transduced with GFP or soluble Jagged118 using PINCO retroviruses (provided by Cathrin Brisken, Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland).
Immunoblotting and Immunohistochemistry
Whole cell extracts were prepared as described.8 Immunoblot analyses used anti-V5 (Invitrogen), anti-β-actin, anti-SMA, anti-hemagglutinin, and anti-FLAG (Sigma). For immunostaining, cells were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton X-100/PBS. Primary antibodies were diluted 1:400, followed by detection using tetramethylrhodamine B isothiocyanate (TRITC)-conjugated secondary antibody.
RT-PCR and Quantitative RT-PCR
Total RNA was extracted, treated with RNase-free DNAase1 (Promega), and reverse-transcribed using 20 pmol/L oligo(dT) with AMV reverse transcriptase (Promega). Quantitative RT-PCR was performed using the iCycler (Bio-Rad) using SYBR green (Bio-Rad) with 20 ng of cDNA as template, in triplicate. Threshold cycle numbers were calculated at log phase of amplification and normalized to cyclophilin. Primer sequences are provided in Table I of the online data supplement, available at http://circres.ahajournals.org.
Transient Transfection and Luciferase Assay
HASMCs were plated at 20 000 cells per well in a 24-well plate and transduced with adenovirus (100 TCID50 virus particles per cell), 0.25 µg reporter plasmid, 0.75 µL of Gene Juice (Invitrogen), and 25 ng of Renilla luciferase plasmid per well. Two days after transfection, cells were collected for luciferase assay.8 All experiments were repeated at least 3 times, and representative results shown. Human SMA-S promoter fragments were generated by PCR from genomic DNA, ligated into pGL3 basic vector, and sequenced. PCR primers amplify 157 bp (−124/+32) of the SMA-S promoter.21
Coimmunoprecipitation Assay
Cell lysates (300 µg) were incubated with 1.5 µg of antibody, followed by the addition of protein A/G agarose beads. Samples were washed to remove unbound proteins, boiled in sample buffer, centrifuged, and removed from beads. Immunoprecipitated proteins were subjected to SDS-PAGE, and immunoblotting was performed with anti-V5, anti-Flag, or anti–CBF-1 antibodies.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described.8 Cross-linked chromatin was immunoprecipitated with anti-Flag (M2, Sigma), anti-V5 (Invitrogen), normal mouse IgG (Santa Cruz Biotechnology), anti–CBF-1 (H-50, Santa Cruz Biotechnology), or normal rabbit IgG (Santa Cruz Biotechnology). Input DNA and immunoprecipitated chromatin were subjected to PCR using primers encompassing the CBF-1–binding site from the SMA promoter.
Statistical Analysis
Statistical analyses were performed using Student’s t test, with a significant difference determined as P<0.05. Data are presented as means±SD.
Results
Expression of NotchICD and HRTs in HASMCs
Initial studies characterized expression conditions in human primary cells. All adenoviral constructs were titered over an 8-fold concentration to determine optimal protein (immunoblot analysis) with minimal cytotoxicity (Figure 1). We achieved high nuclear expression of each construct (Figure 1A and 1B), and NotchICD induced transactivation of a CBF-1 binding element (not shown). HRTs are transcriptional targets of Notch signaling in SMCs.3,5,8 In HASMCs, HRT1 and HRT2 transcripts were significantly increased with NotchICD transduction compared with control (Figure 1C). In contrast, CBF-1 mRNA levels were not different in any of the transfectants (Figure 2A).
Figure 1. Expression and localization of NotchICD and HRT in human primary SMCs.
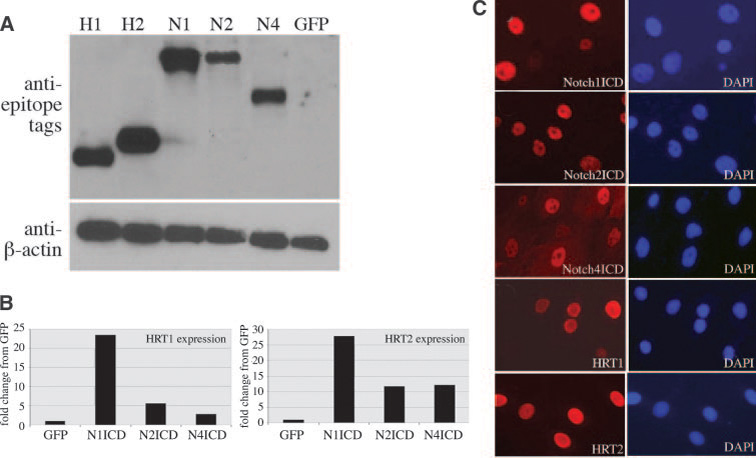
Cells were adenovirally transduced with Notch1ICD (N1), Notch2ICD (N2), Notch4ICD (N4), HRT1 (H1), or HRT2 (H2). A, Cell lysates were collected after 3 days and immunoblotted using antibodies against the epitope tags: flag for HRT1 and HRT2, V5 for N1 and N2, and HA for N4. The control was GFP, and total protein levels were evaluated with anti–β-actin. B, Immunofluorescence was used to detect the epitope tags at 72 hours after adenoviral transduction (left). DAPI staining (right) allowed for quantification of transduction efficiency. C, Quantitative PCR was performed using HRT1 and HRT2 primers to detect transcript levels 72 hours after NotchICD transduction. Graphed are fold changes compared with GFP-transduced cultures.
