Abstract
Interleukine-3 (IL-3) binds its receptor and initiates a cascade of signaling processes that regulate the proliferation and differentiation of hematopoietic cells. To understand the detailed mechanisms of IL-3 induced receptor activation, we generated a homology model of the IL-3:receptor complex based on the closely related crystal structure of the GM-CSF:receptor complex. Model-predicted interactions between IL-3 and its receptor are in excellent agreement with mutagenesis data, which validate the model and establish a detailed view of IL-3:receptor interaction. The homology structure reveals an IL-3:IL-3 interaction interface in a higher-order complex modeled after the dodecamer of the GM-CSF:receptor complex wherein an analogous GM-CSF:GM-CSF interface is also identified. This interface is mediated by a proline-rich hydrophobic motif (PPLPLL) of the AA′ loop that is highly exposed in the structure of isolated IL-3. Various experimental data suggest that this motif is required for IL-3 function through receptor-binding independent mechanisms. These observations are consistent with structure-function studies of the GM-CSF:receptor complex showing that formation of the higher-order cytokine:receptor complex is required for signaling. However, a key question not answered from previous studies is how cytokine binding facilitates the assembly of the higher-order complex. Our studies here reveal a potential cytokine–cytokine interaction that participates in the assembly of the dodecamer complex, thus linking cytokine binding to receptor activation.
Introduction
IL-3 and the closely related short-chain cytokine GM-CSF and IL-5 are a subclass of the cytokines/growth factors secreted largely by activated T cells [1]. These cytokines regulate the survival, proliferation and differentiation of hematopoietic cells, while GM-CSF has also been shown to regulate the function of dentritic cell and T cells [2], [3]. Dysfunction of this family of cytokines has been implicated in a variety of pathologies including rheumatoid arthritis and leukemia [4]–[8], whereas treatment with recombinant cytokine of this family showed beneficial effect in a number of clinical conditions [9]–[12], making them important targets of therapeutic development.
IL-3, GM-CSF and IL-5 function by binding to their cognate receptors expressed on the surface of cells in the hematopoietic system. The receptors for IL-3, GM-CSF and IL-5 consist of a cytokine-specific alpha subunit and a common beta subunit (βc) that are both required for function [13]–[17]. The mechanism by which cytokine binds and activates its receptor has been the subject of extensive structural and functional studies [18]. As first revealed in the human growth hormone receptor complex structure [19], a general theme from these studies is that the binding of cytokine induces dimerization of receptor subunits, which brings signaling molecules (e.g. kinases) associated with the cytoplasmic region of receptor subunits in close proximity for trans-activation. However, higher-order cytokine:receptor complexes have been implicated in receptor-mediated signaling [20], and it becomes increasingly clear that each cytokine:receptor pair has unique features in binding and activation mechanism. Comparison of related cytokine:receptor complexes have been a powerful way to explore the mechanisms of cytokine signaling [20]–[23].
The binding of IL-3 to its receptor has been investigated extensively using various approaches, including constructing interspecies chimera [24], [25], mapping the epitopes of neutralizing antibodies [24], [26], and mutagenesis [26]–[32]. These studies provided rich biochemical and functional data on sites of IL-3 that are important for receptor binding and signaling activation. A model of IL-3:receptor complex was also generated based on the human growth hormone hGH:receptor complex structure [19], [33]. While this model provided satisfactory interpretations for some of the mutagenesis data, its accuracy is limited by the low homology between the IL3:receptor complex and the hGH:receptor complex. The crystal structure of a GM-CSF:receptor complex has recently been determined [34]. IL-3 has a higher homology to GM-CSF than hGH. Moreover, the receptors for IL-3 and GM-CSF share a common beta subunit. Thus the structure of the GM-CSF:receptor complex offers a much better template for building a more accurate model of the IL-3:receptor complex.
The mechanism by which IL-3, GM-CSF and IL-5 activate their receptors has been a longstanding puzzle. Although the cytoplasmic domain of the receptor alpha subunit is required for function, the signaling kinase (JAK2) seems to bind only the beta subunit that adopts an extended structure. Thus the classical model of cytokine-induced dimerization of receptor subunits cannot explain signal activation for this family of cytokines. Although it remains controversial about which JAK kinases associate with the receptor subunits and whether signaling is initiated by homo- or hetero dimerization, the crystal structure of the GM-CSF:receptor complex reveals a dodecameric assembly that provides a plausible mechanism for receptor activation wherein the tyrosine kinase JAK2 associated with the beta subunit are brought together for trans phosphorylation [15]–[17]. Mutations on the receptor beta subunit designed to disrupt the dodecamer assembly interface diminished the activity of the receptor, providing functional evidence that the higher-order GM-CSF:receptor complex is required for signaling [34].
The dodecameric assembly of cytokine:receptor complex has also been proposed as part of the receptor activation mechanism for IL-3 and IL-5 [34]. Indeed, beta subunits containing mutations that presumably disrupt dodecamer assembly showed decreased ability to mediate IL-3-dependent signaling [34]. However, an intriguing question about this model of receptor activation is the role of the cytokine itself. The protein interface in the GM-CSF:receptor dodecamer shown to be functionally important by mutagenesis is mediated entirely by the beta subunit. How the cytokine itself controls the formation of the dodecamer complex and signaling activation is not clear.
To address this question, we generated a homology model of the IL-3:receptor complex based on the crystal structure of the GM-CSF:receptor complex and took advantage of the vast amount of biochemical data on the IL-3:receptor complex. The excellent agreement between the model and previously published mutagenesis data suggest that the homology structure represents a reliable model for IL-3:receptor interaction. We identified a cytokine-cytokine interaction interface in the modeled structure of the IL-3:receptor complex and in the crystal structure of the GM-CSF:receptor complex that participates in the assembly of the dodecamer. In IL-3 this interface is mediated by a proline-rich and exposed hydrophobic motif (PPLPLL) in the AA′ loop. Some modifications of this motif by mutagenesis and by the binding of neutralizing antibodies have been shown to affect the activity of IL-3 without significantly changing its binding affinity for the receptor [35]. These observations are in agreement with mutagenesis studies of the GM-CSF:receptor dodecamer showing that high affinity binding of cytokine to the receptor is not sufficient for activation and that the formation of the higher-order complex is an obligate step to initiate signaling. Our findings not only provide further support for the dodecamer model of receptor activation in the IL-3:receptor complex, but more importantly, reveal a cytokine-cytokine interaction interface that participates in the assembly of the dodecamer and hence signal initiation. The structural model generated by this study will serve as a basis for further studying the functional mechanisms of the IL-3:receptor complex and for the development of therapeutics.
Materials and Methods
Selection of templates
The crystal structures of the GM-CSF:receptor complex (3.3 Å resolution) [34] and the IL-4:IL-4R:IL-13RA ternary complex (3.02 Å resolution) [23]were used as templates to develop a homology model of human IL-3 (hIL-3) bound to its receptor (the hIL-3:receptor complex). The coordinates for both structures were taken from the Brookhaven Protein Data Bank using the accession numbers 3CXE and 3BPN, respectively. The NMR structure of a functionally active hIL-3 variant SC-65369 (PDB accession number 1JLI) was also taken from the Protein Data Bank to build a structural model for the wild type hIL-3 (14–125). The amino acid sequence of hIL-3 receptor α-chain was obtained from ExPASy Proteomics Server (protein accession number P26951). To select templates for modeling the IL-3 receptor α-chain, we searched for its homologous sequences using BLAST (http://www.ncbi.nlm.nih.gov/blast) against PDB database. It turns out that the two domains of the IL-3 receptor α-chain align optimally to two different protein sequences. The best template (with the highest sequence homology) for domain 1 of IL-3 receptor α-chain is chain C of 3BPN.pdb, whereas that for domain 2 is chain C of 3CXE.pdb.
