Abstract
Recent advances in single molecule manipulation methods offer a novel approach to investigating the protein folding problem. These studies usually are done on molecules that are naturally organized as linear arrays of globular domains. To extend these techniques to study proteins that normally exist as monomers, we have developed a method of synthesizing polymers of protein molecules in the solid state. By introducing cysteines at locations where bacteriophage T4 lysozyme molecules contact each other in a crystal and taking advantage of the alignment provided by the lattice, we have obtained polymers of defined polarity up to 25 molecules long that retain enzymatic activity. These polymers then were manipulated mechanically by using a modified scanning force microscope to characterize the force-induced reversible unfolding of the individual lysozyme molecules. This approach should be general and adaptable to many other proteins with known crystal structures. For T4 lysozyme, the force required to unfold the monomers was 64 ± 16 pN at the pulling speed used. Refolding occurred within 1 sec of relaxation with an efficiency close to 100%. Analysis of the force versus extension curves suggests that the mechanical unfolding transition follows a two-state model. The unfolding forces determined in 1 M guanidine hydrochloride indicate that in these conditions the activation barrier for unfolding is reduced by 2 kcal/mol.
In recent studies, both optical tweezers (1, 2) and the cantilever of a scanning force microscope (SFM) (3–6) have been used to induce, record, and characterize the mechanical unfolding of individual protein molecules. These techniques rely on tethering a single polymeric molecule between movable surfaces. A crucial issue in these experiments is to find conditions that facilitate the tethering of the molecule to the surfaces, while keeping the strength of nonspecific interactions between these surfaces themselves to a minimum. This requirement is not difficult to fulfill with long polymers such as DNA (7, 8) or polysaccharides (9), for which the surfaces are too far apart to interact significantly with each other throughout the ranges of forces and extensions of interest. In contrast, manipulating globular proteins or tertiary folded RNAs is prone to error because their small dimensions (typical diameter 1.5–6 nm) require the tethering surfaces to come sufficiently close to interact. For this reason, all mechanical unfolding studies done to date have used proteins that naturally occur as tandem arrays of globular domains (1–5). Naturally occurring polymers, however, are not ideal to investigate protein folding because they often are nonuniform, being composed of globular domains of different size, structure, and stability. Moreover, the tertiary structures of some of these domains have not been characterized, and in some cases even their primary structures are still unknown. Finally, mutants of different structure and stability are not easily available.
Experimental Procedures
Synthesis of Polymers.
To minimize the interactions between the tethering surfaces and simultaneously avoid the inhomogeneity of naturally occurring protein polymers, we have synthesized polymers of bacteriophage T4 lysozyme in the solid state. T4 lysozyme was chosen to demonstrate the method because its structure is known, and its folding-unfolding behavior both for the wild type and a large number of mutants has been extensively investigated by bulk solution methods. After identifying positions at which adjacent molecules contact each other in the crystal lattice, a site-directed mutant was generated in which the corresponding residues were replaced by cysteines while all other cysteines were substituted by structurally neutral residues, such as alanine. This construct is referred to as the CC-mutant, which has the overall designation T21C/C54T/C97A/K124C, and it crystallizes in space group P3221 (10). The refined structure of the mutant reveals that the sulfur atom in the thiol group of residue 21 of a given molecule is 3.5 Å from the sulfur atom in residue 124 of an adjacent molecule related by a 21 screw axis (Fig. 1A). By simple bond rotations these two atoms can be brought to within 2.6 Å. This distance is compatible with disulfide bridge formation because a slight local deformation can place the two sulfur atoms within the distance of 2 Å that is required for a covalent bond. Oxidation of the thiol groups yields polymers in which the lysozyme molecules, connected through disulfide bonds, are arranged in a head-to-tail orientation (Fig. 1B). This procedure circumvents the problems associated with in vivo expression of genes coding for long tandem repeats of globular proteins and produces a higher degree of polymerization than other methods, such as using crosslinking agents (11) or genetic protein engineering (5).
Figure 1.