Figure 2. Notch and HRT differentially regulate SMC markers such as SMA.
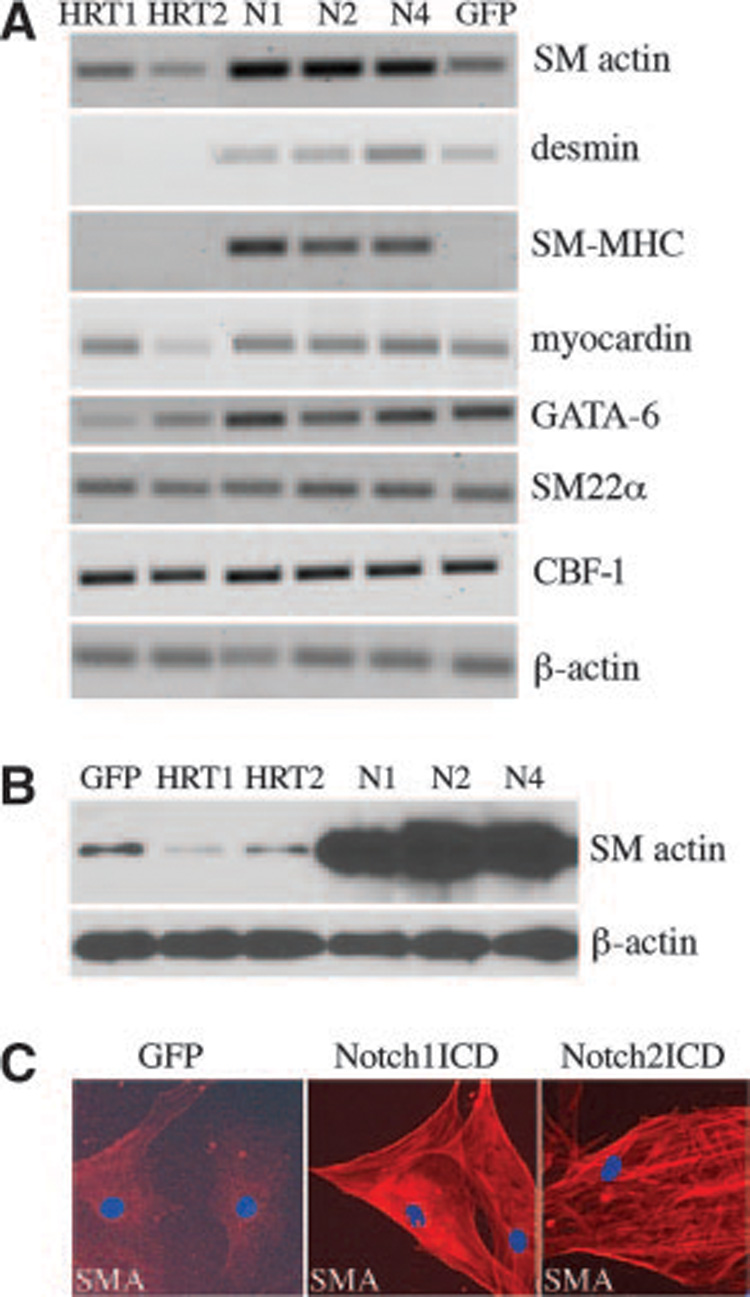
HASMCs were transduced with GFP, NotchICD, or HRTs. A, Total RNA was collected after 72 hours and reverse-transcribed. PCR amplification was performed for indicated transcripts. B, Cell lysates were collected and immunoblotted for SMA and β-actin. C, Immunofluorescence staining was performed to detect SMA localization within GFP or Notch-activated HASMCs (red fluorescence). Cells were also stained with DAPI to detect nuclei (blue fluorescence).
Notch and HRT Differentially Regulate SMC Marker Gene Expression
Activation of Notch through Jagged1 induces SMA transcription in a CBF-1–dependent manner.15 We found that activation of Notch1, Notch2, or Notch4 also increased SMA transcripts and also induced SM myosin heavy chain (SM-MHC), while having minimal effects on transcript levels of desmin, myocardin, GATA-6, and SM22α, other markers of SMC differentiation (Figure 2A). By contrast, HRT1 or HRT2 did not mimic the effects of NotchICD but suppressed transcripts for SMA, desmin, SM-MHC, and GATA-6 (Figure 2A). By immunoblot (Figure 2B), we confirmed that SMA was highly elevated following Notch activation and suppressed by HRT1 or HRT2. The Notch-induced increase of SMA in HASMCs was also evident by immunofluorescence staining (Figure 2C).
Because Notch regulates SMA transcriptionally, we analyzed the SMA gene. There are 2 potential transcripts for SMA, although they both encode for the same protein sequence with the translational start site in exon 2 (Figure 3A). The difference in these 2 transcripts is the size of intron 1, which is 3777 and 41 985 bp for the short (SMA-S) and long (SMA-L) forms, respectively. The SMA-S promoter contains a CBF-1 consensus binding site TGGGAA at position −64 relative to the transcriptional start site,15 and a less conserved CBF-1 motif at −1309 (Figure 4A and supplemental Table II). The SMA-L promoter contains 2 consensus CBF-1 sites and 3 consensus HRT-binding sites. We designed primers, SMA-S, to detect the short intron 1 transcript, another pair of primers, SMA-L, to detect the long intron transcript, and a third pair of primers, SMA-W, to detect both. Although both transcripts were detectable in primary HASMCs, the SMA-S form was expressed at a 10-fold higher level (Figure 3B and Figure 4). Activation of Notch1, Notch2, or Notch4 significantly increased SMA-S transcript, (Figure 4B), whereas HRT1 or HRT2 decreased transcripts. In contrast, activation of Notch did not affect SMA-L, although HRT1 or HRT2 strongly suppressed expression from 15– to 20-fold compared with controls (Figure 4C). Notch-induced elevation of SMA protein is likely attributable to increased SMA-S transcript, thus we focused on SMA-S for subsequent experiments.
