Abstract
A method was developed for characterizing immobilization sites on a protein based on stable isotope labeling and MALDI-TOF mass spectrometry. The model for this work was human serum albumin (HSA) immobilized onto silica by the Schiff base method. The immobilized HSA was digested by various proteolytic enzymes in the presence of normal water, while soluble HSA was digested in 18O-enriched water for use as an internal standard. These two digests were mixed and analyzed, with the 18O/16O ratio for each detected peptide then being measured. Several peptides in the tryptic, Lys-C, and Glu-C digests gave significantly higher 18O/16O ratios than other peptides in the same digests, implying their involvement in immobilization. Analysis of these results led to identification of the N-terminus and several lysines as likely immobilization sites for HSA (e.g., K4, K41, K190, K225, K313 and K317). It was also possible from these results to quantitatively rank these sites in terms of the relative degree to which each might take part in immobilization. This method is not limited to HSA and silica but can be used with other proteins and supports.
INTRODUCTION
The covalent immobilization of proteins to supports and surfaces has been used for many years in applications such as immunoassays, affinity chromatography, enzyme reactors and protein microarrays.1-9 It is generally desirable in these applications to have an immobilized protein that closely retains the structure and activity of the same protein in its native form. However, there is often little information available on the structure and orientation of proteins after they have been immobilized. This is particularly true when using an amine-based coupling method, in which a protein usually has many possible coupling sites. There have been some previous attempts to examine the immobilization sites of proteins. This has included the use of peptic digests to examine the immobilized regions of antibodies10 and the conversion of lysine to homoarginine to examine protein immobilization in the cyanogen bromide method.11 However, these previous methods have provided only qualitative information on immobilization and have been limited to specific types of proteins or coupling methods.
In this study a more general, quantitative approach for examining protein immobilization is described that is based on recent advances in mass spectrometry and isotopic labeling. This approach makes use of the 18O/16O labeling method;12-14 this is a global labeling method used in quantitative proteomics which involves the digestion of a protein by a proteolytic enzyme in the presence of normal or 18O-enriched water, as shown in Figure 1(a). In the case of enzymes like trypsin, Lys-C or Glu-C, this labeling procedure results in one or more isotopic tags being placed at the C-terminus of a peptide that is formed during the digestion process.13,15,16 A major advantage of this approach is it can, in theory, allow all regions of a protein to be examined, with the exception of the region immediately next to the protein's C-terminus. Other attractive features include the fact that this method combines protein digestion and labeling into a single step and uses a labeling process that is resistant to further reactions that could cause loss of the isotopic label. In addition, 18O- and 16O-labeled peptides tend to have similar retention properties in solid-phase extraction and liquid chromatography, which is useful in their isolation after digestion and labeling.13,14,15
Figure 1.
(a) Reactions involved in the 18O/16O labeling method for proteins and (b) the use of this labeling method in quantitative studies of immobilization sites on a protein.
Figure 1(b) shows the general scheme employed in this study when using 18O/16O labeling for the quantitative analysis of protein immobilization sites. In this approach, the immobilized protein is digested with a proteolytic enzyme in the presence of normal water while a soluble form of the same protein is digested in 18O-enriched water. After these digestion reactions have been quenched, the soluble fractions of the digests are combined, fractionated, and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). This gives two sets of peaks for each detected peptide: a set of predominantly 16O-labeled peptides that were generated from the immobilized protein and a set of 18O-labeled peptides from the same protein in its soluble form (the latter of which are used as internal standards). A comparison of the relative abundances of the 16O- and 18O-labeled forms of each peptide is then used to identify which regions of the protein are involved in its immobilization. This is possible since peptides in the digest that are coupled to a support (i.e., that are generated from sections at or near an immobilization site) will not enter the soluble fraction and will not be measured in the final combined digest. This, in turn, will alter the relative intensities measured for the 18O- versus 16O-labeled peaks for this peptide when compared to peptides from other regions of the protein that were not involved in immobilization.
This approach will be tested by using it to study the immobilization of human serum albumin (HSA) to silica. HSA is frequently coupled to silica for use in chromatographic columns during drug-protein binding studies 17,18 or chiral separations.19,20 This immobilization is usually accomplished through an amine-based coupling technique such as the Schiff base method.21 Useful features of HSA for this particular study include the fact that it has a known primary sequence and crystal structure. In addition, it is composed of a single peptide chain with a large number of amine groups (i.e., 59 lysines and the N-terminus) that could potentially take part in an amine-based coupling method. Although it is known that not all regions of this protein are equally susceptible to immobilization effects in amine-based coupling methods,22-25 no specific information is available on the particular regions of HSA that take part in its attachment to supports. This study will use 18O/16O labeling and MALDI-TOF MS to provide more details on this immobilization process. The extension of this approach to other proteins and supports will also be considered.
THEORY
When a protein like HSA is treated by trypsin or another proteolytic enzyme, it is possible that up to two 18O atoms can be incorporated into each peptide that is formed in the digest. As shown in Figure 1(a), one of these 18O labels can be added during cleavage of the peptide's amide bond (i.e., 16O-to-18O1 exchange). Another 18O label can be incorporated through later exchange of the terminal carboxyl oxygens (i.e., 18O1-to-18O2 exchange).26 However, incomplete isotope incorporation can occur during either of these processes. For example, it has been noted that high mass peptides tend to show a lower efficiency for 18O labeling than lower mass peptides due to the slower 18O1-to-18O2 exchange in the high mass peptides.14,15 Although methods have been developed in the past to correct for the possibility of incomplete 18O1-to-18O2 exchange during the determination of 18O/16O peptide ratios,12,13,26 incomplete 16O-to-18O1 exchange has not yet been considered in such work. This second type of incomplete incorporation might occur due to the presence of some 16O-labeled water in a 18O-enriched preparation of water or it might occur through the reverse reaction of 16O-to-18O1 exchange.
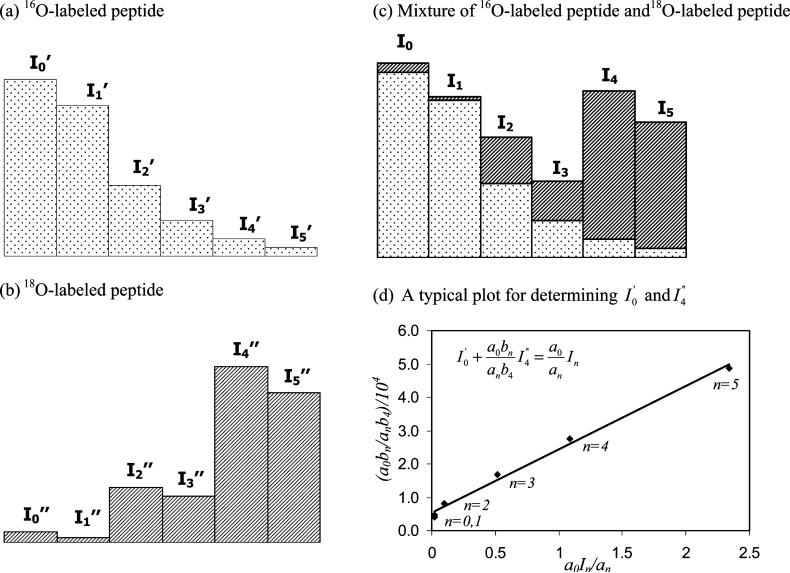
In this paper the following method was used to calculate 18O/16O peptide ratios with a correction being made for incomplete isotope incorporation due to either 16O-to-18O1 or 18O1-to-18O2 exchange effects. The first step in this approach involved determining the relative intensities of the first six isotope peaks for separate preparations of the same peptide form in digested that occurred in normal water (predominantly 16O-water) or 18O-enriched water. The relative intensities measured in these initial studies will be referred to here by using the terms a0, a1, a2, a3, a4 and a5 for the monoisotopic peak (M+1) through the M+6 peptide peaks in the 16O-labeled sample and by using b0, b1, b2, b3, b4 and b5 to refer to the same peaks in the 18O-labeled sample. When these two peptide samples are combined, the intensity of each isotope peak in the mixture (In) should be equal to the sum of intensities from the isotope peptide peaks produced in the 16O-water digest () and the 18O-water digest (), as illustrated in Figure 2. This relationship can be expressed for any peak in the isotopic series by using eq 1.
| (1) |
Figure 2.