Homology modeling
Homology models of hIL-3 receptor α-chain domain 1 (Pro103-Ile207) and domain 2 (Leu208-Glu292) were developed based on the crystal structure of IL-13 receptor α-chain (IL-13RA) and GM-CSF receptor (GMR) α-chain domain 2 respectively, using the program MODELLER [36]. The corresponding sequence alignments are shown in Figure S1 and Figure S2. Domain 1 of GMR α-chain is poorly defined in the crystal structure of the GM-CSF:receptor complex[34]. To get the relative inter-domain orientation, a combined model of hIL-3 receptor α-chain (including both domain 1 and 2) was also developed based on IL-13RA (sequence alignment is shown in Figure S3). Because domain 1 and domain 2 of IL-3 receptor α-chain were modeled on different templates, the linker region (residues Phe202 to Pro210) was rebuilt with plausible backbone conformation and side chain rotamers using program O [37]. The wild type hIL-3 model was generated by in silico mutations from its mutant NMR structure using the program O. A total of 14 substitutions were introduced in the IL-3 NMR structure. The side chains of newly introduced amino acid residues were manually adjusted to remove local clashes and to allow potential hydrogen bonding and electrostatic interactions. All the models were then put through energy minimization in MODELLER with fixed backbones.
The hIL-3:receptor complex model
The structure of isolated GM-CSF is similar to its receptor bound form [34], [38]. We assume that the structure of hIL-3 bound to its receptor is also similar to its free from. We therefore docked IL-3 as a rigid body on the GM-CSF:receptor complex by superimposing its structure with that of GM-CSF (Figure 1A). The superposition is based on the backbone of the four conserved helices (A–D) that shows an RMSD of 2.7 Å for 50 C-α atoms (Figure S4). The homology model of hIL-3 receptor α-chain is docked onto the GM-CSF:receptor complex by superimposing its domain 2 with that of GM-CSF receptor α-chains (Figure S5A). Domain 1 of hIL-3 receptor α-chain, which is constructed based on the template of IL-13RA domain1 (Figure S5), is fitted into the ternary complex based on the inter-domain orientation of IL-13RA with minor manual adjustments (see below). GM-CSF and its receptor alpha chain were then removed from the combined coordinates. With the remaining common β-chain, hIL-3 and the receptor alpha chain were therefore placed together using the molecular framework defined by the crystal structure of the GM-CSF:receptor complex (Figure 1B). The complex model was subject to energy minimization with fixed backbone using MODELLER. A few local clashes in the final model were removed manually in O by choosing different side chain rotamers. Throughout the modeling process, only domain 1 and domain 4 from two different hIL-3 receptor β-subunits that are in contact with IL-3 were included in the model refinement procedures. A multiple sequence alignment among IL-3, GM-CSF, and IL-5 is shown in Figure 1C with residues discussed in the text highlighted. Overall, the homology model is based on the identical beta subunit and closely related cytokine structures between IL-3 and GM-CSF. Although the accuracy of the detailed hIL-3 receptor α-chain structure may be limited by the modest sequence identity, the overall arrangement of its two domains in the final complex can be assigned with high confidence, so the role of the receptor α-chain in the assembly of the dodecamer complex can be assessed with reasonable certainty (see below).
Figure 1. Modeling of the IL-3:receptor complex.
(A) The structure of IL-3 (red) is superimposed on that of GM-CSF (yellow). The IL-3 receptor alpha chain domain 2 (α2, blue) is superimposed on the corresponding domain of GM-CSF receptor alpha chain (light magenta). The position of IL-3 receptor alpha chain domain 1 (α1, blue) is fitted as described in text. Domain 1 of one beta subunit (β1) and domain 4 of the other (β4′) are shown in green. The rest of the beta dimer is not shown. The three interfaces of the complex discussed in the text are labeled. The color scheme is used throughout the illustration unless otherwise indicated; (B) The resulting model of the IL-3:receptor complex after removing GM-CSF and its receptor alpha subunit, shown in ribbon covered with transparent surface; (c)sequence alignment of IL-3, GM-CSF and IL-5. Helices in IL-3 are indicated by horizontal bar in red. Residues discussed in the text are highlighted in different colors: green shade, residues interacting with the receptor β-subunit in IL-3 and GM-CSF; blue shade, residues of IL-3 that interact with the receptor α-subunit; magenta shade, residues of GM-CSF that interact with the receptor α-subunit; red font, functionally important residues from mutagenesis studies. Boxed residues are involved interface V interaction. Residue numbering above the sequence is for IL-3.
Results
Overall structure of the IL-3:receptor complex
The canonical mode of cytokine/receptor interaction
IL-3 is bound to the receptor through interfaces with the alpha subunit (Interface I) and beta subunit (interface II). Upon binding to their cytokine ligand, the alpha and beta subunit also interact with each other through a binding interface termed interface III (Figure 1A and 1B). The buried solvent accessible surface area between IL-3 and its receptor is 2267 Å2, which is comparable to that of the GM-CSF:receptor complex. Domain 2 of IL-3Rα shows notable difference from the corresponding domain of GMRα (Figure 1A). Although most of the residues inside the beta sandwich core are conserved (Figure S5A), the loop region (residues Gln241–Gln247) involved in binding to the beta subunit and several other loops (residues Arg229-Lys235 and residues Arg275-Phe281, here residues are numbered according to hIL-3Rα) show significant divergence from GMRα. These loops of IL-3Rα are substantially shorter than their counter parts in GMRα (Figure 1A, Figure S5A), their structure and trajectory are highly constrained by the beta sandwich core. Thus, the roles of these loops in binding with the beta subunit and the cytokine can be assessed with reasonable certainty. Domain 1 of IL-3Rα also likely has some differences from the corresponding domain of GMRα. In the crystal structure of the GM-CSF:receptor complex, domain 1 of GMRα is poorly defined, suggesting its relatively high flexibility in the complex. In the homology model of the IL-3:receptor complex, the placement of hIL-3Rα domain 1 is guided by the inter domain orientation of IL-13Rα The final position of hIL-3Rα domain 1 is also slightly adjusted as a rigid body to account for potential IL-3:IL-3Rα interactions defined by mutagenesis studies (see below).