Crystal structure of T4 lysozyme. (A) The vicinity of the engineered intermolecular disulfide bridge. The atoms shown in the lower half of the figure are from one lysozyme molecule. The atoms at the top (labeled with *) are from a neighboring molecule in the crystal. The superimposed electron density is from a 1.9-Å resolution map with coefficients (2Fo-Fc). Fo corresponds to the observed structure amplitudes. The calculated structure factors, Fc, and phases were from the model refined, assuming that there is no disulfide bridge between Cys-21 and Cys-124*. As can be seen, the electron density suggests that Cys-21 occupies two alternative positions, one of which corresponds to the oxidized and the other to the reduced form. Additional density extending from Cys-124* was modeled in the refinement as a water molecule (shown in red) but could represent a partial adduct with β-mercaptoethanol. The double mutant T21C/K124C/WT* was obtained as described (17) except that 0.01 M β-mercaptoethanol was present in the column and storage buffers. The protein crystals were grown by vapor diffusion at 5°C as hanging drops as described (18) except that 0.14 M β-mercaptoethanol was present during crystallization. X-ray data to 1.8-Å resolution (88% complete) were collected from 5-day-old crystals. The structure was refined to an R factor of 18.5% with bond length and angle discrepancies of 0.019 Å and 2.6°, respectively. The coordinates have been deposited in the Protein Data Bank, ID code 1B6I. (B) Arrangement of three T4 lysozyme molecules within the crystal lattice, related to each other by a 21 screw axis.
Characterization of Polymers.
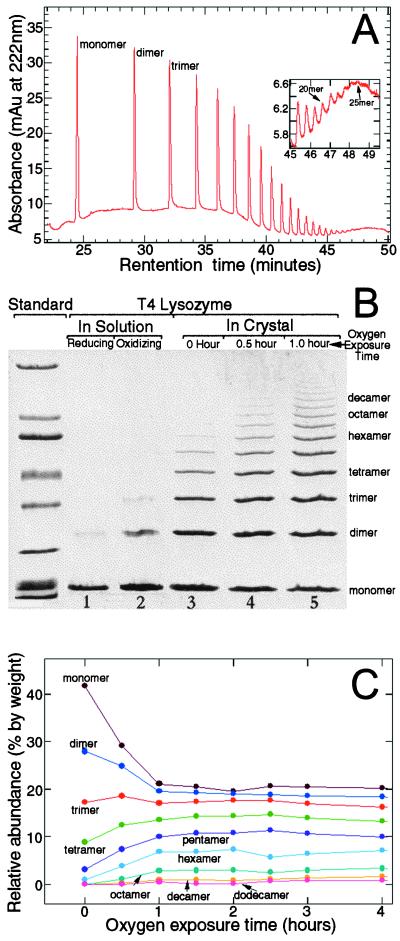
After oxygenation, the polymers were isolated by dissolving the crystals in a low pH buffer (50 mM ammonium acetate, pH 3.0) and removing the unpolymerized monomers. Different techniques then were used to characterize the polymers. Using capillary electrophoresis, polymers up to 25 molecules long have been identified (Fig. 2A). Fig. 2B shows the results of a polyacrylamide gel in which the degrees of polymerization achieved under various conditions are compared. In solution, a reductive environment induces few dimers (Fig. 2B, lane 1); and an oxidative environment induces only a low concentration of dimers and trimers (Fig. 2B, lane 2). In contrast, the favorable arrangement of the lysozyme molecules in the crystal greatly facilitates polymerization, and polymers of different sizes are synthesized (Fig. 2B, lanes 3–5). We have determined the percentage of the initial monomers in the crystal that have been incorporated into various polymeric species for different times of oxygen exposure (Fig. 2C). With the exposure time, the amounts of lysozyme in the monomeric and dimeric forms decrease, whereas the amounts in tetramer, pentamer, and higher oligomers increase. Interestingly, the amount of lysozyme in the trimer remains constant throughout the course of the reaction, indicating that the rate of formation of this species is approximately equal to the rate at which it is used to form longer polymers. The yields of various species reach plateau values within the first 2 hr of oxygen exposure. That a significant amount of monomers remains unreacted after hours and even days of exposure to oxygen can be explained if the internal stresses and strains generated in the crystal during polymerization function as the termination step of the polymerization reaction. The observation that crystals become less transparent and diffract less well as polymerization proceeds is consistent with this interpretation. We also imaged the T4 lysozyme molecules with an SFM. Polymers of T4 lysozyme appear as long, multiglobular features, whereas the monomers are seen as smaller globular structures, as shown in Fig. 3.