Nomenclature used to describe the isotopic distribution for a peptide digested in (a) 16O-enriched water, (b) 18O-enriched water or a (c) a mixture of these two digests, and (d) a typical plot based on eq 3 for analyzing such data. The plot in (d) was generated using data for a peptide peak with a MH+ mass of 2650.26 in the tryptic digest of HSA. The best-fit line shown in (d) was y = 1.89 (± 0.08) % 104 × + 0.54 (± 0.09) % 104, with a correlation coefficient of 0.9964 (n = 6); according to eq 3, the values of and for the given peptide were then determined from the intercept and slope of this plot.
If the relative intensities of the isotope peaks for the separate 16O- and 18O-labeled samples are known, as measured in the first step of this approach, eq 1 can be rewritten in the form shown below.
| (2) |
In this particular study the terms of , a0, , and b4 were selected for use in eq 2 since they represented the most intense peaks in the 16O- or 18O-labeled isotope clusters; equivalent relationships to eq 2 could be derived using other peaks in these clusters. Eq 2 can next be rearranged into the form shown in eq 3.
| (3) |
Through this process it is possible to obtain a set of six equations (for n = 0 through 5) with two unknowns ( and ). According to eq 3, a plot of a0bn/anb4 versus a0In/an for a given isotopic peak should give a linear relationship. In addition, the values of and can then be obtained from the intercept and slope of this line. An example of such a plot is also shown in Figure 2, where each point in this plot represents the results obtained at a given value of n and a0In/an.
Once and have been determined from such a plot, and can also be calculated from the isotopic pattern of the 18O-labeled peptide by using eqs 4 and 5.
| (4) |
| (5) |
In this case, the peptides formed in 18O-water can be a mixture of peptides that have one, two or no 18O-labels. Such a mixture will give overlapping isotopic patterns, as shown in Figure 2. A peak intensity that has been corrected for this overlap, as represented here by the term I”n(corr) for peak n, can be obtained by using relationships like those shown in eqs 6-7 for the M+2 and M+4 peaks in an isotopic series.
| (6) |
| (7) |
Finally, the 18O/16O ratio for the desired peptide in a mixture of soluble and immobilized protein preparations that have been separately digested in 16O- or 18O-enriched water can be calculated by using these results in eq 8.
| (8) |
In this report this corrected ratio was then used to determine the relative extent to which a given region on the protein was involved in its immobilization to a solid support.
EXPERIMENTAL SECTION
Reagents
The Nucleosil Si-100 (7 μm particle diameter, 100 Å pore size) was from Macherey Nagel (Düren, Germany). The HSA (approximately 99% pure, fatty acid free, essentially globulin free), trypsin (sequencing grade), endoproteinase Lys-C (Lys-C, sequencing grade), and endoproteinase Glu-C (Glu-C, sequencing grade) were from Sigma-Aldrich (St. Louis, MO). The α-cyano-4-hydroxycinnamic acid (CHCA, >99% pure), 2,5-dihydroxybenzoic acid (DHB, 98% pure), des-Arg-bradykinin (99% pure), angiotensin I (97% pure, acetate salt), Glu-fibrinopeptide B (97% pure), 18O-enriched water (95 atom% 18O) were also from Sigma-Aldrich. Reagents for the bicinchoninic acid (BCA) protein assay were from Pierce (Rockford, IL). All other chemicals were reagent grade or better. All aqueous solutions were prepared using water from a Nanopure system (Barnstead, Dubuque, IA) and filtered using 0.22 μm nylon filters from Fisher (Pittsburgh, PA).
Apparatus
The BCA protein assay was performed using a UV-160A spectrophotometer (Shimadzu, Kyoto, Japan). Samples were dried using a Savant SpeedVac from ThermoQuest (San Jose, CA). ZipTipsμ-C18 (5.0 μg capacity) were obtained from Millipore (Billerica, MA). The Slide-A-Lyzer dialysis cassettes (7 kDa MW cutoff, 0.5−3 mL capacity) were from Pierce. The overhead transparency film for sample/matrix mixing in the MALDI studies was from C-line Products (Des Plaines, IL). The MALDI-TOF MS experiments were performed using a Voyager 6184 system (Applied Biosystems, Foster City, CA) operated in a positive-ion delayed extraction reflecton mode. The instrument settings for this analysis were as follows: accelerating voltage, 20 kV; grid voltage, 76% of the accelerating voltage; guide wire voltage, 0.008% of the accelerating voltage; delay time, 100 ns.
HSA immobilization & pretreatment
Nucleosil Si-100 silica was converted into a diol-bonded form according to a previous procedure.27 HSA was immobilized onto this diol-bonded silica by the Schiff base method.21 The protein content of the resulting HSA support was determined in triplicate by a BCA assay,28 using HSA as the standard and diol-bonded silica as the blank. The final protein content of the support tested in this work was found to be 51 (± 4) mg HSA per gram of silica (±1 S.D.).
A total of 5 mg soluble HSA or approximately 85 mg of HSA silica (i.e., a sample containing roughly 5 mg immobilized HSA) was denatured, reduced and alkylated according to a previous described procedure.29 To remove excess chemicals before digestion, the immobilized HSA was washed three times with water, while the soluble HSA was dialyzed three times against fresh portions of 500 mL water for 4 h at room temperature. These soluble and immobilized HSA samples were then adjusted to the same total volume (i.e., approximately 1800 μL), with the samples being divided into several 18 μL aliquots (each containing roughly 50 μg HSA).
After their pretreatment and prior to their digestion with trypsin, one aliquot of the immobilized HSA sample and one aliquot of the soluble HSA sample were dried separately in microcentrifuge tubes. The dried samples were reconstituted in 120 μL of pH 7.8, 50 mM ammonium bicarbonate buffer prepared in standard deionized water (99.8% H216O) for the immobilized HSA or in 18O-enriched water (97% H218O) for the soluble HSA. Both aliquots were combined with 2.0 μg trypsin (giving a substrate-to-enzyme ratio of 30:1), with these mixtures then being incubated at 37 °C for 18 h.
Prior to digestion with Glu-C, portions of the pretreated soluble HSA and immobilized HSA samples were dried and reconstituted in the same manner as described for trypsin digestion. These samples was combined with 5 μg Glu-C (giving a substrate-to-enzyme ratio of 10:1) and incubated at 37°C for 18 h. An additional 2.5 μg Glu-C (giving a new substrate-to-enzyme ratio of 20:1) was later added and incubated with the samples for another 8 h at 37°C.
Pretreated HSA samples for digestion with Lys-C were dried and reconstituted in 120 μL 75 mM Tris-HCl buffer that had been prepared in standard deionized water or 18O-enriched water. A 2.5 μg portion of a Lys-C solution (giving a substrate-to-enzyme ratio of 20:1) was added to both sets of samples, which were then incubated at 37°C for 18 h.
After their incubation with trypsin, Glu-C or Lys-C, the digests for the immobilized HSA samples were centrifuged to remove the support particles and peptides attached to these particles. The soluble fractions of the digests for the soluble and immobilized HSA samples were then combined with 5 μL concentrated formic acid to adjust their pH to a value less than 4.0. For each type of enzyme used for these digests, 5 μL of the soluble HSA solution that had been digested in 18O-enriched water or 5 μL of the immobilized HSA solution that had been digested in 16O-water were combined and mixed prior to further treatment.