Figure 3. Two SMA transcripts are expressed in HASMCs.
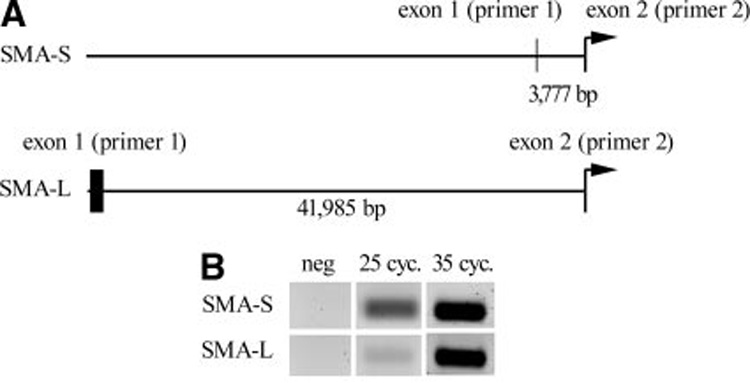
A, Schematic representation of the SMA gene. The SMA-S form is transcribed from an exon 1 that is ≈3.8 kb upstream of the translational start site in exon 2. Exon 1 of the SMA-L form is located ≈42 kb upstream of exon 2. Primers to amplify the alternate forms were generated within exons 1 and 2. B, HASMCs were used for RT-PCR for amplification of the SMA-S or SMA-L forms. The SMA-S primers amplify a ≈100-bp product, and the SMA-L primers amplify a ≈200-bp product. Although both transcripts are expressed in HASMCs, SMA-S was more abundant. Samples were PCR-amplified for 25 or 35 cycles.
Figure 4. The SMA transcripts are regulated differently by Notch.
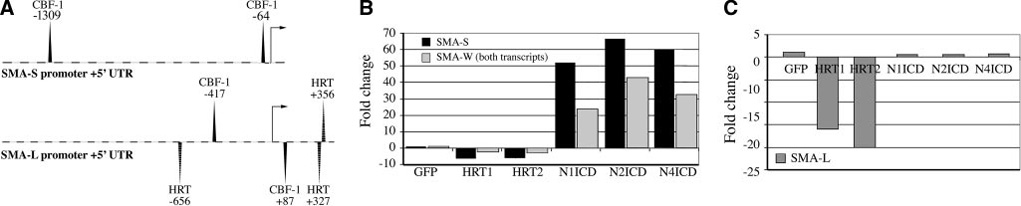
A, Diagrammed are the short (SMA-S) and the long (SMA-L) promoter sequences with relation to the transcriptional start site (arrow). The 2000-bp stretch of the upstream promoter sequence was analyzed for transcription factor motifs using Genomatix weight matrices for transcription factor binding sites (MatInspector40), and significant consensus sites for CBF-1 binding and HRT binding are indicated, relative to the transcriptional start site. Quantitative PCR was performed using primers for SMA-S and SMA-W (recognizing all SMA mRNA forms) (B) or SMA-L primers (C) under different HASMC transduction conditions. Data were compared with expression with control GFP transduction. SMA-S is expressed at an ≈10-fold higher quantity than SMA-L, although both are detectable in HASMCs. Notch activation selectively increased SMA-S transcript, and HRTs suppress levels of both SMA-S and SMA-L. UTR indicates untranslated region.
Notch Induction of SMA Is Dependent on CBF-1 and Antagonized by HRTs
Because Notch has CBF-1–independent functions, we tested whether CBF-1 was required for SMA regulation. We used the DN-CBF-1 (R218H)20 and found a dose-dependent inhibition of Notch-induced SMA expression (Figure 5A) showing a requirement for CBF-1 activity. The same was true whether Notch1, Notch2, or Notch4 was activated.
Figure 5. HRTs antagonize Notch induction of SMA expression.
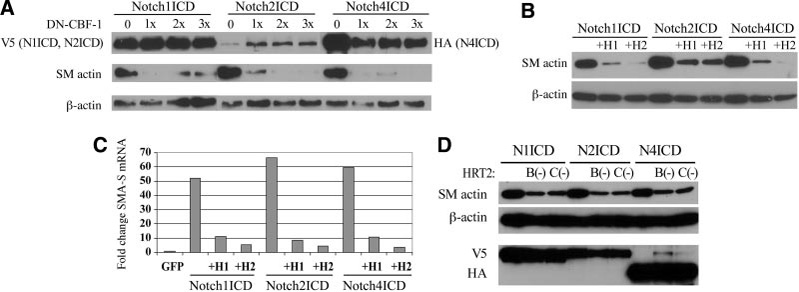
HASMCs were transfected as indicated, and total cell lysates collected 3 days after transduction. A, Cells were cotransfected with NotchICD and increasing amounts (3-fold range) of DN-CBF-1. Cell lysates were immunoblotted using antibodies against NotchICD, SMA, and β-actin. B, Cells were cotransfected with NotchICD and either HRT1 (H1) or HRT2 (H2), and cell lysates were immunoblotted for SMA and β-actin. C, Quantitative PCR was performed using SMA-S primers. Data are expressed as fold changes in relation to GFP-transduced cells. All NotchICD significantly increased SMA-S transcripts, which was antagonized by either HRT1 or HRT2 coexpression. D, This experiment was repeated using the HRT2 mutants. Both HRT2 B(−) and HRT2 C(−) suppressed Notch-induced SMA protein levels.