Higher-order structure of the IL-3:receptor complex
We applied symmetry operations derived from the GM-CSF:receptor crystal to generate both hexamer and dodecamer of the hIL-3:receptor complex (Figure 2A and 2B). The resulting higher-order complex shows that the components of the dodecamer fit nicely with each other. A unique feature of the receptors for GM-CSF, IL-3 and IL-5 is that the beta subunit forms an intertwined dimer [39]. In vitro binding studies suggest that domain 1 and domain 4 from different beta subunits act together to bind the cytokine ligand [40], [41]. The crystal structure of the GM-CSF:receptor complex reveals that this is indeed the case. Domain 1 of one beta subunit and domain 4 of the other form a composite surface to bind GM-CSF at interface II, such that two cytokines and two GMRα chains are cross linked by the beta dimer so as to form a 2∶2∶2 hexamer (Figure 1 in Hansen et al.[34]). In the crystal of the GM-CSF:receptor complex, two hexamers pack head-to-head together to form a dodecamer wherein domain 4 of two adjacent beta subunits contact each other through an extensive protein interface (Figure 3 in Hansen et al. [34]). This interface, referred to as interface IV, has been shown by mutagenesis and neutralizing antibodies to be important for receptor activation. The same interface is also present in the modeled IL-3:receptor complex (Figure 3A). As discussed below, we identified another protein-protein interaction interface involved in the assembly of the GM-CSF:receptor dodecamer (Figure 3B). This interface, termed interface V, is mediated by the loop region between helix A and helix B of GM-CSF bound to two different hexamers. This region of GM-CSF contains a short alpha helix that forms the major part of the interface. In the modeled hIL-3:receptor complex, the corresponding region of IL-3, the AA′ loop, is similarly positioned to form an analogous interaction interface in the dodecamer (Figure 3A and 3C).
Figure 2. Higher-order structure of the IL-3:receptor complex.
(A) Assembly of one trimeric IL-3 (red), IL-3Rα (blue), and IL-3Rβ (green) cytokine/receptor complex with the other (pink, marine and forest) into the hexamer. The dodecameric partners at each end are shown in magenta (IL-3), orange (IL-3Rβ) and cyan (IL-3Rα, domain 1 is omitted in this view), with the interaction interface indicated by a dashed box. (B) Side view of the complex showing the arched structure. Only the foreground structure is shown in this view.
Figure 3. A cytokine–cytokine interaction interface in the dodecamer.
(A) A ribbon/surface presentation of interface IV and V (dashed boxes) in the modeled IL-3:receptor complex. Only domain 4 of the interaction beta subunit and IL-3 are shown in this view. The two residues at interface IV (Glu359 and Lys362) that engage in potential charge repulsion are indicated. The arrows indicate the two fold symmetry axis that runs through both interface IV and V. (B) A top view of interface V in the crystal structure of the GM-CSF:receptor complex (dashed box) and its relative position with respect to other components of the complex. The receptor alpha subunit domain 1 showed only partial density of a few beta strands that are represented by backbone trace here. (C) A top view of interface V (dashed box) mediated by the AA′ loop in the modeled IL-3:receptor complex.
Comparing the IL-3:receptor structure model with biochemical data
In the following sections, we will analyze the modeled structure in the context of biochemical data to gain insights into the molecular details of IL-3:receptor interactions. We also seek to evaluate the dodecamer model of receptor activation through structure-function analysis of the higher-order IL-3:receptor complex.
Protein interfaces involved in IL-3 binding
IL-3 binds the IL-3R alpha subunit with modest affinity through interface I but not to the beta subunit alone. Upon IL-3 binding to the alpha subunit, the beta subunit dimer is recruited to the complex through interfaces II and III (Figure 1A and 1B). These interfaces act cooperatively to stabilize the overall complex, resulting in high affinity binding of the cytokine ligand. We will discuss interface II first because it is constructed with minimal model manipulation. This interface is defined by the common beta subunit and by the experimentally derived IL-3 structure superimposed on GM-CSF as a rigid body. At interfaces I and III, on the other hand, loops of IL-3R alpha subunits required local adjustments to dock onto IL-3 and the beta subunit.
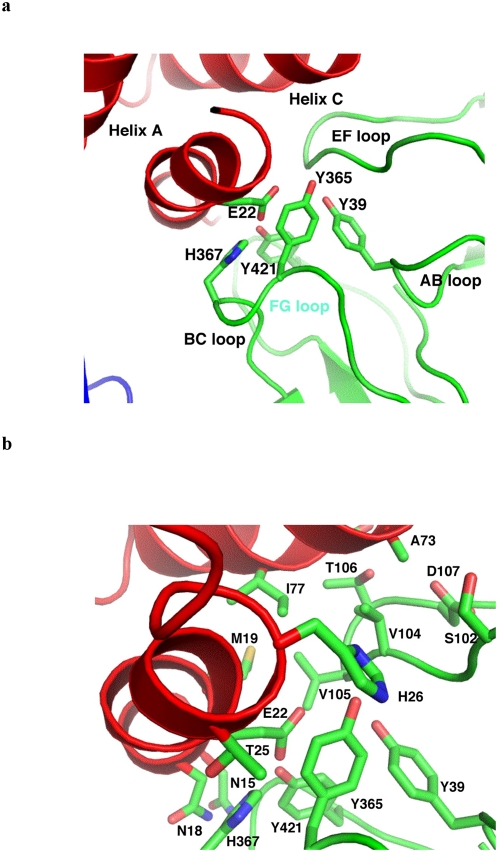
At interface II (Figure 4), helix A and C of IL-3 bind a composite protein surface formed by the AB and EF loops of domain 1 of one beta subunit and the BC and FG loops of domain 4 of the other. A prominent feature of interface II is the intimate interaction between Glu22 (residues in IL-3 are italicized throughout the text) of IL-3 and a group of aromatic residues of the beta subunit, including Tyr39, Tyr365, His367 and Tyr421 (Figure 4A). In addition to extensive van der Waals contacts, the carboxylate group of Glu22 is in position to form a hydrogen bond with the phenolic hydroxyl group of Tyr421. An adjacent patch of interactions involves Met19, Ser76, Ile77 and Ala73 of IL-3 that pack against Ser102, Val104, Val105 and Thr106 on the beta subunit. Asn15, Asn18, Thr25, and His26 of IL-3 are also in close proximity to contact the beta subunit (Figure 4B). The detailed interactions at interface II are similar to those seen in the GM-CSF:receptor complex but also show some differences. For example, a salt bridge between Lys72 of GM-CSF and Asp107 of the beta subunit is missing at interface II of the IL-3:receptor complex, whereas Ile77 of IL-3 may bind the hydrophobic pocket formed by Ser102, Val104, Val105 and Thr106 of the beta subunit better than the proline counterpart (Pro76) in GM-CSF. The structural features of interface II are consistent with mutagenesis data on the functional roles of interface residues (Figure 1C). For example, the tight packing interaction at the center of the interface is highly sensitive to mutations of Glu22 of IL-3 and Tyr421 of the beta subunit [30], [31], [33], [42], [43]. Even subtle changes, such as substituting Tyr421 with phenylalanine in the beta subunit, abolish the high affinity binding of IL-3 and GM-CSF to the receptor.
Figure 4. Protein–protein interactions at interface II.
(A) A zoom-out view of the interface showing the secondary structural elements of IL-3 and the receptor involved in the reaction. The interaction between Glu22 of IL-3 and a number of aromatic residues from the receptor beta subunit (Tyr39, Tyr365, His367, Tyr421) is shown. (B) A zoom-in view showing additional details of interface II, including the interactions between Met19, Ile77 and Ala73 of IL-3 and Ser102, Val104, Val105 and Thr106 on the receptor beta subunit.