The peptides in the combined samples were next fractionated using ZipTipμ-C18 pipette tips, as described in a previous paper.29 To determine the isotopic pattern of a given 18O- or 16O-labeled peptide, 10 μL of digests from the immobilized HSA sample and 10 μL of digests from the soluble HSA sample were fractionated in the same way as the combined sample. Soluble HSA control samples were pretreated in the same fashion as the immobilized HSA/soluble HSA mixtures but used soluble HSA for both the 18O- and 16O-labeling steps.
Mass spectrometric analysis
The matrix used for the MALDI-TOF MS analysis of HSA digests was a mixture of α-cyano-4-hydroxycinnamic acid and 2,5-dihydroxybenzoic acid, as prepared according to previous procedures.29,30 A 0.5 μL aliquot of this matrix and 0.5 μL of a digested HSA sample (about 15 pmol protein) were placed on a transparency film and mixed together with a pipette tip. The final mixture was then aspirated and applied by digital pipette onto a MALDI plate.
A stock solution of standard peptides was prepared for MALDI-TOF MS analysis with the following composition: 18.4 μL of 1 μg/μL des-Arg-bradykinin, 33.6 μL of 1 μg/μL angiotensin I, and 408 μL of 0.1 μg/μL Glu-fibrinopeptide B plus 7540 μL of a 50:50 (v/v) mixture of acetonitrile and water. This stock solution was divided into 10 μL aliquots and stored at −80°C until use. A 4 μL portion of this stock solution was mixed with 96 μL of the MALDI matrix (see previous paragraph), giving a final concentration for each standard peptide of approximately 1.0−1.3 pmol/μL. A 1 μL portion of this standard mixture was spotted on each well of every other row on the MALDI plate. The spotted plate was allowed to air dry for 15−20 min before analysis. The mass scale of the mass spectrum was adjusted by the calibration curve data obtained with this standard peptide mixture.
The spot for each HSA digest and standard peptide sample on the MALDI plate was examined using 250 laser shots, giving ions that were analyzed over a mass range of 500−3500 Da. A total of three to five spectra were acquired for each spot. The masses of the peptide peaks detected in the HSA digests were compared to those predicted by PeptideMass.31 The following parameters were used in this program for predicting the theoretical mass for a peptide: 1) all cysteines were assumed to be treated with iodoacetamide; 2) the oxidation of methionines was allowed, 3) monoisotopic masses were used for all amino acid residues, and 4) the maximum number of allowed missed cleavage sites was two. All detected peptides that could be matched to a mass within 50 ppm of a value for a predicted peptide were selected for further study. The sequence coverage for HSA was calculated by using the fraction of all amino acids in HSA that occurred in the detected peptides and that could be identified as arising from a specific portion of this protein's primary sequence.29
For the quantitative analysis of peptides, the ratio of each 18O and 16O-labeled peptide pair in the digest mixtures was determined as described in the THEORY. The pKa values for the lysine residues in HSA were calculated using PROPKA.32 The fractional solvent accessible surface area for the side chain of each residue was calculated using VADAR and a solvent probe radius of 1.4 Å.33 The regions determined to be at or near possible immobilization sites were mapped onto the 3-D structure of HSA by using Protein Explorer 2.45 Beta.34
RESULTS AND DISCUSSION
General considerations in method development
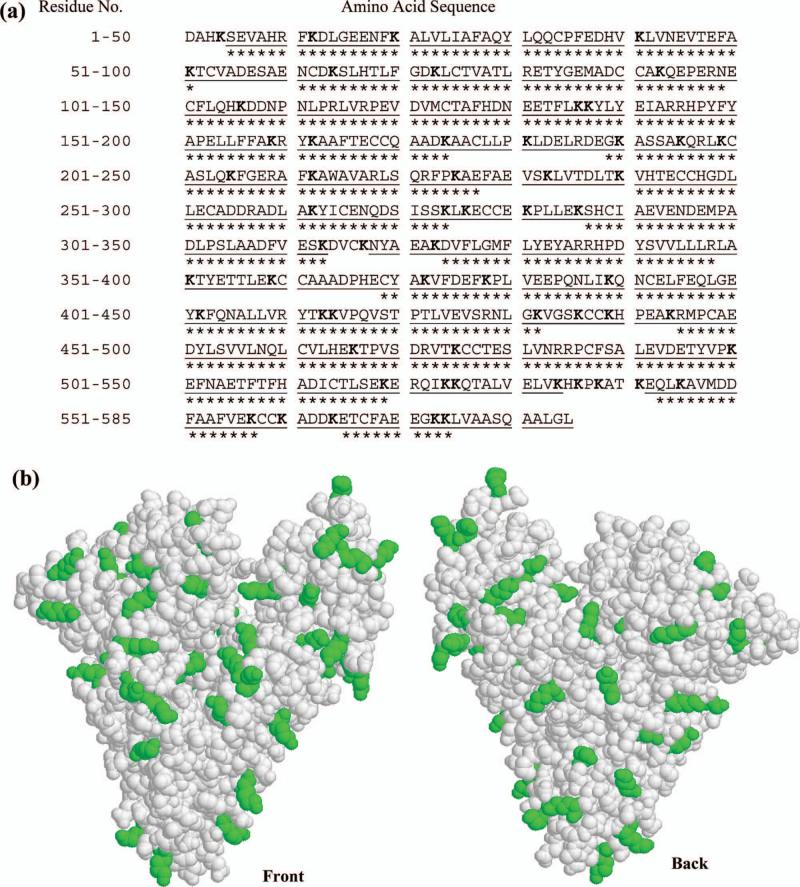
An amine-based coupling method for a protein generally involves the reaction of the N-terminus or lysine residues on such a ligand. The locations of the 59 lysines and N-terminus of HSA are shown in Figure 3. These sites are distributed in a relatively uniform pattern throughout both the primary sequence and tertiary structure of HSA. Although it might appear at first that all of these residues would have an equal probability of taking part in an amine-based coupling method, these sites can have differences in their pKa values, accessibilities to other molecules, and other features that may cause them to differ in their reactivity. This work sought to study this large number of possible coupling sites by using a previous protocol developed to maximum the sequence coverage of HSA when it is examined by MALDI-TOF MS.29 This involved the use of multiple proteolytic enzymes (i.e., trypsin, Lys-C and Glu-C) and peptide fractionation prior to MALDI-TOF MS analysis. When using this approach in qualitative studies, the sequence coverage of HSA has been reported to be 97.4%.29 However, not all of the peptides detected in this technique have sufficient intensities to allow their use in quantitative proteomics and measurements of 18O/16O ratios. This additional requirement was found in this current study to lower the effective sequence coverage of HSA to 76.9%; however, this level of coverage was still sufficient for examining most of the possible immobilization sites on HSA. In this case, 42/60 (or 70%) of the possible immobilization sites on this protein were within or adjacent to a measurable peptide sequence and another 13/60 (or 22%) were within six residues of such a sequence.
Figure 3.