Hes1 and HRT2 have negative autoregulation activity,5,22 and we asked whether HRT had any effect on Notch-mediated SMA expression. We used cotransfection of Notch-ICD with HRT1 or HRT2 in HASMCs. As described, all of the NotchICDs strongly induced SMA expression (Figure 5B). When HRT1 or HRT2 were simultaneously expressed, however, induction of SMA by NotchICD was dramatically decreased. Importantly, neither HRT1 nor HRT2 affected CBF-1 expression (Figure 2A). These findings suggest that HRT1 and HRT2 inhibit Notch-induced SMA expression by functionally antagonizing NotchICD/CBF-1 activity, not by modulation of CBF1 expression. Quantitative PCR showed that HRTs repressed NotchICD-mediated SMA expression at the transcript level (Figure 5C). We also took advantage of HRT mutant constructs to query selected HRT structural domains. When cells were cotransfected with NotchICD and the HRT2 B(−) or HRT2 C(−) mutant, both HRT2 mutants decreased SMA protein levels compared with NotchICD control (Figure 5D). There are at least 2 possible mechanisms by which HRTs antagonize Notch regulation of SMA expression. First, HRT may interfere with complex formation between NotchICD and CBF-1, or, secondly, HRT may inhibit NotchICD/CBF-1 complex binding to its recognition site in the SMA promoter.
HRT Suppression of SMA Is Independent of Histone Deacetylase Activity
HRT interacts with the corepressors mSin3 and SIRT1, which have histone deacetylase (HDAC) activity.23,24 To determine whether HRT activity in the context of SMA expression requires recruitment of HDACs, we treated HASMCs with trichostatin A, a potent HDAC inhibitor. Even high concentrations of 500 ng/mL trichostatin A were ineffective in altering HRT activity in suppressing SMA expression (Figure 6A). Likewise, the expression of SMA following Notch activation with HRT coexpression was not significantly altered by the presence of trichostatin A (Figure 6B). These data suggest that HRT does not need to recruit HDACs to inhibit SMA expression and antagonize Notch.
Figure 6. HRT inhibition of SMA expression is not dependent on histone deacetylase activity.
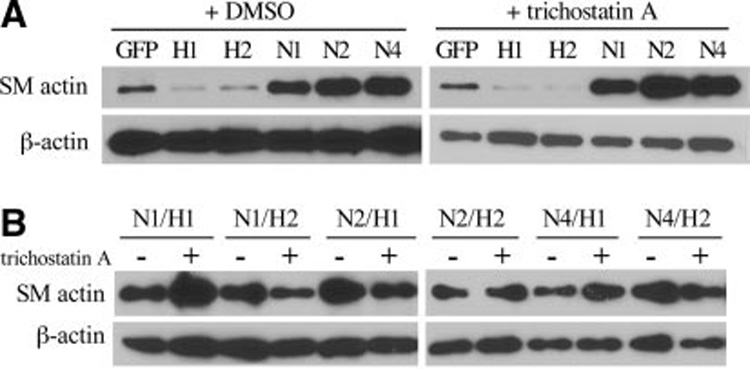
HASMCs were transduced as indicated, and after 48 hours, either DMSO (control,−) or trichostatin A were added for 24 hours before total cell lysate collection (3 days after transduction). A, SMA levels, in comparison with GFP controls, was decreased in HRT1 (H1)- or HRT2 (H2)- expressing cells and increased in NotchICD-expressing cells. B, Cells expressing ICD forms of Notch1 (N1), Notch2 (N2), or Notch4 (N4) in combination with either HRT1 (H1) or HRT2 (H2) were analyzed for SMA levels. There were no major differences in the presence or absence of trichostatin A.
HRT Does Not Disrupt NotchICD/CBF-1 Complex Formation
HRT and HES can complex with CBF-1 in Cos1 cells25; thus, we tested whether HRTs bind CBF-1 in HASMCs. This is a potential mechanism by which HRT can disrupt NotchICD/CBF-1 activity at the SMA promoter. HASMCs were transduced with constructs separately or in combination, and immunoprecipitation of total cell lysates were performed, followed by immunoblot to detect coimmunoprecipitating proteins (Figure 7A). When NotchICD was expressed alone (lanes 1 to 2) and immunoprecipitated with anti-V5, endogenous CBF-1 was undetectable by immunoblot. However, coexpression of NotchICD and CBF-1 led to coimmunoprecipitation of the complex (lanes 3 to 4). When NotchICD and CBF-1 were coexpressed with either HRT 1 (lanes 5 and 7) or HRT2 (lanes 6 and 8), there was no difference in the ability to immunoprecipitate NotchICD and detect CBF-1. Under no conditions were HRT1 or HRT2 (Flag) detected following immunoprecipitation of NotchICD. Likewise, we found no evidence of HRT binding to CBF-1 (not shown). Our findings show that HRTs do not disrupt the NotchICD/CBF-1 complex and do not bind to the complex. Similar results were obtained using Notch4ICD (not shown).