Figure 2.
Characterization of T4 lysozyme polymers from dissolved crystals. (A) Polymers as detected by capillary electrophoresis. After exposure to oxygen for 23 days, protein crystals were dissolved in CE-SDS Protein Kit Sample Buffer (Bio-Rad). Samples were loaded with pressure (50 mbar × 50 sec) and run at constant voltage (−20 kV) at a nominal temperature of 20°C. Detection was by means of absorbance at 220 nm. (B) Polyacrylamide-SDS gel (4–15% gradient) showing degrees of polymerization of T4 lysozyme molecules obtained under different conditions. Lane 1 shows the polymerization achieved in solution under reducing conditions (0.55 M NaCl, 0.1 M Na3PO4, 0.02% Na3N, and 0.05 M DTT, pH 6.5). The protein concentration was 0.75 mg/ml. Lane 2 shows the polymerization obtained in solution under oxidizing conditions (0.55 M NaCl, 0.1 M NaPO4, 0.02% Na3N, pH 6.5, exposed to pure oxygen for 5 weeks). Lanes 3–5 show the degree of polymerization in the crystals after different times of oxygen exposure. To expose the crystals to oxygen, the hanging drops and well buffer were transferred to individual vials after crystallization. With the coverslips ajar, these vials then were placed in an oxygen-filled glass desiccator at room temperature. Pure oxygen gas was supplied to the desiccator valve at a flow rate of about 0.5 cc/min after being humidified by passage through 500 ml of well buffer. The oxygen flow removed the mercaptoethanol from the drops and the oxygen/mercaptoethanol mixture was allowed to escape through a paper gasket between the ground glass mating surfaces of the desiccator lid and body. SDS/PAGE broad range molecular weight standards (Bio-Rad) were used. (C) The relative abundance of different polymer species as a function of the oxygen exposure time. For clarity, not all polymer species are shown. The gels were scanned into a computer and the volume of each band was determined with imagequant (Molecular Dynamics). The relative abundance is the ratio of the volume of a particular band to the total volume of all the bands in a lane.
Figure 3.
Images of T4 lysozyme molecules obtained with the SFM. The overall size of each image field is 500 nm × 500 nm. (A) Polymers of T4 lysozyme molecules from dissolved crystals (exposed to oxygen for 30 days). The dimensions of the extended features indicate that they are polymers with lengths up to 30 monomers. (B) Monomeric T4 lysozyme molecules. The globular features have typical diameters of ≈8 nm and heights of ≈2 nm, which are consistent with that of a folded T4 lysozyme molecule because the lateral dimensions of the molecule are overestimated in the image because of the finite size of the SFM tip. If a T4 molecule is approximated as a sphere with a radius of 1.8 nm, its image will have radii of 4 nm in the lateral dimensions when imaged with a tip with a radius of curvature of 10 nm. In these experiments, 20 μl of protein solution (in PBS buffer) was deposited on freshly cleaved mica and allowed to adsorb for 10 min. The sample then was washed with the same buffer and dried with nitrogen gas. Imaging was performed in air by using a Nanoscope SFM (Digital Instruments, Santa Barbara, CA) operating in the tapping mode.