The (a) primary sequence and (b) crystal structure of HSA. The sequence in (a) shows the regions of the primary sequence that could be identified qualitatively when using each peptide digest ( ), along with those sections that could be used for quantitative determination of 18O/16O ratios (***). The lysine residues in HSA are given in bold in (a) and as darkened regions in (b). The plot is (b) is based on PDB file 1AO6.39
), along with those sections that could be used for quantitative determination of 18O/16O ratios (***). The lysine residues in HSA are given in bold in (a) and as darkened regions in (b). The plot is (b) is based on PDB file 1AO6.39
Another item considered in this report was the effect immobilization might have on ability of proteolytic enzymes to digest regions on HSA that were not directly involved in its immobilization. For instance, steric hindrance from the pores of a support could prevent a proteolytic enzyme from effectively reaching and digesting part of an immobilized protein. This effect was avoided in this study by using a support with a nominal pore size of 100 Å, which is roughly twice the diameter of HSA (molecular weight, 66.5 kDa) and even larger than any of the enzymes used here to digest this protein (molecular weights, 22−29 kDa). In addition, both the immobilized and soluble HSA were denatured before digestion, giving greater accessibility of proteolytic enzymes to this protein's primary sequence. Under these conditions statistically equivalent 18O/16O ratios were seen for the majority of peptides (83% to 87% depending on the enzyme used) in mixtures containing immobilized versus soluble HSA and in control samples containing only digests of soluble HSA for both labels. This similarity indicated that steric hindrance effects were not a significant factor during the digestion of immobilized HSA in this study.
The reproducibility of the MALDI-TOF MS analysis was examined early in this work by using a mixture of 18O and 16O-labeled peptides from tryptic digests of soluble HSA. In one such study, a peptide MH+ peak with a mass of 927.4939 had a relative standard deviation of ! 5% in its 18O/16O ratio for spectra collected from several spots (i.e., spot-to-spot precision) while the same peptide had a within-spot precision of ! 3%. A similar trend was noted for other peptides in the same digest, with the within-spot precision being slightly better than the spot-to-spot precision. Thus, all further work in this study used spectra obtained from the same spot when determining 18O/16O ratios. This gave an average overall precision in the final results of ! 24% (range, 10−46%) for peptides with 18O/16O ratios below 1.0, ! 16% (range, 6−36%) for peptides with 18O/16O ratios between 1.0 and 2.0, and ! 11% (range, 3−20%) for peptides with 18O/16O ratios above 2.0. As will be shown later, these levels of precision were more than sufficient for comparing the 18O/16O ratios for peptides from immobilized versus soluble HSA.
Analysis of peptides in tryptic digests of HSA
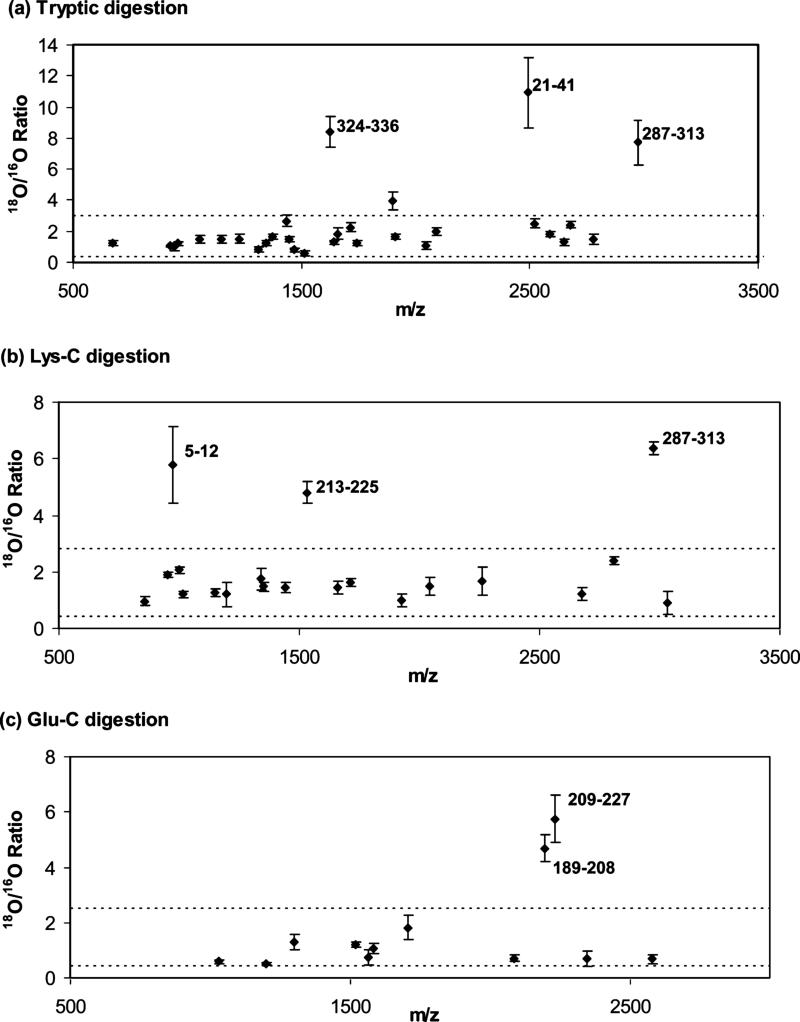
Table 1 shows the monoisotopic masses, sequences and 18O/16O ratios determined for peptides in mixtures of soluble and immobilized HSA that had been digested with trypsin. A total of 30 peptides from HSA were identified under these conditions and measured for their 18O/16O ratios. These peptides were from regions comprising 55.9% of the primary sequence of HSA. As shown in Figure 4(a), most of these peptides (26/30, or 87%) gave 18O/16O ratios less than 3.0, with ratios ranging from 0.4−3.0 and with an average of 1.5 (± 0.5) (1 SD). These results were equivalent to those obtained in control experiments using only soluble HSA for both the 18O and 16O-labeled digests, which gave 18O/16O ratios ranging from 0.3−3.0 and with an average of 1.4 (± 0.5). The fact that no measurable difference was found between these peptides and those in the soluble HSA control meant that amine groups within or near these particular regions were not involved to any significant extent in the immobilization of HSA by the Schiff base method.
Table 1.