Figure 7. HRT antagonizes NotchICD/CBF-1 at the level of SMA promoter binding and activation.
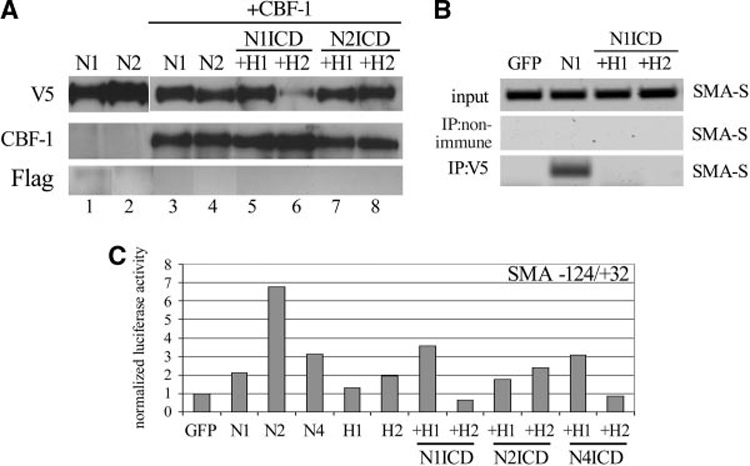
HASMCs were transduced as indicated. A, Cell lysates were immunoprecipitated using anti-V5 (NotchICD) and immunoblotted with antibodies against V5, CBF-1, or Flag (HRTs). A NotchICD/CBF-1 complex was clearly detectable (lanes 3 and 4) and was not inhibited by the addition of HRT1 (lanes 5 and 7) or HRT2 (lanes 6 and 8). Endogenous levels of CBF-1 were not sufficient to be detected by immunoblot after immunoprecipitation (lanes 1 to 2). HRTs (Flag epitope) were never detectable in the NotchICD-immunoprecipitated lysates. B, Protein/DNA complexes in transduced cells were cross-linked, and the DNA was sheared. Lysates were used for PCR amplification of SMA-S using specific primers (input). Lysates were immunoprecipitated with anti-V5 to pull down protein/DNA complexes containing NotchICD. HRT1 (H1) or HRT2 (H2) expression inhibited immunoprecipitation of a NotchICD/DNA complex containing SMA-S amplifiable sequence. C, Luciferase promoter transactivation assays were performed with a construct containing the −124/+32 SMA sequence.
HRTs Interfere With NotchICD/CBF-1 Binding to the SMA Promoter
To test whether HRT1 and HRT2 dissociate CBF-1 from its binding site in SMA-S (−64 in relation to the transcriptional start), we performed chromatin immunoprecipitation (Figure 7B). HASMCs were transduced with GFP, Notch1ICD, or Notch1ICD in combination with either HRT1 or HRT2. Protein/DNA complexes were cross-linked, and lysates were tested for the amplification of SMA-S (input). Immunoprecipitation was performed using either normal IgG or anti-V5 to detect NotchICD. Following specific immunoprecipitation, we amplified SMA-S in Notch1ICD-expressing cells but not in the GFP group. However, coexpression of HRT1 or HRT2 eliminated immunoprecipitation of SMA-S with NotchICD, suggesting that HRTs were inhibiting NotchICD/CBF-1 complex binding to the SMA promoter. The functional consequence of this inhibition was tested using an SMA reporter construct encompassing the sequence −124/+32. Activation of Notch, particularly Notch2, was effective in increasing reporter transactivation compared with GFP-expressing cells (Figure 7C). The activity of HRTs varied in this assay but, in general, was effective in inhibiting NotchICD-induced SMA promoter transactivation.
Activation of Endogenous Notch Signaling in SMCs via Coculture With Endothelial Cells Induces HRT-Sensitive Expression of SMA
Notch activation facilitated molecular studies of the SMA promoter and analysis of protein complex formation. Our next question was whether placing HASMCs under conditions to mimic physiological interactions would regulate gene expression similarly. We previously demonstrated that endothelial denudation in vivo induced reciprocal expression of Jagged1 and Notch receptors in endothelium and SMC, respectively.26 The regenerating endothelial wound edge expressed localized, high levels of Notch ligands, and adjacent SMCs had elevated Notch expression, supporting the hypothesis that heterotypic Notch stimulation from endothelial cells affects SMC behavior. We established a coculture system of primary human endothelial cells in contact with HASMCs. Using RT-PCR and immunoblot, we verified that HAECs and HUVECs express both Jagged1 and Delta-like1, and Notch1, Notch2, and Notch3 are expressed endogenously in HASMCs (not shown). We established monolayers of endothelium expressing GFP (Figure 8A) and then overlaid the monolayer with HASMCs transfected with the CBF-1 luciferase reporter construct to test for Notch activation in heterotypic cell culture. HASMCs alone have low levels of endogenous CBF-1 activation that increased 7-fold when HASMCs were in contact with endothelial cells (Figure 8B). This induction was significantly inhibited when endothelial cells were expressing dominant negative sJagged1.18 Using this system, we tested HAECs and HUVECs in coculture with HASMCs to determine effects on SMA. Although SMA is expressed in HASMCs, protein levels were enhanced in coculture with HAECs (Figure 8C) or HUVECs (Figure 8D). Similar to its effects on CBF-1 reporter activation, sJagged1 expression in HAECs blocked the increase in SMA induced by coculture (Figure 8C), showing specificity of this effect to Notch signaling. Similarly, γ-secretase inhibitor decreased SMA levels in cocultures of endothelial/SMCs (Figure 8E). These results show that coculture of endothelial cells with HASMCs induces Notch signaling in SMCs, leading to increased SMA production.