The enzymatic activity of the polymers was determined at 20°C by using a CD-based assay described previously (12). Using cell wall fragments, the natural substrate for T4 lysozyme, these assays revealed that the activity, on a per-monomer basis, of a heterogeneous polymer mixture was approximately 10 times lower than that of the nonpolymerized protein. The lower activity of the polymers may result from steric hindrance of the active site by a neighboring monomer, because one of the attachment points (position 21) is at the edge of active-site cleft (Fig. 1B). The arrangement of the monomers in the polymer also may restrict their access to cleavage sites within the peptidoglycan matrix. After reduction of the polymers by incubation with DTT, the enzymatic activity of the polymer mixture increased by a factor of about 5. Partial reduction of the disulfide bonds may have contributed to the incomplete recovery of the enzymatic activity. The stability of both the monomeric and polymeric forms of this mutant was determined from van't Hoff analysis of the thermally induced changes in the CD at 223 nm. As shown in Fig. 4, the melting temperature of the polymer mixture is 9.5°C lower than that of the monomers.
Figure 4.
The thermal stability of monomeric (+, Tm = 49.5oC, ΔH = 110 kcal/mol) and polymeric (○, apparent Tm = 40.0oC, apparent ΔH = 50 kcal/mol) T21C/K124C/WT* T4 lysozyme. Monomeric and polymeric samples were thermally unfolded (18) in 20 mM glycine-HCl, 1 mM H4EDTA, pH 3.05, with protein concentrations of 15 and 10 μM (in residue), respectively. The changes in the CD signal at 223 nm were analyzed as two-state transitions (solid curves). CD signals returned to in excess of 95% of their starting values upon cooling but approximately 10% of the polymeric sample adhered to the wall of the cuvette. The van't Hoff enthalpy at the apparent Tm for the polymeric sample is about 50% of the value expected on the basis of enthalpy changes with Tm of other T4 lysozyme mutants. The low value of ΔH for the polymeric sample likely reflects both heterogeneity inherent in a mixture as well as a loss of solubility of some of the material. The polymeric sample is less stable than the monomer. The change in ΔΔG relative to monomeric T21C/K124C/WT* is −3.5 kcal/mol with uncertainty likely ±1 kcal/mol.
Mechanical Unfolding of T4 Lysozyme.
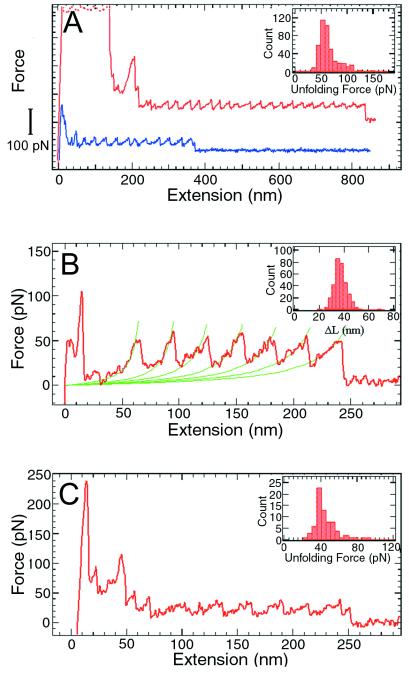
Next, we characterized the mechanical unfolding of T4 lysozyme by stretching the polymers with an SFM (3) specifically modified for these experiments. At the beginning of an experiment, the tip first was pressed against the sample surface to induce nonspecific attachment of the molecules, and then pulled away to generate tension in the polymer (3–6). Fig. 5A shows two of the force vs. extension curves thus obtained. Each peak in the sawtooth pattern corresponds to the unfolding of an individual lysozyme molecule between residues 21 and 124. At a pulling speed of 1,000 nm/sec, the unfolding force for a T4 lysozyme domain is 64 ± 16 pN (mean ± SD). This value is significantly smaller than the unfolding forces of 150–300 pN measured for titin Ig domains under similar conditions (3), and most likely reflects the structural differences between the β-barrel motif of titin Ig domains and the largely α-helical structure of T4 lysozyme (6). The variation in the unfolding forces (Fig. 5A Inset) reflects the stochastic nature of the protein unfolding process in the time scale of the current experiments. Thermal and instrumental noises also contribute to the spread of the unfolding force distribution. In Fig. 5B, successive stretching parts of a force curve are fitted to the worm-like-chain model (3, 4, 13, 14). The adjustable parameters in the fitting are the persistence length and the contour length of the polymer (3, 4). By fitting multiple force curves, a persistence length of 0.65 ± 0.25 nm for the unfolded polypeptide and a contour length increase of 36 ± 5 nm for each unfolding event were obtained. The expected contour length increment value for the 103 residues between the attachment points is 32 nm. The distribution of the contour length increments (Fig. 5B Inset) indicates that no shorter intermediates are present in the force curves; thus, the mechanical unfolding of T4 lysozyme is essentially a two-state process under the conditions used in these experiments.