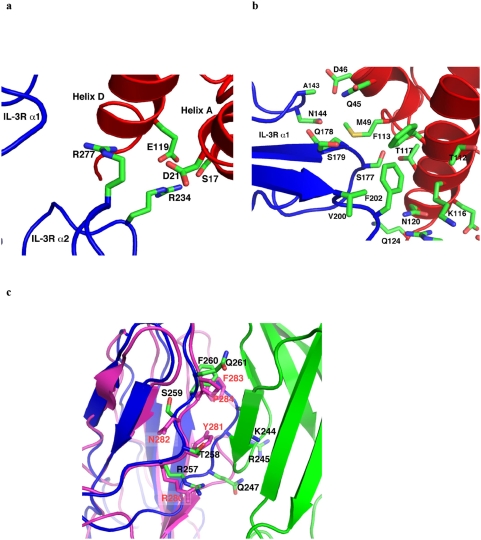
At interface I, helix A, A′ and D of IL-3 bind domain 1 and domain 2 of IL-3R alpha through two distinct patches of protein-protein interactions, one dominated by electrostatic interactions while the other rich in hydrophobic/van der Waals contacts. The charge interaction patch is formed between helix A of IL-3 and domain 2 of IL-3R alpha, where Asp21 and Glu119 of IL-3 are in position to engage in electrostatic interaction with Arg277 and Arg234 of IL-3R alpha, respectively (Figure 5A). Arg234 of IL-3R alpha is also in position to interact with Ser17 of IL-3 through hydrogen bonding. For the hydrophobic patch, residues located on various loops of IL-3R alpha domain 1, including Val200, Phe202, Ser177–Ser179, and Ala143-Arg146, project toward the cytokine to interact with Gln124, Asn120, Thr117, Lys116, Phe113, Gly42, Gln45, Asp46, and Met49 on helix D and A′ of IL-3. Most notably, Phe202 of IL-3R alpha is poised to insert into a hydrophobic pocket formed by Phe113, Thr112 and the aliphatic side chain of Lys116 on IL-3 (Figure 5B). Consistent with the structure model, a number of IL-3 residues located at interface I, including Ser17, Asn18, Asp21, Arg108, Phe113, Lys116 and Glu119 have been shown to be important for receptor binding and activation by mutagenesis [29], [30], [32] (Figure 1C). The loss of function for mutations Ser17Lys, Asn18Lys, Thr25Arg and Glu119Arg of IL-3 could be explained by charge repulsion with Arg234 and Arg277 of IL-3R alpha [32], while that for Phe113A could be explained by diminished hydrophobic interactions with Phe202 of IL-3R alpha. Interestingly, a number of IL-3 mutations, such as Lys116Val, Gln45Val, Lys116Trp, and Thr112Arg, showed enhancement in receptor binding and/or cellular activities [29], [31], [32]. Remarkably, these residues are located around Phe113 in the folded structure of IL-3. Substitution of these hydrophilic residues with hydrophobic residues or residues with longer aliphatic side chain could therefore augment the hydrophobic interaction at interface I and hence enhance the activity of mutated IL-3.
Figure 5. Protein–protein interactions at interfaces I and III.
(A) A charged interaction patch between helix A of IL-3 and domain 2 of IL-3R alpha. Here Asp21 and Glu119 of IL-3 engage in electrostatic interaction with Arg277 and Arg234 of IL-3R alpha, respectively. (B) A hydrophobic interaction patch between IL-3 and IL-3R alpha domain 1. Note that Phe202 of IL-3R alpha inserts into a hydrophobic pocket formed by Phe113, Thr112 and the aliphatic side chain of Lys116 on IL-3. (C) Foreground: the loop region between Arg257 and Gln261 of IL-3R alpha (blue) is very similar to the corresponding region of GMR alpha (magenta) in sequence, structure and interaction with beta-subunit (green). Background: residues Lys244, Arg245 and Gln247 of IL-3R alpha also contact the beta subunit.
At interface III, the loop region of Arg257-Gln261 in IL-3R alpha, which forms a major part of the binding site of beta domain 4, is very similar to the corresponding region of GMR alpha in sequence, structure and interaction with beta-subunit (Figure 5C). On the other side of the beta sandwich (Figure 5C), residues Lys244, Arg245 and Gln247 of IL-3R alpha are also involved in beta subunit binding. This part of interface III in the IL-3:receptor complex is different from that of the GM-CSF:receptor complex because the loop region following the β-strand Lys235-Leu240 is much shorter in IL-3Rα than in GMRα (Figure S5A). Overall, interface III in the IL-3:receptor and GM-CSF:receptor complexes show significant similarity as well as some differences.
Protein–protein interactions involved in the assembly of the dodecamer
A dodecamer complex is formed in the crystal of the GM-CSF:receptor complex [34]. Interactions between domain 4 of two adjacent beta subunits belonging to two different hexamers constitute a significant part of the protein interface (interface IV) in the assembly of the dodecamer. Residues on several beta strands of domain 4, including Gln346 - Asp352, Arg357, Glu359, Lys362 and Arg432-Asp435, are in position to contact each other at this interface [34]. These interactions are also conserved in the IL-3:receptor dodecamer complex (Figure 3A), which may also be involved in receptor activation. Consistent with this hypothesis, extensive mutations at interface IV not only reduced the activity of the GM-CSF:receptor complex but also that of the IL-3:receptor complex [34].
In the crystal structure of the GM-CSF:receptor complex, Asn261 of the α subunit domain 2 of one hexamer makes a small contact to Glu430 and Arg 432 of the beta subunit domain 4 of the other hexamer (Figure S6). The contribution of this interaction to the dodecamer assembly is likely small due to the limited contact. Moreover, it is absent in the modeled hIL-3:receptor dodecamer complex because IL-3R alpha has a much shorter loop in this region than GMR alpha (Figure S5A). These observations raise the question on how cytokine controls the assembly of the dodecamer.
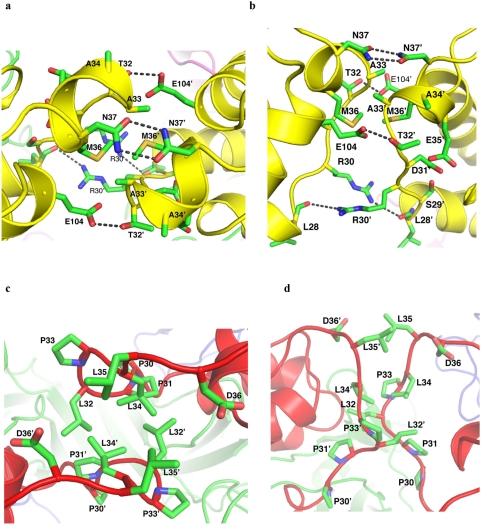
Further examination of the GM-CSF:receptor crystal reveals that another protein-protein interaction interface, mediated by the AB loop of GM-CSF, forms an integral part of the dodecamer (Figure 3B, Figure 6A and 6B). This interface, termed interface V, is located directly above interface IV and shares the same twofold symmetry axis with interface IV (Figure 3A). These structural features suggest that interface V and IV may form concertedly and act cooperatively in the assembly of the dodecamer. Interface V in the GM-CSF:receptor complex buries about 646 Å2 solvent accessible surface area that includes residues Leu28-Asn37 and Glu104 from two interacting GM-CSF molecules. The interface is mediated by two short helices and has a hydrophobic core consisting of Ala33, Met36, Ala33′ and Met36′ (the prime sign denotes residues from symmetry partner), which is surrounded by extensive van der Waals contacts and hydrogen bond interactions (Figure 6A). For example, Arg30 forms a hydrogen bond with the main chain carbonyl of Leu28′, Thr32 forms a hydrogen bond with Glu104′, while Asn37 form a bidendate hydrogen bond with Asn37′ (Figure 6B). These structural features suggest that interface V has excellent shape and chemical complementarity and likely makes significant contribution to the dodecamer assembly. It is interesting to note that interface V in GM-CSF is bounded by two glycosylation sites, Asn27 and Asn37, suggesting that the sugar chain attached at the periphery of interface V could augment the protein-protein interaction and hence the dodecamer stability [44], [45].