18O/16O Ratios for peptides in tryptic digests for mixtures of soluble and immobilized HSAa
| Mass of MH+ ionb | Peptide sequence |
18O/16O ratio for peptides collected at a given % acetonitrile (ACN) in water |
Average 18O/16O Ratio | ||||
|---|---|---|---|---|---|---|---|
| 5% ACN | 10% ACN | 20% ACN | 30% ACN | 50% ACN | |||
| 673.3785 | 467−472 | 1.21 (± 0.15) | 1.21 (± 0.15) | ||||
| 927.4939 | 138−144 | 1.06 (± 0.02) | 1.04 (± 0.01) | 0.99 (± 0.11) | 1.03 (± 0.07) | ||
| 940.4488 | 107−114 | 1.02 (± 0.25) | 1.02 (± 0.25) | ||||
| 960.5630 | 403−410 | 1.19 (± 0.06) | 1.24 (± 0.12) | 1.16 (± 0.10) | 1.20 (± 0.09) | ||
| 1055.5889 | 137−144 | 1.48 (± 0.26) | 1.47 (± 0.27) | 1.47 (± 0.24) | |||
| 1149.6155 | 42−51 | 1.54 (± 0.41) | 1.46 (± 0.13) | 1.49 (± 0.26) | |||
| 1226.6057 | 11−20 | 1.40 (± 0.13) | 1.63 (± 0.32) | 1.52 (± 0.26) | |||
| 1311.7424 | 338−348 | 0.80 (± 0.17) | 0.80 (± 0.17) | ||||
| 1342.6352 | 546−557 | 1.22 (± 0.18) | 1.22 (± 0.18) | ||||
| 1371.5673 | 163−174 | 1.67 (± 0.18) | 1.67 (± 0.18) | ||||
| 1434.5339 | 82−93 | 2.67 (± 0.36) | 2.67 (± 0.36) | ||||
| 1443.6425 | 263−274 | 1.48 (± 0.20) | 1.48 (± 0.20) | ||||
| 1467.8435 | 337−348 | 0.78 (± 0.10) | 0.83 (± 0.14) | 0.81 (± 0.12) | |||
| 1511.8433 | 415−428 | 0.56 (± 0.18) | 0.56 (± 0.18) | ||||
| 1623.7881 | 324−336 | 8.44 (± 0.96) | 8.44 (± 0.96) | ||||
| 1639.9382 | 414−428 | 1.22 (± 0.08) | 1.29 (± 0.14) | 1.36 (± 0.20) | 1.28 (± 0.14) | ||
| 1657.7531 | 390−402 | 1.73 (± 0.33) | 2.01 (± 0.43) | 1.85 (± 0.37) | |||
| 1714.7971 | 94−106 | 2.18 (± 0.33) | 2.35 (± 0.19) | 2.26 (± 0.27) | |||
| 1742.8946 | 146−159 | 1.25 (± 0.16) | 1.25 (± 0.16) | ||||
| 1898.9957 | 145−159 | 3.94 (± 0.60) | 3.94 (± 0.60) | ||||
| 1910.9322 | 484−500 | 1.56 (± 0.18) | 1.70 (± 0.17) | 1.63 (± 0.18) | |||
| 2045.0958 | 373−389 | 1.14 (± 0.36) | 1.08 (± 0.11) | 1.11 (± 0.25) | |||
| 2086.8380 | 241−257 | 1.86 (± 0.15) | 2.04 (± 0.18) | 1.94 (± 0.41) | 1.96 (± 0.24) | ||
| 2490.2854 | 21−41 | 10.92 (± 2.26) | 10.92 (± 2.26) | ||||
| 2518.2143 | 446−466 | 2.50 (± 0.30) | 2.50 (± 0.30) | ||||
| 2585.1669 | 241−262 | 1.84 (± 0.15) | 1.84 (± 0.15) | ||||
| 2650.2645 | 115−136 | 1.28 (± 0.24) | 1.28 (± 0.24) | ||||
| 2674.3155 | 445−466 | 2.41 (± 0.22) | 2.41 (± 0.22) | ||||
| 2778.3594 | 115−137 | 1.47 (± 0.32) | 1.47 (± 0.32) | ||||
| 2974.3449 | 287−313 | 7.73 (± 1.44) | 7.73 (± 1.44) | ||||
The numbers in parentheses represented a range of ±1 SD, with 3−5 spectra being were acquired for each spot and peptide; the 18O/16O ratios in bold represent the values that were significantly higher than those obtained in a control sample based on only soluble HSA.
The given masses of the MH+ ions were calculated from the sequence of HSA. All experimental values were within 50 ppm of these masses.
Figure 4.
Measured 16O/18O ratios for mixtures of peptides from immobilized and soluble HSA prepared by digestion with (a) trypsin, (b) Lys-C or (c) Glu-C. The error bars represent a range of ± 1 S.D. The horizontal dashed lines represent a cutoff ratio that was used to discriminate between immobilized and non-immobilized peptides, as determined by labeling studies using only soluble HSA.
There were, however, three peptides in the tryptic digests that gave significantly higher 18O/16O ratios than those seen for the soluble HSA control. These three peptides had 18O/16O ratios in the range of 7.7 to 10.9, which were different at the 95% confidence level from other peptides in the mixed digest or soluble control (note: there was one other peptide with a 18O/16O ratio above 3.0; however, its ratio of 3.94 was not significantly different versus the soluble HSA control or peptides with ratios at or below 3.0 in the mixed digest). These high 18O/16O ratios indicated that their corresponding peptides had a higher occurrence for soluble HSA in the combined digest than the same peptides from immobilized HSA. This, in turn, suggested that these peptides remained on the support during the digestion of immobilized HSA and, thus, were at or near an immobilization site for this protein. Furthermore, the relative size of these 18O/16O ratios should be related to the probability of these regions being involved in immobilization, with the highest ratios representing those parts of HSA which were most likely to be involved in the coupling of this protein to the support.
As is shown in Table 1 and has been noted previously,29 the fractionation of peptides after digestion greatly increases (i.e., by approximately two-fold) the number of the peptides that can be analyzed for HSA. However, peptide fractionation did not appear to affect any of the measured 18O/16O peptide ratios. For example, peptides with protonated monoisotopic masses (MH+ peaks) at 927.4939, 960.5630, 1639.9382 and 2086.8380 were detected in three different acetonitrile fractions, but all of these peptides gave consistent 18O/16O ratios in each fraction (see Table 1). This confirmed that the 16O and 18O-labeled peptides had equivalent elution and retention behavior in the fractionation method used in this study. This also meant that data from each of these fractions could be combined to obtain more precise estimates of a given peptide's 18O/16O ratio, as was used later in this study.
As mentioned earlier, many peptides that undergo labeling with 18O-enriched water during digestion can incorporate two 18O labels per peptide,26 but the efficiency of this process can vary from one peptide to the next.14 An example of this situation was a peptide with a MH+ mass of 1226.6057 that was observed in the tryptic digest. This peptide was found to incorporate only one 18O label while most other peptides in this mass range had two such labels. However, these variations in labeling efficiency did not affect the calculation of 18O/16O ratios in this study since, as stated in the THEORY, the method used for this calculation allowed for the possibility of having incomplete labeling and either one or two 18O labels per peptide.
Analysis of peptides in Lys-C digests of HSA
Table 2 shows the monoisotopic masses, sequences and 18O/16O ratios that were obtained for peptides in mixtures of soluble and immobilized HSA that had been digested with endoproteinase Lys-C. In this digest Twenty peptides were identified in this digest and used in the measurement of 18O/16O ratios, representing 48.7% of primary sequence for HSA. As is illustrated in Figure 4(b), most of these peptides (17/20, or 85%) had 18O/16O ratios in the same range as those seen for a soluble HSA control, with a range and average of 0.5−2.6 and 1.5 (± 0.4) versus a range of 0.4−2.8 and an average of 1.5 (± 0.5) for the control. This similarity again indicated that these particular regions of HSA were not involved to any significant extent in the immobilization of this protein by the Schiff base method.
Table 2.
18O/16O Ratios for peptides found in Lys-C digests for mixtures of soluble and immobilized HSAa
| Mass of MH+ ionb | Peptide sequence |
18O/16O Ratio for peptides collected at a given % acetonitrile (ACN) in water |
Average 18O/16O Ratio | ||||
|---|---|---|---|---|---|---|---|
| 5% ACN | 10% ACN | 20% CAN | 30% ACN | 50% ACN | |||
| 854.4524 | 206−212 | 0.90 (± 0.09) | 1.04 (± 0.20) | 0.97 (± 0.16) | |||
| 951.4423 | 13−20 | 1.89 (± 0.10) | 1.89 (± 0.10) | ||||
| 973.5219 | 5−12 | 5.78 (± 1.35) | 5.78 (± 0.35) | ||||
| 1002.5583 | 467−475 | 2.06 (± 0.09) | 2.06 (± 0.09) | ||||
| 1017.5369 | 65−73 | 1.20 (± 0.08) | 1.20 (± 0.13) | 1.20 (± 0.10) | |||
| 1149.6155 | 42−51 | 1.18 (± 0.15) | 1.33 (± 0.07) | 1.25 (± 0.13) | |||
| 1198.5414 | 565−574 | 1.20 (± 0.44) | 1.20 (± 0.44) | ||||
| 1342.6352 | 546−557 | 1.75 (± 0.38) | 1.75 (± 0.38) | ||||
| 1352.769 | 403−413 | 1.63 (± 0.15) | 1.34 (± 0.09) | 1.43 (± 0.04) | 1.47 (± 0.16) | ||
| 1443.6425 | 263−274 | 1.46 (± 0.18) | 1.46 (± 0.18)c | ||||
| 1529.8704 | 213−225 | 4.81 (± 0.39) | 4.81 (± 0.39) | ||||
| 1657.7531 | 390−402 | 1.42 (± 0.14) | 1.47 (± 0.28) | 1.45 (± 0.21) | |||
| 1714.7971 | 94−106 | 1.64 (± 0.09) | 1.60 (± 0.18) | 1.62 (± 0.14) | |||
| 1924.0867 | 415−432 | 0.99 (± 0.23) | 0.99 (± 0.23) | ||||
| 2045.0958 | 373−389 | 1.63 (± 0.30) | 1.21 (± 0.05) | 1.49 (± 0.32) | |||
| 2260.0232 | 501−519 | 1.67 (± 0.49) | 1.67 (± 0.49) | ||||
| 2674.3155 | 445−466 | 1.22 (± 0.22) | 1.22 (± 0.22) | ||||
| 2807.4713 | 138−159 | 2.47 (± 0.07) | 2.33 (± 0.12) | 2.39 (± 0.12) | |||
| 2974.3449 | 287−313 | 6.37 (± 0.23) | 6.37 (± 0.23)c | ||||
| 3030.4123 | 476−500 | 0.89 (± 0.41) | 0.89 (± 0.41) | ||||
The numbers in parentheses represented a range of ±1 SD, with 3−5 spectra being were acquired for each spot and peptide; the 18O/16O ratios in bold represent the values that were significantly higher than those obtained in a control sample based on only soluble HSA.