Figure 5.
Force vs. extension curves obtained by stretching single T4 lysozyme polymers between a silicon nitride probe and a gold surface. (A) Two force curves obtained in 10× PBS (1,260 mM NaCl, 72 mM Na2HPO4, 30 mM NaH2PO4, adjusted to pH 7.0 with HCl). The curves are shifted in the vertical direction for clarity. Each curve has its own zero force level, which is the rightmost part of the curve where the polymer is either detached or broken. The high force features at the beginning were caused by nonspecific interactions between the probe and the surface. Inset, histogram of the unfolding forces (n = 443). (B) The force curve with the stretching parts fitted to the worm-like-chain model (3, 4). The contour length increments from the unfolding of a single lysozyme molecule were obtained from the fitting. (B Inset) Histogram (n = 328) of contour length increments, ΔL = 36 ± 5 nm. When the protein unfolds following the “two-state” model, each unfolding event will lengthen the polymer by an amount equal to the unfolded (contour) length between the two attachment points (i.e., the two linking cysteine residues) minus the distance of the two points in the folded protein. This contour length increase is not equal to the distance between adjacent peaks in the force curve because the polymer is never fully stretched. For T4 lysozyme, there are 103 aa between the two attachment points at positions 21 and 124. The distance between adjacent α-carbon atoms is 0.38 nm, but the tetrahedral geometry reduces the maximal extension of a polypeptide chain to about 0.34 nm per residue (i.e., in a fully extended β-sheet conformation). In the folded protein, the distance between sites 21 and 124 is 3.15 nm. Thus the contour length increase upon unfolding of one lysozyme molecule is (103 × 0.34) − 3.15 ≅ 32 nm. (C) A force curve obtained in 1 M GuHCl. (C Inset) Histogram of the unfolding forces (n = 73, pulling speed = 1,000 nm/sec). When a polymer is stretched in the absence of GuHCl, one monomer unfolds every 0.03 sec (average distance between adjacent peaks/pulling speed = 30 nm/1,000 nm per sec = 0.03 sec), corresponding to a rate constant of 33.3 sec−1. The free energy that has to be supplied from pulling is ΔΔG = RT ln(k1/k2), where k2 is the spontaneous rate constant of unfolding and k1 is the rate constant when pulled. In the absence of GuHCl, k2 = 10−4 sec−1 (19) and k1 = 33.3 sec−1, so ΔΔG = RT ln(k1/k2) = 7.6 kcal⋅M−1. Using the relation ΔΔG = F Δx (3), and the average unfolding force of F = 64 pN, we get Δx = ΔΔG/F = 0.81 nm. In 1.0 M GuHC, the average unfolding force is F = 44 pN. Assuming that Δx is the same, the reduction in the unfolding barrier is: (62–44) pN × 0.81 nm = 2.1 kcal⋅M−1.