Figure 6. Detailed protein–protein interactions at interfaces V.
(A) Detailed GM-CSF:GM-CSF interactions in the crystal structure of the GM-CSF:receptor dodecamer (top view); (B) side view of (A); (C) Detailed IL-3:IL-3 interactions in the modeled structure of the IL-3:receptor dodecamer (top view); (D) side view of (C).
In the modeled IL-3:receptor complex, a similar interface is formed between the AA′ loop of IL-3 bound to two opposing hexamers (Figure 3A and 3C). Both IL-3 and GM-CSF have an extended loop between helix A and B but share little sequence conservation in this region (Figure 1C), suggesting that interface V can form in the two family members but not between them. The AA′ loop of IL-3 contains a characteristic PPLPLL motif (residues 30–35 in human IL-3). The exposed and hydrophobic nature of this motif suggests that the AA′ loop has a high tendency to bind other hydrophobic protein surface. Here in the IL-3:receptor dodecamer complex, the PPLPLL motifs from two opposing IL-3 molecules are poised to interact with each other (Figure 6C and 6D). Although the detailed interactions at interface V of IL-3 await direct experimental determination, our modeling suggest that Leu32, Leu34 and Leu35 are in positions to interact with their counterparts in the symmetric partner, much like the leucine zipper interaction observed in many protein complexes. However, unlike classical leucine zippers where leucine residues are presented by alpha helix, the conformation of the AA′ loop of IL-3 seems to be partially restricted by a series of proline residues, which may be required to reduce non-specific hydrophobic interactions.
Though far away from the receptor binding sites, the PPLPLL motif (Pro30-Leu35) has been shown to be a functionally important site on IL-3 by a number of experiments. In mapping functional sites of IL-3 using monoclonal antibodies that bind IL-3 and inhibit its activity, Lokker et al. showed that the epitopes of two neutralizing antibodies were located between Leu32 and Asp36 [26], [35]. Deletion of a significant portion of this region, such as Pro30-Leu33 (del-Pro30-Leu33) and Pro30-Leu34 (del-Pro30-Leu34), diminished the activity of IL-3. Site-specific mutations of residues in this region affected the activity of IL-3 both positively and negatively. While the Leu34Glu mutation showed little effect on the function of IL-3, the Leu34Gly mutant showed much reduced activity than the wild type IL-3. On the other hand, substitution of Pro33 with Asn (Pro33Asn) and Gly (Pro33Gly) enhanced the specific activity of IL-3. Gain-of-function mutations of IL-3 have also been identified in the PPLPLL motif in a saturation mutagenesis study [31], where Leu32Arg, Leu34Ser and Leu34Met showed increased activity. Many of the loss-of-function mutations such as del-Pro30-Leu33, del-Pro30-Leu34, and Leu34Gly also showed decreased affinity for the receptor [35]. How these modifications affect the binding of IL-3 to its receptor is not clear since the AA′ loop does not make direct contact to either subunits of the receptor in the modeled structure (Figure 3A and 3C). While these negative results seem to contradict the model of interface V mediated signal activation through dodecamer assembly, it must be kept in mind that loss-of-function of mutations often involve complex but trivial mechanisms, such as protein mis-folding or aggregation, especially considering the hydrophobic nature of the AA′ loop (see discussion below). By contrast, the gain-of-function mutations can often be much more mechanistically revealing. Here these mutations enhanced the specific activity of IL-3 substantially with little to modest effect on the binding affinity for the receptor. Most notably, the Pro33Gly mutant showed 14-fold increase in activity but virtually the same receptor affinity as compared with the wild type protein in cell-based assays, indicating that modifications in the PPLPLL motif of the AA′ loop can affect the function of IL-3 through mechanisms independent of receptor binding. Animal studies of an IL-3 triple mutant (Pro33Gly/Trp104Glu/Asn105Asp) also showed that modifications of the PPLPLL motif and its nearby regions could enhance the specific activity of IL-3 significantly under physiological conditions (Kunmei Ji and Zhigang Liu, data not shown). Mutations on interface IV have also been shown to disrupt the function of the receptor without affecting the binding of the cytokine [34]. These observations suggest that interface V identified here contribute to the activation of the receptor through a similar mechanism to interface IV, namely the assembly of the dodecameric cytokine:receptor complex (Figure 7).
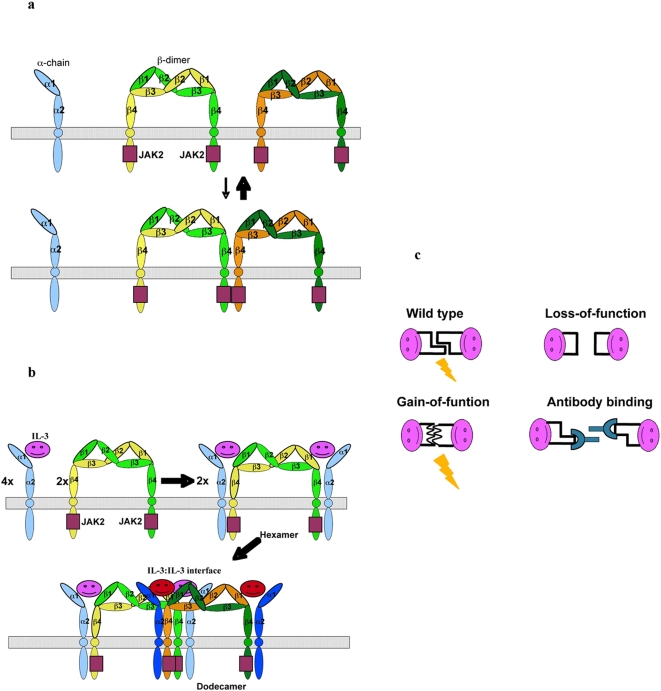
Figure 7. Activation mechanism of the IL-3:receptor complex.
(A) In the absence of cytokine, the receptor subunits are scattered on the cell surface, where the beta dimer, which binds JAK2 through its cytoplasmic region, form transient tetramer through interface IV that may contribute to residual phosphoryaltion of JAK2. (B) In the presence of IL-3, the cytokine and receptor subunits assembly into a dodecamer that is stabilized by IL-3:IL-3 interaction, leading to robust signal activation through JAK2 trans-phosphorylation. (C) A variety of experimental data showed that modifications of the AA′ loop of IL-3, including deletion, loss-of-function mutations, and antibody binding, diminished the activity IL-3, whereas gain-of-function mutations are also frequently observed in this region that enhanced the IL-3 without affecting the receptor binding affinity.