The given masses of the MH+ ions were calculated from the sequence of HSA. All experimental values were within 50 ppm of these masses.
Only two spectra were collected and used to determine the average ratios for these peptides.
There were three peptides in the Lys-C digest with significantly higher ratios than those in the soluble HSA control. These peptides corresponded to the 5−12, 213−225 and 287−313 regions of HSA, which gave 18O/16O ratios of 5.78, 4.81 and 6.37, respectively. All of these ratios were higher at the 95% confidence level versus those in the soluble HSA control. This indicated that amines in or near these regions were taking part in the coupling of HSA by the Schiff base method.
Although higher mass peptides usually have a lower extent of incorporation for a second 18O label than lower mass peptides, it was noted in the Lys-C digest that some low mass peptides contained only one 18O label. This was found to be the case for the peptide with an MH+ peak at a mass of 1002.56 in the 5% acetonitrile fraction. As mentioned earlier, this single label did not create any difficulties in this study since the method used to calculate 18O/16O ratios allowed for the presence of either one or two labels in such peptides.
Analysis of peptides in Glu-C digests of HSA
The results obtained for the Glu-C digests of soluble and immobilized HSA are shown in Table 3. A total of 12 peptides were identified and used in these digests for the measurement of 18O/16O ratios, representing 30.6% of the primary sequence of HSA. Figure 4(c) shows that ten of these peptides (10/12, or 83%) had 18O/16O ratios within a range of 0.3−2.2 and with an average of 0.9 (± 0.4). This was comparable to the results found for the soluble HSA control, which gave a range of 18O/16O ratios of 0.4−2.5 and an average ratio of 1.1 (± 0.4).
Table 3.
18O/16O Ratios for peptides found in Glu-C digests for mixtures of soluble and immobilized HSAa
| Mass of MH+ ionb | Peptide sequence |
18O/16O Ratio for peptides collected at a given % acetonitrile (ACN) in water |
Average 18O/16O Ratio | ||||
|---|---|---|---|---|---|---|---|
| 5% ACN | 10% ACN | 20% ACN | 30% ACN | 50% ACN | |||
| 1031.4508 | 369−376 | 0.59 (± 0.06) | 0.59 (± 0.06) | ||||
| 1197.5791 | 496−505 | 0.52 (± 0.05) | 0.52 (± 0.05) | ||||
| 1300.6649 | 7−17 | 1.18 (± 0.10) | 1.46 (± 0.36) | 1.30 (± 0.27) | |||
| 1519.7809 | 142−153 | 1.26 (± 0.14) | 1.20 (± 0.08) | 1.19 (± 0.04) | 1.22 (± 0.09) | ||
| 1566.6204 | 120−132 | 0.73 (± 0.27) | 0.65 (± 0.30) | 0.69 (± 0.34) | 0.94 (± 0.12) | 0.74 (± 0.26) | |
| 1583.7883 | 543−556 | 1.08 (± 0.19) | 1.08 (± 0.19) | ||||
| 1704.9477 | 154−167 | 1.82 (± 0.43) | 1.82 (± 0.43) | ||||
| 2086.0642 | 377−393 | 0.70 (± 0.12) | 0.70 (± 0.12) | ||||
| 2194.1766 | 189−208 | 4.68 (± 0.47) | 4.68 (± 0.47) | ||||
| 2232.2517 | 209−227 | 5.77 (± 0.86) | 5.75 (± 0.86) | ||||
| 2348.1933 | 101−119 | 0.64 (± 0.29) | 0.58 (± 0.07) | 0.69 (± 0.28) | |||
| 2578.2757 | 61−82 | 0.67 (± 0.13) | 0.69 (± 0.24) | 0.68 (± 0.17) | |||
The numbers in parentheses represented a range of ±1 SD, with 3−5 spectra being were acquired for each spot and peptide; the 18O/16O ratios in bold represent the values that were significantly higher than those obtained in a control sample based on only soluble HSA.
The given masses of the MH+ ions were calculated from the sequence of HSA. All experimental values were within 50 ppm of these masses.
There were two peptides in the mixed digest that had significantly higher 18O/16O ratios (i.e., at the 95% confidence level) than those found for the soluble HSA control. These peptides were from the 189−208 and 209−227 regions of HSA and gave 18O/16O ratios of 4.68 and 5.75, respectively. Thus, it was concluded that that amines in or near these regions were taking part in the Schiff base immobilization of HSA.
There were two peptides in the Glu-C digests found to have different labeling behavior from the other peptides. One of these peptides was in the 5% acetonitrile fraction and had a MH+ mass of 1300.66. This peptide incorporated only one 18O label during Glu-C digestion, as noted previously for some other peptides in the tryptic and Lys-C digests. Another peptide with unique behavior had a MH+ mass of 1326.80 and gave the same isotopic pattern for digests in normal or 18O-enriched water. This particular peptide was from the C-terminus of HSA (residues 572−585) and is believed to have not undergone Glu-C catalyzed 16O-to-18O exchange that lead to the lack of this label.
Another challenge encountered in examining peptides in the Glu-C digests concerned peptides with overlapping clusters of peaks. An example occurred in the 5% acetonitrile fraction of the Glu-C digest, which contained two peptides that had overlapping isotopic peaks. These two peptides had MH+ masses of 1699.84 and 1704.95. In the case of this particular fraction, direct calculation of the 18O/16O ratios for these peptides was not performed because of this peptide-peptide interference. However, only the 1704.95 peptide was seen in 30% and 50% acetonitrile fraction of the same digest, which made it possible to obtain its 18O/16O ratio without interference from the 1699.84 peptide. Another example of situation occurred for a peptide with a MH+ mass of 1519.78, which had no any interferences in the 5% acetonitrile fraction of Glu-C digest but did overlap in the 30% acetonitrile fraction with a peptide that had a MH+ mass of 1518.77.
Identification of immobilization sites
The results in Tables 1-3 and Figure 4 were combined and compared to determine the specific residues in HSA that were taking part in its immobilization. These results are summarized in Table 4. This includes a comparison of the measured 18O/16O ratios for the various peptides used in this study, as well as the use of established computational methods to estimate the pKa of each lysine in these peptides and this lysine's fractional accessible surface area (ASA).32,33 The locations of these peptides in the structure of HSA are indicated in Figure 5.
Table 4.