The effects of guanidine hydrochloride (GuHCl) on the mechanical unfolding of T4 lysozyme also were investigated. As shown in Fig. 5C, the characteristic sawtooth pattern was observed in 1 M GuHCl, with an unfolding force of 44 ± 13 pN (Fig. 5C Inset). In 2 M GuHCl, no reproducible unfolding events were seen. In higher concentrations (3–6 M) of GuHCl, only curves consistent with pulling an unfolded protein chain were observed. Like most globular proteins, monomeric T4 lysozyme shows a linear decrease in its free energy of folding as the concentration of GuHCl is increased. Similarly, the logarithm of the apparent first-order rate constant for unfolding increases linearly with increasing GuHCl. The decrease in the unfolding force observed in GuHCl corresponds to a reduction of the unfolding activation barrier by about 2.1 kcal/mole (see Fig. 5 legend). This value is somewhat larger than the value (≈1.5 kcal/mole) obtained with bulk solution methods and may reflect the differences in the transition states visited in the two approaches.
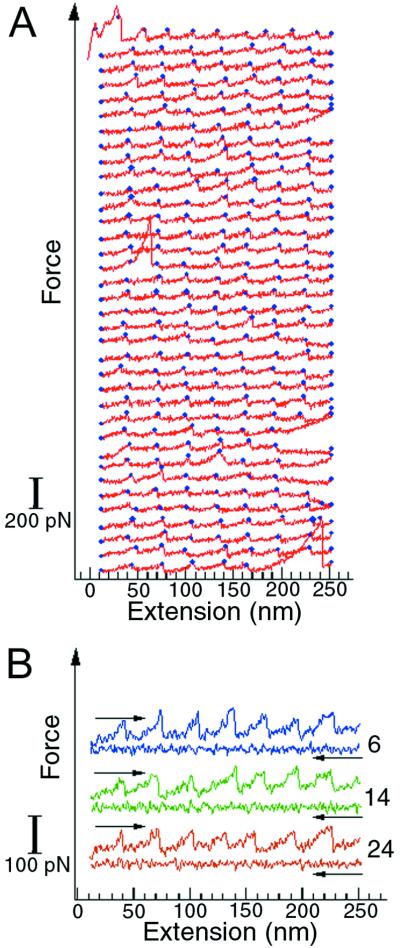
T4 lysozyme polymers were stretched and relaxed multiple times to investigate the reversibility of the folding-unfolding transition. The unfolding-refolding cycles were observed up to 50 times before the polymer broke or detached (Fig. 6). The unfolding forces were similar for all the unfolding events, indicating that the protein molecules refolded properly in each cycle. The refolding process was fast, completed within 1 sec after relaxation. The lysozyme molecules were still able to fold with the same efficiency after many repeated unfolding-refolding cycles, thus the molecular “wearing out” previously described for titin (1) was not apparent for this protein. This observation is of interest because T4 lysozyme is an α-helical protein whereas the globular domains of titin are almost 100% β-sheet structures. The difference in refolding efficiency between T4 lysozyme and titin may reflect a fundamental distinction in the accessibility of β-sheet and α-helical structures from the unfolded state (15). Because α-helices involve local, sequential interactions along the protein backbone, their organization can occur, at least in part, before the collapse toward the tertiary folded state takes place. In contrast, the organization of β-sheet structures requires mainly nonlocal, nonsequential interactions and must necessarily occur during or soon after the collapse of the molecule into the folded state. Thus, the formation of these structures may involve completely different kinetic barriers to the attainment of the folded state.
Figure 6.
Repeated unfolding-refolding of T4 lysozyme molecules in a polymer. The curves were obtained by stretching and relaxing the same polymer multiple times. (A) The unfolding traces of the unfolding-refolding cycles. The blue dots mark the positions where unfolding occurs. Three complete unfolding and refolding cycles are shown in B. The numbers on the right indicate the order of the curves in the sequence displayed in A. The force curves in these plots are shifted vertically for clarity, so they do not share a common zero force level.
Discussion
The observation that T4 lysozyme can still refold efficiently in the polymeric form may shed new light on the dynamics of the protein folding process. The molecular configurations accessible to the T4 lysozyme molecules in a polymer are greatly reduced because of the constraints imposed on the two linking cysteine residues by the neighboring molecules. T4 lysozyme molecules do not naturally exist in the polymerized form, as is the case for titin or tenascin domains (1–4) that may have evolved to fold despite the constraints imposed by the polymerization. Thus, it is unlikely that a polymeric lysozyme molecule and a monomeric molecule would go through the same series of conformational microstates during their refolding transitions. The efficient and rapid refolding observed for T4 lysozyme in the polymer strongly supports the existence of multiple folding pathways by which the protein can reach its native state and highlights the robustness of the folding process.