Discussion
The homology model of the IL-3:receptor complex generated in this study provides a molecular framework to analyze a large body of mutagenesis data on the roles of specific residues in IL-3:receptor complex formation and function. These analyses reveal three prominent features at the IL-3:receptor interface (Figures 1, 4, and 5). First is the tight interaction between Glu22 of IL-3 and a group of aromatic residues on the receptor beta subunit. This patch of interaction is highly sensitive to mutations, suggesting its critical role in the binding of IL-3 to the receptor [30], [31], [33], [42], [43]. The second feature is a perfectly matched charge interface between Asp21 and Glu119 of IL-3 and Arg277 and Arg234 of IL-3R alpha. This feature is consistent with a series of mutational studies showing that Ser17Lys, Asn18Lys, Thr25Arg and Glu119Arg can severely disrupt the function of IL-3, presumably by introducing charge repulsion with Arg277 and Arg234 of IL-3R alpha [29], [30], [32]. The third feature is a hydrophobic interface formed between Phe202 of IL-3R alpha and a hydrophobic pocket lined by Phe113, Thr112 and the aliphatic side chain of Lys116 on IL-3. This feature provides potential interpretations for the loss-of-function mutation of Phe113Ala as well as the gain of function mutations of Lys116Val, Gln45Val, Lys116Trp, and Thr112Arg, which may decrease or increase the hydrophobic interaction, respectively [29], [31], [32].
A large number of IL-3 residues, including some at the IL-3:receptor interface, show high tolerance to mutations [31], [33]. This fact does not necessarily indicate poor quality of the structure of the IL-3:receptor complex generated by homology modeling. It is very common that interactions observed in experimentally derived structures may or may not have energetic contribution to binding and function. As shown in this study, structure-based analysis of specific mutations that alter the binding and function with defined chemical changes is needed to understand the functional mechanism of protein-protein interactions between IL-3 and its receptor. Overall, the homology model of the IL-3:receptor complex is in excellent agreement with previously published mutagenesis data, thus providing a reliable framework for analyzing the IL-3:receptor interaction.
The beta subunit of IL-3, GM-CSF, and IL-5 receptors forms a stable dimer [34], [39], where a cytokine-binding site is located at each end of the extended dimer structure. Such an unusual structural feature leads to the assembly of the cytokine:receptor hexamer when two cytokine:receptor alpha subunit complexes are recruited to the beta dimer (Figures 2 and 7). Although the cytoplasmic domain of the receptor alpha chain is required for function [13], [14], it does not appear to be associated with any signaling kinases [15]–[17], [34]. The cytoplasmic region C-terminal to domain 4 of the beta subunit, on the other hand, has been shown to bind JAK2 that is capable of initiating the signaling cascade [34]. However, the membrane proximal domain (domain 4) of the beta subunit in the 2∶2∶2: hexamer is 120 Å apart, raising the question on how trans-phosporyaltion of JAK2 and signal activation are achieved in the cytokine:receptor complex.
The crystal structure of the GM-CSF:receptor complex provides a potential answer to this question. In the crystal lattice, two GM-CSF:receptor hexamers stack against each other to form a dodecamer, which brings domain 4 of two beta subunits from different hexamers in close proximity. Initial support for this model of dodecamer-dependent activation came from mutagenesis analysis of a protein interface (interface IV) between domain 4 of two adjacent beta subunits (Figure 3A). However, a puzzling aspect of this model is how the assembly of the dodecamer, which is apparently required for receptor activation, is coupled to cytokine binding to the receptor, which must be the initiation event of signaling. Interface IV is mediated entirely by the beta subunit, suggesting that it could form stochastically and transiently without the binding of cytokine. In this regard it is interesting to note that low level phosphorylation of JAK2 does occur in the absence of cytokine (see Figure 4C in Hansen et al. [34]) (Figure 7). The protein-protein interactions at interface IV are largely polar without significant chemical complementarity in hydrogen bonding and electrostatic interaction. In fact, a number of charged residues such as Glu359 and Lys362 seem to engage in electrostatic repulsion (Figure 3A), which may play a role in reducing background signaling. These structural features suggest that interface IV will not form stably in solution by itself. However, its structural integrity is still important for function as extensive but not limited mutations at this interface diminished the activity of the receptor. In the event of cytokine-mediated signaling, interface IV will most likely work with other protein interaction interfaces that provide the driving force for the dodecamer assembly and signaling activation.
One such interface is a cytokine-cytokine interaction identified in this study, which we termed interface V. In the crystal structure of the GM-CSF:receptor complex, the AB loop of GM-CSF bound to adjacent hexamers contact each other through a significant binding interface (Figure 3B, Figure 6A and 6B). Although no functional data is currently available for the AB loop of GM-CSF, this interface could contribute to the dodecamer assembly and receptor activation. In the modeled IL-3:receptor complex, the AA′ loop of two adjacent IL-3 molecules are similarly positioned to form an interaction interface through the PPLPLL motif (Figure 3C, Figure 6C and 6D). Here a large body of data is available to assess the functional roles of this motif in IL-3. While the importance of the PPLPLL motif in IL-3 has long been demonstrated by neutralizing antibodies and by mutagenesis [26], [35], the observation that a number of mutations in this region can increase the specific activity of IL-3 without changing the binding affinity of the receptor has been puzzling [35]. In the dodecamer of the GM-CSF:receptor complex, mutations at interface IV have also been shown to disrupt the function of the receptor without changing the binding of IL-3 and GM-CSF [34]. These observations suggest that interface V, which is mediated entirely by cytokine-cytokine interactions, could contribute to the receptor activation through the assembly of the dodecamer (Figure 7).
The AA′ loop of IL-3 is conserved between human, chimapanzee and gibbon but shows variations across a range of other species including rhesus, tamarin, marmoset, sheep, bovin, mouse and rat. This region of IL-3 is also tolerant to a series of mutations [31], suggesting that sequence conservation in this region is not absolutely required for maintaining interface V interactions. Because the AA′ loop makes few contacts to the main body of the IL-3 structure, it is highly exposed and flexible. Interactions between the AA′ loop from two adjacent IL-3 molecules most likely involve co-folding. Such a mechanism of interaction will make interface V more tolerant to mutations than protein-protein interaction mediated by rigid structures, as long as the mutations, which will occur simultaneously on both sides of the interface, permit the co-folding of the modified AA′ loop. This mechanism of interaction may also explain why a large number of gain-of-function mutations could be generated by modifying the AA′ loop (Figure 1C and Figure 7C). In the context of natural cytokine:receptor complex, sequence divergence and the co-folding mechanism at interface V would provide a way to confer specie-specific activity of IL-3 or member-specific function in the IL-3, GM-CSF, IL-5 family. In this regard, it is interesting to note that IL-5 forms a stable dimer [46]. Although the IL-5 dimer interface is incompatible with interface IV observed in the GM-CSF:receptor dodecamer [34], it may represent a cytokine-cytokine interaction that has evolved into a stable form for function, probably through the assembly of a different type of higher-order cytokine:receptor complex.
In summary, our analysis of the IL-3:receptor complex by homology modeling and published mutagenesis data provides further evidence for the dodecamer-model of receptor activation proposed for GM-CSF and IL-3 [34]. Our studies suggest that the AA′ loop in IL-3 and the analogous region in GM-CSF participate in the assembly of the dodecamer. Such a revised model of dodecamer-dependent activation solves the paradox in the previous model and provides a mechanism by which cytokines control the activation of receptor-mediated signaling (Figure 7).
Supporting Information
Sequence alignment between domain 1 of IL-3 receptor alpha chain (IL3R-α1) and the corresponding region in IL-13 (IL13R-α). The arrows above the sequence denote regions of beta strand. Identity and similarity are indicated by standard conventions below the sequence. These conventions are used throughout the illustration. The sequence between residue 176 and 177 in IL-3 corresponds a large loop insertion in IL-13R alpha chain (residues192–199).