Figure 8. Activation of endogenous Notch signaling by endothelial cell and SMC coculture activates SMA, which is sensitive to HRT.
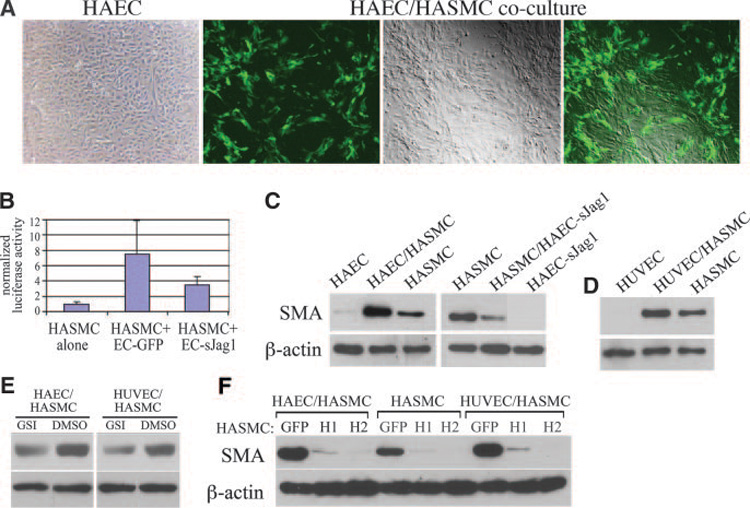
A, We established a coculture system of fluorescently labeled (GFP) monolayer endothelial cells overlaid with HASMCs. Shown are HAECs alone (left) and HAECs/HASMC coculture (next 3 images) with HAECs retrovirally transduced with GFP. The images show cells under fluorescence, phase, or combined microscopy. B, Endothelial cells were transduced with GFP or sJagged1 and cocultured with HASMCs expressing a CBF-1 luciferase reporter construct and assayed after 48 hours. C through F, Endothelial cell/SMC cocultures were performed with control or transduced cells, and protein lysates were collected after 3 days for immunoblot analysis for SMA and β-actin. C, HAEC/HASMC coculture induced SMA expression, which was inhibited in coculture with sJagged1-expressing endothelial cells. D, HUVEC/HASMC coculture also increased SMA levels. E, Coculture was performed in the presence of γ-secretase inhibitor or DMSO (vehicle). F, HASMCs were transduced with GFP, HRT1 (H1), or HRT2 (H2) and plated alone or in coculture with HAECs or HUVECs as indicated.
To determine whether HRT can impact SMA expression in our coculture system, HRT1 or HRT2 were expressed in HASMCs before coculture with endothelial cells (Figure 8F). In the absence or presence of endothelial cells, HRT expression was a strong suppressor of SMA accumulation. Taken together, our data show that HRT is a major inhibitor of SMA expression in SMCs, whether it is basal expression induced by constitutive Notch signaling or induced via endogenous Notch signaling following coculture with endothelial cells.
Discussion
Phenotypic modulation of SMCs occurs during embryonic development, vascular injury, and cardiovascular disease. Transcriptional pathways regulating SMC genes include serum response factor and myocardin,27 and accumulating evidence suggests that Notch is an additional pathway critical for SMC differentiation.6,10,15,28 Functional roles of Notch/HRT signaling in the cardiovasculature in vivo were uncovered by extensive mouse genetic studies.29 Notch signaling components are temporally and spatially expressed following injury in vivo,8,30 and HRT2 is present in human atherosclerotic lesions.28 Mutations of the Notch pathway in Alagille syndrome and CADASIL emphasize the clinical importance of these pathways in human diseases.31
We studied human primary SMCs and their coculture with endothelial cells. One SMC marker, SMA, is exquisitely sensitive to Notch and HRT signaling. Although distinct Notch receptors may provide variable signals,8,32–34 all Notch pathways we tested (N1ICD, N2ICD, N4ICD) had conserved activity in the induction of SMA. HRT1 and HRT2 have similar functions in repressing basal SMA-S and SMA-L transcript levels and antagonizing Notch induction of SMA-S. The positive (Notch) and negative (HRT) balance in the regulation of SMA is a novel finding that provides a mechanism by which SMC differentiation genes can be sensitively fine tuned. Indeed, vascular injury corresponds to induction of Notch ligands and receptors,8,30 and we previously noted a reciprocal relationship between endothelial cell expression of ligand and SMC expression of Notch receptors in vivo26 following injury. We propose that HRT is a candidate suppressor of mature SMC markers, including SMA, which are downregulated following vascular injury in vivo.35 The physiological relevance of our work is highlighted by the conserved regulation of SMA in endothelial/SMC coculture, a condition that activates Notch signaling in SMCs. Our work thus adds to our understanding of the positive and negative regulators of Notch signaling that are important in modulating Notch responsiveness in vascular cells.36
There are still discrepancies about whether Notch induces or inhibits differentiation, which may be partly attributable to different experimental systems and species, as well as phenotypic readouts. For example, there is an established crossregulation of myocardin target genes by Notch/HRT2,17 but other SMC gene regulatory pathways may be differentially impacted by Notch signaling. In vitro studies showed that Notch signaling induces SMC differentiation and identified SMA and SM-MHC as NotchICD/CBF-1 targets.10,15 However, Proweller et al17 and Morrow et al37 reported that HRT2-mediated Notch signaling inhibits SMC differentiation, and Doi et al28 reported that HRT repressed myocardin-induced SMC differentiation. These apparent discrepancies should be considered in light of our findings that although NotchICD directly activates markers such as SMA and SM-MHC, concomitant expression of HRT1 or HRT2 is sufficient to turn off these signals. We envision a regulatory loop that in vivo would allow for temporally restricted Notch signaling and induction of target gene expression until HRT production rises above a threshold needed to antagonize the signal.