The present results show that it may be possible to extend the newly developed single molecule manipulation techniques from the limited class of naturally occurring polymeric proteins to globular proteins in general. The polymerization method presented here is quite general, requiring only that the CC-mutant proteins crystallize in a suitable form and the symmetry and packing of the crystal be compatible with the generation of extended polymers rather than dimers (16) or other closed assemblies. Many protein crystals should meet these requirements. It also should be possible to extend this approach to cocrystallize and copolymerize stability mutants with wild-type protein, so that the latter can be used as internal standards in the same pulling experiment, thus facilitating their comparison and minimizing the variability between experiments. Finally, by replacing alternative intermolecular contact points with cysteines, it will be possible to generate polymers with different topological connectedness, to systematically investigate the effect of pulling direction on the mechanical unfolding of the protein.
Acknowledgments
We thank S. Smith, D. Keller, M. Hegner, and B. Mooers for their comments, and T. Lowther, H. Xiao, and L. Gay for technical assistance. This work was supported in part by National Institutes of Health Grants GM-32543 to C.B., GM-21967 to B.W.M., and GM-57766 to F.W.D., a National Science Foundation grant (MBC 9118482) to C.B., and a National Institutes of Health training grant (GM-07759) to the Institute of Molecular Biology at the University of Oregon.
Abbreviations
- SFM
scanning force microscope
- GuHCl
guanidine hydrochloride
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1B6I).
References
- 1.Kellermayer M S, Smith S B, Granzier H L, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 2.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 3.Rief M, Gautel M, Oesterheld F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 4.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 5.Carrion-Vazquez M, Oberhauser A F, Fowler S B, Marszalek P E, Broedel S E, Clarke J, Fernandez J M. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rief M, Pascual J, Saraste M, Gaub H E. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 7.Smith S, Finzi L, Bustamante C. Science. 1992;258:1122–1297. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 8.Rief M, Clausen-Schaumann H, Gaub H E. Nat Struct Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 9.Rief M, Oesterheld F, Heymann B, Gaub H E. Science. 1997;275:1295–1297. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 10.Weaver L H, Matthews B W. J Mol Biol. 1987;193:189–199. doi: 10.1016/0022-2836(87)90636-x. [DOI] [PubMed] [Google Scholar]
- 11.Barnikol W K. Artif Cells Blood Substitutes Immobilization Biotechnol. 1994;22:725–731. doi: 10.3109/10731199409117904. [DOI] [PubMed] [Google Scholar]
- 12.Gassner N G, Baase W A, Lindstrom J D, Shoichet B K, Matthews B W. In: Techniques in Protein Chemistry VIII. Marshak D R, editor. London: Academic; 1997. pp. 851–863. [Google Scholar]
- 13.Bustamante C, Marko J, Siggia E, Smith S. Science. 1994;265:1599–1601. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 14.Marko J, Siggia E. Macromolecule. 1995;28:8759–8770. [Google Scholar]
- 15.Plaxco K W, Simons K T, Baker D. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 16.Heinz D W, Matthews B W. Protein Eng. 1994;7:301–307. doi: 10.1093/protein/7.3.301. [DOI] [PubMed] [Google Scholar]
- 17.Poteete A R, Sun D P, Nicholson H, Matthews B W. Biochemistry. 1991;30:1425–1432. doi: 10.1021/bi00219a037. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson A E, Baase W A, Matthews B M. J Mol Biol. 1993;229:747–769. doi: 10.1006/jmbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 19.Chen B L, Baase W A, Nicholson H, Schellman J A. Biochemistry. 1992;31:1464–1476. doi: 10.1021/bi00120a025. [DOI] [PubMed] [Google Scholar]