(1.56 MB TIF)
Sequence alignment between domain 2 of IL-3 receptor alpha chain (IL3R-α2) and the corresponding region in GM-CSF receptor alpha chain (GMR-α2).
(1.56 MB TIF)
Sequence alignment between the full length IL-3 receptor alpha chain (IL3R-α) and IL-13 receptor alpha chain (IL13R-α).
(1.56 MB TIF)
The structure of IL-3 (red) is superimposed on that of GM-CSF (yellow) as a rigid body. Representative residues involved in the core packing of the helix bundle are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled. The AA′ loop of IL-3 and the AB loop of GM-CSF are indicated.
(1.56 MB TIF)
(A) Superposition of domain 2 of IL-3 receptor alpha chain (IL-3R-α2, blue) with the corresponding domain of GM-CSF receptor alpha chain (GMR-α2, magenta). Representative residues involved in the packing of the beta sandwich core are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled. The two long loops in GMR-α2 located at the end of the beta barrel correspond to much shorter loops in IL-3. (B) Superposition of domain 1 of IL-3 receptor alpha chain (IL-3R-α1, blue) with the corresponding domain of IL13 receptor alpha chain (IL13Rα, pink). Representative residues involved in the packing of the beta sandwich core are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled.
(1.55 MB TIF)
In the crystal structure of the GM-CSF:receptor complex, Asn261 of the α subunit domain 2 of one hexamer makes a small contact to Glu430 and Arg 432 of the beta subunit domain 4 of the other hexamer.
(1.56 MB TIF)
Acknowledgments
The authors thank Reza Kalhor, Dr. Aidong Han, and Dr. Yongheng Chen for helpful discussion.
The coordinates of the IL-3:receptor homology model are available from the authors upon request.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported in part by research funding from National Institute of Heart, Lung and Blood to LC (HL076334) and the National Natural Science Foundation of China to KJ and ZL (No. 30471505). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Guthridge MA, Stomski FC, Thomas D, Woodcock JM, Bagley CJ, et al. Mechanism of activation of the GM-CSF, IL-3, and IL-5 family of receptors. Stem Cells. 1998;16:301–313. doi: 10.1002/stem.160301. [DOI] [PubMed] [Google Scholar]
- 2.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 3.Barouch DH, Santra S, Tenner-Racz K, Racz P, Kuroda MJ, et al. Potent CD4+ T cell responses elicited by a bicistronic HIV-1 DNA vaccine expressing gp120 and GM-CSF. J Immunol. 2002;168:562–568. doi: 10.4049/jimmunol.168.2.562. [DOI] [PubMed] [Google Scholar]
- 4.Korzenik JR, Dieckgraefe BK, Valentine JF, Hausman DF, Gilbert MJ. Sargramostim for active Crohn's disease. N Engl J Med. 2005;352:2193–2201. doi: 10.1056/NEJMoa041109. [DOI] [PubMed] [Google Scholar]
- 5.Cook AD, Braine EL, Campbell IK, Rich MJ, Hamilton JA. Blockade of collagen-induced arthritis post-onset by antibody to granulocyte-macrophage colony-stimulating factor (GM-CSF): requirement for GM-CSF in the effector phase of disease. Arthritis Res. 2001;3:293–298. doi: 10.1186/ar318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnbaum RA, O'Marcaigh A, Wardak Z, Zhang YY, Dranoff G, et al. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 7.Ramshaw HS, Bardy PG, Lee MA, Lopez AF. Chronic myelomonocytic leukemia requires granulocyte-macrophage colony-stimulating factor for growth in vitro and in vivo. Exp Hematol. 2002;30:1124–1131. doi: 10.1016/s0301-472x(02)00903-7. [DOI] [PubMed] [Google Scholar]
- 8.Dirksen U, Hattenhorst U, Schneider P, Schroten H, Gobel U, et al. Defective expression of granulocyte-macrophage colony-stimulating factor/interleukin-3/interleukin-5 receptor common beta chain in children with acute myeloid leukemia associated with respiratory failure. Blood. 1998;92:1097–1103. [PubMed] [Google Scholar]
- 9.Sun X, Hodge LM, Jones HP, Tabor L, Simecka JW. Co-expression of granulocyte-macrophage colony-stimulating factor with antigen enhances humoral and tumor immunity after DNA vaccination. Vaccine. 2002;20:1466–1474. doi: 10.1016/s0264-410x(01)00476-5. [DOI] [PubMed] [Google Scholar]
- 10.Fleetwood AJ, Cook AD, Hamilton JA. Functions of granulocyte-macrophage colony-stimulating factor. Crit Rev Immunol. 2005;25:405–428. doi: 10.1615/critrevimmunol.v25.i5.50. [DOI] [PubMed] [Google Scholar]
- 11.Gianni AM, Siena S, Bregni M, Di Nicola M, Peccatori F, et al. Recombinant human interleukin-3 hastens trilineage hematopoietic recovery following high-dose (7 g/m2) cyclophosphamide cancer therapy. Ann Oncol. 1993;4:759–766. doi: 10.1093/oxfordjournals.annonc.a058661. [DOI] [PubMed] [Google Scholar]
- 12.Biesma B, Willemse PH, Mulder NH, Sleijfer DT, Gietema JA, et al. Effects of interleukin-3 after chemotherapy for advanced ovarian cancer. Blood. 1992;80:1141–1148. [PubMed] [Google Scholar]
- 13.Muto A, Watanabe S, Itoh T, Miyajima A, Yokota T, et al. Roles of the cytoplasmic domains of the alpha and beta subunits of human granulocyte-macrophage colony-stimulating factor receptor. J Allergy Clin Immunol. 1995;96:1100–1114. doi: 10.1016/s0091-6749(95)70195-8. [DOI] [PubMed] [Google Scholar]
- 14.Sakamaki K, Miyajima I, Kitamura T, Miyajima A. Critical cytoplasmic domains of the common beta subunit of the human GM-CSF, IL-3 and IL-5 receptors for growth signal transduction and tyrosine phosphorylation. Embo J. 1992;11:3541–3549. doi: 10.1002/j.1460-2075.1992.tb05437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brizzi MF, Zini MG, Aronica MG, Blechman JM, Yarden Y, et al. Convergence of signaling by interleukin-3, granulocyte-macrophage colony-stimulating factor, and mast cell growth factor on JAK2 tyrosine kinase. J Biol Chem. 1994;269:31680–31684. [PubMed] [Google Scholar]
- 16.Quelle FW, Sato N, Witthuhn BA, Inhorn RC, Eder M, et al. JAK2 associates with the beta c chain of the receptor for granulocyte-macrophage colony-stimulating factor, and its activation requires the membrane-proximal region. Mol Cell Biol. 1994;14:4335–4341. doi: 10.1128/mcb.14.7.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lilly MB, Zemskova M, Frankel AE, Salo J, Kraft AS. Distinct domains of the human granulocyte-macrophage colony-stimulating factor receptor alpha subunit mediate activation of Jak/Stat signaling and differentiation. Blood. 2001;97:1662–1670. doi: 10.1182/blood.v97.6.1662. [DOI] [PubMed] [Google Scholar]
- 18.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 19.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 20.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 22.Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA. Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc Natl Acad Sci U S A. 2006;103:2788–2793. doi: 10.1073/pnas.0511161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaushansky K, Shoemaker SG, Broudy VC, Lin NL, Matous JV, et al. Structure-function relationships of interleukin-3. An analysis based on the function and binding characteristics of a series of interspecies chimera of gibbon and murine interleukin-3. J Clin Invest. 1992;90:1879–1888. doi: 10.1172/JCI116065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorssers LC, Burger H, Wagemaker G, De Koning JP. Identification of functional domains of interleukin-3 by construction of primate interspecies chimera. Growth Factors. 1994;11:93–104. doi: 10.3109/08977199409001051. [DOI] [PubMed] [Google Scholar]