Summary of regions in immobilized HSA with high 18O/16O ratios
|
Region in HSA (18O/16O ratio) |
Possible immobilization sitesa |
Estimated pKa |
Estimated fractional solvent accessible surface area |
||
|---|---|---|---|---|---|
|
Tryptic digest |
Lys-C digest |
Glu-C digest |
|||
| 21−41 (Ratio, 10.92) |
K12 | 10.43 | 0.59 | ||
| K20 | 10.43 | 0.32 | |||
| |
|
K41 |
10.15 |
0.56 |
|
| 324−336 (Ratio, 8.44) |
K317 | 10.36 | 0.80 | ||
| |
|
K323 |
10.50 |
0.54 |
|
| 287−313 (Ratio, 7.73) |
287−313 (Ratio, 6.37) |
K281 | 10.50 | 0.52 | |
| K286 | 10.29 | 0.20 | |||
| K313 | 10.50 | 0.80 | |||
| |
K317 |
10.36 |
0.80 |
||
| 5−12 (Ratio, 5.78) |
N-terminus | 7.6−8.0b | Undeterminedb | ||
| K4 | 7.79 | Undeterminedb | |||
| |
|
K12 |
10.43 |
0.59 |
|
| 213−225 (Ratio, 4.81) |
209−227 (Ratio, 5.75) |
K205 | 10.43 | 0.58 | |
| K212 | 10.43 | 0.40 | |||
| |
K225 |
9.80 |
0.44 |
||
| 189−208 (Ratio, 4.68) | K190 | 6.23 | 0.42 | ||
| K195 | 10.76 | 0.50 | |||
| K199 | 7.47 | 0.16 | |||
| K205 | 10.43 | 0.58 | |||
| K212 | 10.43 | 0.40 | |||
The residues in bold are the suspected immobilization sites determined in this work for HSA.
The pKa and fractional solvent accessible surface area of the N-terminus could not be calculated by PROPKA and VADAR because this portion of HSA does not have a well-defined crystal structure. The pKa value given here was instead obtained from the literature41.
Figure 5.
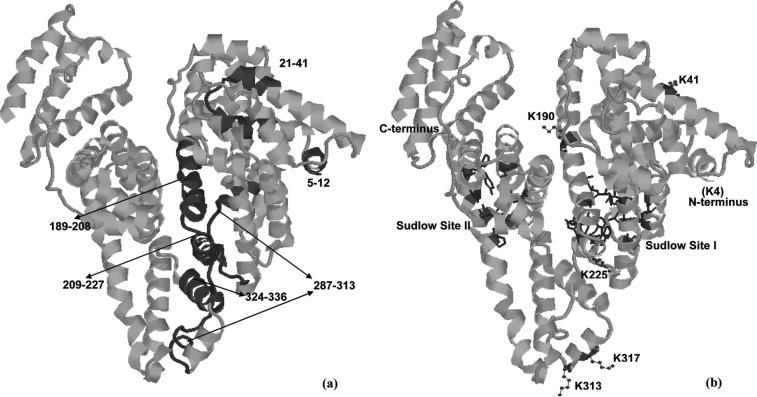
The three dimensional structure of HSA, showing the locations of peptides with high 18O/16O ratios (a) and the proposed immobilization sites (b). The known locations of Sudlow sites I (in green) and II (in yellow) were also shown in the Figure 4(b).40 These structures are based on PDB file 1AO6.39
The region of HSA that was found in this study to give the highest 18O/16O ratio was that containing residues 21−41 in the tryptic digest. There are three possible attachment sites near or within this region when using an amine-based coupling methods; these sites are K12, K20 and K41. All of these lysines have medium-to-high pKa values (10.15−10.43 versus a range of 6.23−11.11 and average of 10.08 for all lysines in HSA). This feature is important to consider since lysines with low pKa values would have a larger fraction of non-protonated amine groups at pH 6.0 (i.e., the coupling pH used here in the Schiff base method).21,35,36 However, these amine groups must also be accessible to the protein's surroundings for this coupling to occur. It was found that both K12 and K41 have good accessibility to the surrounding solvent (ASA values of 0.56−0.59 versus a range of 0.01−0.96 and average of 0.52 for all lysines on HSA), but the results obtained for several other peptides containing K12 did not show increased 18O/16O ratios. This indicated that K41 was the most likely site for the immobilization of HSA in this region.
The region on HSA with the second highest 18O/16O ratio was that which included residues 324−336. This region was near lysines K323 and K317. Although these two lysines again have medium-to-high pKa values (10.50 and 10.36, respectively), K317 has a higher accessibility to the surrounding solvent (ASA, 0.80). Thus, this lysine was determined to be the most likely residue in this region for the immobilization of HSA.
Another region on HSA that gave a high 18O/16O ratio was the one containing residues 287−313. There are four lysines in this region: K281, K286, K313 and K317. All of these lysines have moderate-to-high pKa values (10.29−10.50), but K313 and K317 have a much higher accessibility to solvent (ASA of 0.80 versus 0.20−0.52 for K281 and K286). This suggests that K313 and K317 were both contributing to the high 18O/16O ratios noted for residues 287−313. The fact that K313 is located in a flexible loop region of HSA further supports the hypothesis that this residue is a likely immobilization site for this protein.
A high 18O/16O ratio was also found for residues near the N-terminus of HSA (i.e., residues 5−12). Possible immobilization sites in or near this region include the N-terminal amine and lysines K4 and K12. As discussed earlier, K12 was already eliminated as a likely site for immobilization since other peptides containing this residue gave low 18O/16O ratios. The estimated pKa values for the remaining possibilities, the N-terminus and K4, are both low compared to other amines on HSA (with values of 7.6−8.0), making these sites favorable for immobilization. The flexibility of the N-terminal region in HSA would also be expected to make both these locations good candidates as immobilization sites.
The 213−225 and 209−227 regions of HSA gave high 18O/16O ratios as well. Possible immobilization sites in or near these regions are lysines K205, K212 and K225. However, residues 206−212 gave a low 18O/16O ratio in the Lys-C digest, indicating that K205 and K212 were not involved in the immobilization of HSA. This fact, plus the location of K225 in a flexible turn of HSA and its low pKa value versus these other lysines (9.80 versus 10.43), indicates that K225 is the most likely immobilization site for HSA in this region.
The final peptide found to have a significantly elevated 18O/16O ratio was from residues 189−208. This peptide contained lysines K190, K195, K199, K205 and K212. As indicated earlier, a low 18O/16O ratio for residues 206−212 ruled out two of these lysines (K205 and K212) as possible immobilization sites. The remaining three lysines are all located in a α-helix, with K190 and K199 having the lowest pKa values in this group (6.23 and 7.47 versus 10.76 for K195). However, K199 is buried deep inside of HSA and has an ASA value of only 0.16, which would give it a small likelihood of coupling to a solid support. Thus, K190 was found to be the most likely immobilization site for HSA in this region.