Members of the Hes and HRT families of bHLH (basic helix–loop–helix) proteins have been characterized as Notch effectors that mediate Notch activity.3,5 In some cases, expression of Hes or HRT alone recapitulate the phenotype of activated Notch signaling,8,38 and some Notch-induced vascular phenotypes are inhibited by blocking Hes/HRT function.6 However, Hes/HRT can participate in specific negative-feedback loops that may limit the extent of Notch/ Hes/HRT signaling. Hes-1 inhibits its own transcription by binding to N boxes (CACNAG) in its promoter,22 providing negative feedback. Although HRT2 also shows characteristics of negative autoregulation of its expression,5 this appears to be a novel mechanism distinct from HRT2 binding to its own promoter. We also observed that expression of HRT2 led to accumulation of HRT2 protein but not HRT2 mRNA, consistent with a negative autoregulatory activity. Another related bHLH transcription factor, Stra13/DEC1/SHARP2, also negatively autoregulates its expression in an HDAC-dependent manner.39 Our study extends this concept to show that other genes such as SMA are also susceptible to negative-feedback regulation by HRT through a mechanism involving disruption of Notch/CBF-1 interaction with the SMA promoter in an HDAC-independent manner. This disruption is not attributable to disassembly of the NotchICD/CBF-1 complex, and extensive coimmunoprecipitation experiments did not show evidence for ternary complex formation with HRTs. The matrix group from MatInspector40 did not identify strong consensus HRT binding sites in the SMA-S promoter, but there are several E box motifs at −258, −330, −1222, and −1742 and HRT consensus motifs in SMA-L. Although the SMA-S luciferase promoter (−124/+32; Figure 7C) showed responsiveness to both Notch activation and accompanying HRT expression with induction and suppression, respectively, the extent of the response was not as dramatic as expected based on the quantitative changes in SMA-S transcript levels. This leaves the likely possibility that important Notch and HRT regulatory sites are upstream of −124 in the SMA-S promoter, and these upstream sequences are currently under investigation. We also need to consider the possibility that HRT is interacting with DNA via another DNA-binding protein or through previously uncharacterized DNA motifs. Experiments to test the direct binding of HRTs to the SMA promoter have been as yet inconclusive, because of the lack of information regarding HRT binding sites in the SMA gene.
In summary, our studies provide the novel findings that HRT1 and HRT2 are repressors of SMA expression and, further, are able to antagonize Notch-induced SMA expression. HRT does not inhibit NotchICD/CBF-1 complex formation, although it decreases NotchICD/CBF-1 binding and activation of the SMA promoter. SMC responsiveness to Notch signaling, therefore, may be limited temporally and spatially by the production of HRT factors, which feed back to inhibit specific Notch target gene activation.
Supplementary Material
Acknowledgments
We are grateful to our colleagues who generously shared reagents. We appreciate the work of Nancy Chandler-Conrey and Jeong Yoon, PhD, of our Viral Vector core for production of adenoviral constructs. We also thank Drs Doug Spicer, Jeong Yoon, and Howard Crawford for critical discussions and expertise.
Sources of Funding
This work was supported by NIH grants R01 HL070865 (to L.L.) and P20RR1555 (to R.E. Friesel and L.L.).
Footnotes
Disclosures
None.