- 26.Lokker NA, Strittmatter U, Steiner C, Fagg B, Graff P, et al. Mapping the epitopes of neutralizing anti-human IL-3 monoclonal antibodies. Implications for structure-activity relationship. J Immunol. 1991;146:893–898. [PubMed] [Google Scholar]
- 27.Lokker NA, Zenke G, Strittmatter U, Fagg B, Movva NR. Structure-activity relationship study of human interleukin-3: role of the C-terminal region for biological activity. Embo J. 1991;10:2125–2131. doi: 10.1002/j.1460-2075.1991.tb07746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorssers LC, Mostert MC, Burger H, Janssen C, Lemson PJ, et al. Receptor and antibody interactions of human interleukin-3 characterized by mutational analysis. J Biol Chem. 1991;266:21310–21317. [PubMed] [Google Scholar]
- 29.Lopez AF, Shannon MF, Barry S, Phillips JA, Cambareri B, et al. A human interleukin 3 analog with increased biological and binding activities. Proc Natl Acad Sci U S A. 1992;89:11842–11846. doi: 10.1073/pnas.89.24.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barry SC, Bagley CJ, Phillips J, Dottore M, Cambareri B, et al. Two contiguous residues in human interleukin-3, Asp21 and Glu22, selectively interact with the alpha- and beta-chains of its receptor and participate in function. J Biol Chem. 1994;269:8488–8492. [PubMed] [Google Scholar]
- 31.Olins PO, Bauer SC, Braford-Goldberg S, Sterbenz K, Polazzi JO, et al. Saturation mutagenesis of human interleukin-3. J Biol Chem. 1995;270:23754–23760. doi: 10.1074/jbc.270.40.23754. [DOI] [PubMed] [Google Scholar]
- 32.Bagley CJ, Phillips J, Cambareri B, Vadas MA, Lopez AF. A discontinuous eight-amino acid epitope in human interleukin-3 binds the alpha-chain of its receptor. J Biol Chem. 1996;271:31922–31928. doi: 10.1074/jbc.271.50.31922. [DOI] [PubMed] [Google Scholar]
- 33.Klein BK, Feng Y, McWherter CA, Hood WF, Paik K, et al. The receptor binding site of human interleukin-3 defined by mutagenesis and molecular modeling. J Biol Chem. 1997;272:22630–22641. doi: 10.1074/jbc.272.36.22630. [DOI] [PubMed] [Google Scholar]
- 34.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 35.Lokker NA, Movva NR, Strittmatter U, Fagg B, Zenke G. Structure-activity relationship study of human interleukin-3. Identification of residues required for biological activity by site-directed mutagenesis. J Biol Chem. 1991;266:10624–10631. [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.Rozwarski DA, Diederichs K, Hecht R, Boone T, Karplus PA. Refined crystal structure and mutagenesis of human granulocyte-macrophage colony-stimulating factor. Proteins. 1996;26:304–313. doi: 10.1002/(SICI)1097-0134(199611)26:3<304::AID-PROT6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 39.Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, et al. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 40.Murphy JM, Ford SC, Wiedemann UM, Carr PD, Ollis DL, et al. A novel functional epitope formed by domains 1 and 4 of the human common beta-subunit is involved in receptor activation by granulocyte macrophage colony-stimulating factor and interleukin 5. J Biol Chem. 2003;278:10572–10577. doi: 10.1074/jbc.M211664200. [DOI] [PubMed] [Google Scholar]
- 41.Murphy JM, Ford SC, Olsen JE, Gustin SE, Jeffrey PD, et al. Interleukin-3 binding to the murine betaIL-3 and human betac receptors involves functional epitopes formed by domains 1 and 4 of different protein chains. J Biol Chem. 2004;279:26500–26508. doi: 10.1074/jbc.M402705200. [DOI] [PubMed] [Google Scholar]
- 42.Woodcock JM, Zacharakis B, Plaetinck G, Bagley CJ, Qiyu S, et al. Three residues in the common beta chain of the human GM-CSF, IL-3 and IL-5 receptors are essential for GM-CSF and IL-5 but not IL-3 high affinity binding and interact with Glu21 of GM-CSF. Embo J. 1994;13:5176–5185. doi: 10.1002/j.1460-2075.1994.tb06848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodcock JM, Bagley CJ, Zacharakis B, Lopez AF. A single tyrosine residue in the membrane-proximal domain of the granulocyte-macrophage colony-stimulating factor, interleukin (IL)-3, and IL-5 receptor common beta-chain is necessary and sufficient for high affinity binding and signaling by all three ligands. J Biol Chem. 1996;271:25999–26006. doi: 10.1074/jbc.271.42.25999. [DOI] [PubMed] [Google Scholar]
- 44.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Structural determinants for alpha-neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels (Austin) 2007;1:234–237. doi: 10.4161/chan.4909. [DOI] [PubMed] [Google Scholar]
- 45.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 46.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, et al. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature. 1993;363:172–176. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment between domain 1 of IL-3 receptor alpha chain (IL3R-α1) and the corresponding region in IL-13 (IL13R-α). The arrows above the sequence denote regions of beta strand. Identity and similarity are indicated by standard conventions below the sequence. These conventions are used throughout the illustration. The sequence between residue 176 and 177 in IL-3 corresponds a large loop insertion in IL-13R alpha chain (residues192–199).
(1.56 MB TIF)
Sequence alignment between domain 2 of IL-3 receptor alpha chain (IL3R-α2) and the corresponding region in GM-CSF receptor alpha chain (GMR-α2).
(1.56 MB TIF)
Sequence alignment between the full length IL-3 receptor alpha chain (IL3R-α) and IL-13 receptor alpha chain (IL13R-α).
(1.56 MB TIF)
The structure of IL-3 (red) is superimposed on that of GM-CSF (yellow) as a rigid body. Representative residues involved in the core packing of the helix bundle are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled. The AA′ loop of IL-3 and the AB loop of GM-CSF are indicated.
(1.56 MB TIF)
(A) Superposition of domain 2 of IL-3 receptor alpha chain (IL-3R-α2, blue) with the corresponding domain of GM-CSF receptor alpha chain (GMR-α2, magenta). Representative residues involved in the packing of the beta sandwich core are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled. The two long loops in GMR-α2 located at the end of the beta barrel correspond to much shorter loops in IL-3. (B) Superposition of domain 1 of IL-3 receptor alpha chain (IL-3R-α1, blue) with the corresponding domain of IL13 receptor alpha chain (IL13Rα, pink). Representative residues involved in the packing of the beta sandwich core are shown to demonstrate the similar folding interactions between the two proteins. The residues are colored according to their host proteins. Only residues of IL-3 are labeled.
(1.55 MB TIF)
In the crystal structure of the GM-CSF:receptor complex, Asn261 of the α subunit domain 2 of one hexamer makes a small contact to Glu430 and Arg 432 of the beta subunit domain 4 of the other hexamer.
(1.56 MB TIF)