CONCLUSIONS
This report examined the use of MALDI-TOF MS and 18O/16O-labeling in identifying immobilization sites on the protein HSA. When data obtained by this method were combined with predicted pKa values and accessible surface areas, it was possible to identify at least seven major immobilization sites for HSA in the Schiff base method. These sites included the N-terminus and lysines 4, 41, 190, 225, 313 and 317. As shown in Figure 5, none of these immobilization regions are located within the major drug binding regions of HSA (i.e., Sudlow sites I and II). This explains why the use of the Schiff base method and other amine-based coupling methods have been observed in previous studies to give HSA with comparable binding behavior to that seen for soluble HSA.21,37,38 The non-uniform distribution of the immobilization sites identified in this report further suggests that local steric hindrance effects, rather than protein inactivation, may be the reason why different binding capacities have been noted for various drugs and other solutes at Sudlow sites I and II on immobilized HSA.21,38
The results of this study clearly indicate that not all of the possible coupling sites on a protein have an equal probability of being involved in its immobilization. It is also interesting to note that calculated parameters such as pKa and ASA values alone are not sufficient in predicting which of these sites will be the most likely to take part in immobilization. For example, residues K564 and K574 in HSA both have high ASA values (0.93 and 0.82), but neither appeared to take part to any appreciable extent in the immobilization of HSA by the Schiff base method (e.g., peptide 565−574 in the Lys-C digest gave a 18O/16O ratio of only 1.20). In addition, some residues that had low pKa values (e.g., K199 with pKa = 6.23) were also shown to not be involved in the immobilization of HSA. It instead appears that a combination of good accessibility and favorable pKa values, as well as some structure flexibility and a favorable microenvironment, is required for a given amine on HSA to take part in its immobilization. An important advantage of the method described in this paper is it allows the overall effect of these factors to be examined simultaneously for a protein by providing quantitative data on a potential immobilization site.
In summary, the results obtained in this report indicate that 18O/16O labeling and MALDI-TOF MS can be successfully employed to provide information on protein immobilization sites. A key advantage of this approach compared to previous methods10,11 is that it gives both qualitative information on the location of an immobilization site and quantitative information on the relative extent to which this site is involved immobilization. This last feature is made possible by the use of stable isotope labeling and methods adapted from quantitative proteoomics, which provides each detected peptide with its own internal standard. This technique requires only a small amount of protein (pmol-to-fmol per analysis) and is relatively easy to perform. In addition, this method can be modified for work with proteins, supports and coupling methods besides those considered in this current study. Such a method should be beneficial to many areas that employ immobilized proteins for analysis or separations, including affinity chromatography, enzyme reactors and protein microarrays.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health under grant R01 GM044931 and by the University of Nebraska Research Council. This work was performed in facilities that were remodeled under NIH grant RR015468-001. The authors thank Dr. Kurt Wulser and Meijian Zhou for their assistance during this project.
REFERENCES
- 1.Hage DS, Bian M, Burks R, Karle E, Ohnmacht C, Wa C. In: Handbook of Affinity Chromatography. Hage DS, editor. CRC Press; Boca Raton, FL: 2006. Chap. 5. [Google Scholar]
- 2.Gallant SR. Methods Mol. Biol. 2004;251:103–109. doi: 10.1385/1-59259-742-4:103. [DOI] [PubMed] [Google Scholar]
- 3.Novick SJ, Rozzell JD. Methods Biotechnol. 2005;17:247–271. [Google Scholar]
- 4.Hage DS, Nelson MA. Anal. Chem. 2001;73:198A–205A. [PubMed] [Google Scholar]
- 5.Vandevyver C, Freitag R. Antibodies. 2004;1:133–168. [Google Scholar]
- 6.Markoglou N, Wainer IW. Handbook Anal. Sep. 2003;4:215–234. [Google Scholar]
- 7.Elia G, Silacci M, Scheurer S, Scheuermann J, Neri D. Trends Biotechnol. 2002;20:S19–S22. doi: 10.1016/s1471-1931(02)00201-x. [DOI] [PubMed] [Google Scholar]
- 8.Hage DS, Chen J. In: Handbook of Affinity Chromatography. Hage DS, editor. CRC Press; Boca Raton, FL: 2006. Chap. 22. [Google Scholar]
- 9.Chen J, Hage DS. Nature Biotechnol. 2004;22:1445–1448. doi: 10.1038/nbt1022. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, Velander WH. J. Mol. Recognit. 1996;9:528–535. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<528::aid-jmr296>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.Wilchek M, Miron T. J. Biochem. Biophys. Methods. 2003;55:67–70. doi: 10.1016/s0165-022x(02)00178-1. [DOI] [PubMed] [Google Scholar]
- 12.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Rapid Commun. Mass Spectrom. 2000;14:1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal. Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 14.Stewart II, Thomson T, Figeys D. Rapid Commun. Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 15.Schnolzer M, Jedrzejewski P, Lehmann WD. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds Kristy J, Yao X, Fenselau C. J. Proteome Res. 2002;1:27–33. doi: 10.1021/pr0100016. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Ohnmacht C, Hage DS. J. Chromatogr. B. 2004;809:137–145. doi: 10.1016/j.jchromb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Ho E, Subramanyam B, Tseng J-L. J. Chromatogr. B. 2004;809:67–73. doi: 10.1016/j.jchromb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Andrisano V, Bertucci C, Cavrini V, Recanatini M, Cavalli A, Varoli L, Felix G, Wainer IW. J. Chromatogr. A. 2000;876:75–86. doi: 10.1016/s0021-9673(00)00195-3. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Hage DS. J. Chromatogr. A. 1997;766:15–25. doi: 10.1016/s0021-9673(96)01040-0. [DOI] [PubMed] [Google Scholar]
- 21.Loun B, Hage DS. J. Chromatogr. 1992;579:225–235. [PubMed] [Google Scholar]
- 22.Zhivkova Z, Russeva V. J. Chromatogr. B. 1998;707:143–149. doi: 10.1016/s0378-4347(97)00586-0. [DOI] [PubMed] [Google Scholar]
- 23.Tweed SA, Loun B, Hage DS. Anal. Chem. 1997;69:4790–4798. doi: 10.1021/ac970565m. [DOI] [PubMed] [Google Scholar]
- 24.Hage DS, Noctor TAG, Wainer IW. J. Chromatogr. A. 1995;693:23–32. doi: 10.1016/0021-9673(94)01009-4. [DOI] [PubMed] [Google Scholar]
- 25.Rollag JG, Hage DS. J. Chromatogr. A. 1998;795:185–198. doi: 10.1016/s0021-9673(97)00975-8. [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal. Chem. 2004;76:2675. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 27.Ruhn PF, Garver S, Hage DS. J. Chromatogr. A. 1994;669:9–19. doi: 10.1016/0021-9673(94)80332-3. [DOI] [PubMed] [Google Scholar]
- 28.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Wa C, Cerny R, Hage DS. Anal. Biochem. 2006;349:229–241. doi: 10.1016/j.ab.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Laugesen S, Roepstorff P. J. Am. Soc. Mass Spectrom. 2003;14:992–1002. doi: 10.1016/S1044-0305(03)00262-9. [DOI] [PubMed] [Google Scholar]
- 31.Wilkins MR, Lindskog I, Gasteiger E, Bairoch A, Sanchez J-C, Hochstrasser DF, Appel RD. Electrophoresis. 1997;18:403–408. doi: 10.1002/elps.1150180314. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Robertson Andrew D, Jensen Jan H. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 33.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martz E. Trends in Biochem. Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
- 35.Hornsey VS, Prowse CV, Pepper DS. J. Immunol. Methods. 1986;93:83–88. doi: 10.1016/0022-1759(86)90436-9. [DOI] [PubMed] [Google Scholar]
- 36.Stults NL, Asta LM, Lee YC. Anal. Biochem. 1989;180:114–119. doi: 10.1016/0003-2697(89)90097-3. [DOI] [PubMed] [Google Scholar]
- 37.Loun B, Hage DS. Anal. Chem. 1994;66:3814–3822. doi: 10.1021/ac00093a043. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Hage DS. J. Chromatogr. 1993;645:241–250. doi: 10.1016/0021-9673(93)83383-4. [DOI] [PubMed] [Google Scholar]
- 39.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 40.Theodore Peters J. In: All About Albumin: Biochemistry, Genetics, and Medical Applications. Theodore Peters J, editor. Academic Press Limited; San Diego, CA: 1996. pp. 36–38. [Google Scholar]
- 41.Natarajan SK, Assadi M;, Sadegh-Nasseri S. J. Immunol. 1999;162:4030–4036. [PubMed] [Google Scholar]