References
- 1.Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann JJ, Iruela-Arispe ML. Notch signaling in blood vessels: who is talking to whom about what? Circ Res. 2007;100:1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 3.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci U S A. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrow D, Scheller A, Birney YA, Sweeney C, Guha S, Cummins PM, Murphy R, Walls D, Redmond EM, Cahill PA. Notch-mediated CBF-1/ RBP-J{kappa}-dependent regulation of human vascular smooth muscle cell phenotype in vitro. Am J Physiol Cell Physiol. 2005;289:C1188–C1196. doi: 10.1152/ajpcell.00198.2005. [DOI] [PubMed] [Google Scholar]
- 7.Sweeney C, Morrow D, Birney YA, Coyle S, Hennessy C, Scheller A, Cummins PM, Walls D, Redmond EM, Cahill PA. Notch 1 and 3 receptor signaling modulates vascular smooth muscle cell growth, apoptosis, and migration via a CBF-1/RBP-Jk dependent pathway. FASEB J. 2004;18:1421–1423. doi: 10.1096/fj.04-1700fje. [DOI] [PubMed] [Google Scholar]
- 8.Havrda MC, Johnson MJ, O’Neill CF, Liaw L. A novel mechanism of transcriptional repression of p27kip1 through Notch/HRT2 signaling in vascular smooth muscle cells. Thromb Haemost. 2006;96:361–370. doi: 10.1160/TH06-04-0224. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Prince CZ, Hu X, Pollman MJ. HRT1 modulates vascular smooth muscle cell proliferation and apoptosis. Biochem Biophys Res Commun. 2003;308:596–601. doi: 10.1016/s0006-291x(03)01453-0. [DOI] [PubMed] [Google Scholar]
- 10.Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J Biol Chem. 2006;281:28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- 11.Sakata Y, Xiang F, Chen Z, Kiriyama Y, Kamei CN, Simon DI, Chin MT. Transcription factor CHF1/Hey2 regulates neointimal formation in vivo and vascular smooth muscle proliferation and migration in vitro. Arterioscler Thromb Vasc Biol. 2004;24:2069–2074. doi: 10.1161/01.ATV.0000143936.77094.a4. [DOI] [PubMed] [Google Scholar]
- 12.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 13.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, Owens GK. Molecular determinants of vascular smooth muscle cell diversity. Circ Res. 2005;96:280–291. doi: 10.1161/01.RES.0000155951.62152.2e. [DOI] [PubMed] [Google Scholar]
- 15.Noseda M, Fu Y, Niessen K, Wong F, Chang L, McLean G, Karsan A. Smooth Muscle alpha-actin is a direct target of Notch/CSL. Circ Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 16.High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proweller A, Pear WS, Parmacek MS. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J Biol Chem. 2005;280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- 18.Small D, Kovalenko D, Kacer D, Liaw L, Landriscina M, Di Serio C, Prudovsky I, Maciag T. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J Biol Chem. 2001;276:32022–32030. doi: 10.1074/jbc.M100933200. [DOI] [PubMed] [Google Scholar]
- 19.Aoki K, Barker C, Danthinne X, Imperiale MJ, Nabel GJ. Efficient generation of recombinant adenoviral vectors by Cre-lox recombination in vitro. Mol Med. 1999;5:224–231. [PMC free article] [PubMed] [Google Scholar]
- 20.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 21.Reddy S, Ozgur K, Lu M, Chang W, Mohan SR, Kumar CC, Ruley HE. Structure of the human smooth muscle alpha-actin gene. Analysis of a cDNA and 5′ upstream region. J Biol Chem. 1990;265:1683–1687. [PubMed] [Google Scholar]
- 22.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 23.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 25.King IN, Kathiriya IS, Murakami M, Nakagawa M, Gardner KA, Srivastava D, Nakagawa O. Hrt and Hes negatively regulate Notch signaling through interactions with RBP-Jkappa. Biochem Biophys Res Commun. 2006;345:446–452. doi: 10.1016/j.bbrc.2006.04.097. [DOI] [PubMed] [Google Scholar]
- 26.Lindner V, Booth C, Prudovsky I, Small D, Maciag T, Liaw L. Members of the Jagged/Notch gene families are expressed in injured arteries and regulate cell phenotype via alterations in cell matrix and cell-cell interaction. Am J Pathol. 2001;159:875–883. doi: 10.1016/S0002-9440(10)61763-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 28.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 29.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Prince CZ, Mou Y, Pollman MJ. Notch3 signaling in vascular smooth muscle cells induces c-FLIP expression via ERK/MAPK activation. Resistance to Fas ligand-induced apoptosis. J Biol Chem. 2002;277:21723–21729. doi: 10.1074/jbc.M202224200. [DOI] [PubMed] [Google Scholar]
- 31.Joutel A, Tournier-Lasserve E. Notch signalling pathway and human diseases. Semin Cell Dev Biol. 1998;9:619–625. doi: 10.1006/scdb.1998.0261. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill CF, Urs S, Cinelli C, Lincoln A, Nadeau RJ, Leon R, Toher J, Mouta-Bellum C, Friesel RE, Liaw L. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am J Pathol. 2007;171:1023–1036. doi: 10.2353/ajpath.2007.061029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu K, Chiba S, Saito T, Kumano K, Hamada Y, Hirai H. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem Biophys Res Commun. 2002;291:775–779. doi: 10.1006/bbrc.2002.6528. [DOI] [PubMed] [Google Scholar]
- 34.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 35.Hendrix JA, Wamhoff BR, McDonald OG, Sinha S, Yoshida T, Owens GK. 5′ CArG degeneracy in smooth muscle alpha-actin is required for injury-induced gene suppression in vivo. J Clin Invest. 2005;115:418–427. doi: 10.1172/JCI22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols JT, Miyamoto A, Weinmaster G. Notch signaling - constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- 37.Morrow D, Sweeney C, Birney YA, Cummins PM, Walls D, Redmond EM, Cahill PA. Cyclic strain inhibits Notch receptor signaling in vascular smooth muscle cells in vitro. Circ Res. 2005;96:567–575. doi: 10.1161/01.RES.0000159182.98874.43. [DOI] [PubMed] [Google Scholar]
- 38.Sun J, Kamei CN, Layne MD, Jain MK, Liao JK, Lee ME, Chin MT. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J Biol Chem. 2001;276:18591–18596. doi: 10.1074/jbc.M101163200. [DOI] [PubMed] [Google Scholar]
- 39.Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


