Abstract
Rationale: Nontypeable Haemophilus influenzae (NTHi) commonly infects patients with cystic fibrosis (CF), especially early in childhood. Bacteria biofilms are increasingly recognized as contributing to bacterial persistence and disease pathogenesis in CF.
Objectives: This study investigated ability of NTHi to form biofilms and its impact on airway epithelia using in vivo and in vitro analyses.
Methods: We evaluated bronchoalveolar lavage fluid from young patients with CF for evidence of NTHi biofilms. To further investigate the pathogenesis of NTHi in respiratory infections, we developed a novel in vitro coculture model of NTHi biofilm formation on polarized human airway epithelial cells grown at the air–liquid interface.
Measurements and Main Results: In bronchoalveolar lavage fluid samples from young, asymptomatic patients with CF, we found morphologic evidence suggestive of NTHi biofilm formation. In addition, 10 clinical NTHi isolates from patients with CF formed biofilms on plastic surfaces. NTHi formed biofilms on the apical surface of cultured airway epithelia. These biofilms exhibited decreased susceptibility to antibiotics and were adherent to epithelial surfaces. Airway epithelial cells remained viable throughout 4 d of coculture, and responded to NTHi with nuclear factor-κB signaling, and increased chemokine and cytokine secretion.
Conclusions: NTHi formed adherent biofilms on the apical surface airway epithelia with decreased susceptibility to antibiotics, and respiratory cells exhibited inflammatory and host defense responses—evidence of a dynamic host–pathogen interaction. The data presented here have implications both for understanding early CF lung disease pathogenesis and for the treatment of early, asymptomatic colonization of patients with CF with H. influenzae.
Keywords: bacterial infections, inflammation, lung diseases
Biofilms are increasingly recognized as contributing to disease pathogenesis in cystic fibrosis (CF) and other respiratory tract diseases associated with chronic bacterial infections. Pseudomonas aeruginosa in CF sputum displays morphologic and physiologic evidence suggestive of biofilm formation in vivo (1, 2). Biofilms enable bacteria to persist in CF pulmonary infections (for a review, see Reference 3). P. aeruginosa in a biofilm state can exhibit a more than 1,000-fold increase in antibiotic resistance (4) and more readily evades host defense factors (5). Nontypeable Haemophilus influenzae (NTHi) has recently been shown to form biofilms in vitro and in otitis media (OM) (6–10); however, there is no information on NTHi biofilm formation in the lung or in CF.
NTHi commonly infects patients with CF, especially early in childhood (11, 12). It is hypothesized that H. influenzae causes inflammation and damage to the airway and acts as a gateway organism paving the way for colonization with P. aeruginosa (13). Supportive of this, most children with P. aeruginosa had prior infection with H. influenzae or Staphylococcus aureus (14), and single-pathogen infections with H. influenzae, S. aureus, or P. aeruginosa showed no statistically significant differences for airway inflammatory markers (15).
We hypothesized that NTHi forms biofilms on airway epithelia, which leads to increased airway epithelial inflammatory responses. We further hypothesized that NTHi biofilms form in the lungs of patients with CF in childhood and may contribute to early lung disease pathogenesis. Some of the results of these studies have been previously reported in the form of abstracts (16, 17).
METHODS
All bronchoalveolar lavage fluid (BALF) and bacterial culture samples used in these studies were obtained using protocols approved by the institutional review board at the University of Iowa.
Human BAL Samples
BALF samples were collected from patients with CF as part of their standard care. Samples were immediately placed on ice before further processing. Quantitative cultures and white blood cell counts with differential were performed by standardized protocols in the Clinical Pathology Laboratory at the University of Iowa. Remaining BALF was processed for transmission electron microscopy (TEM) analysis as follows. BALF was centrifuged at 14,000 × g for 10 min at 4°C, fixed, and embedded in Epon or LR White (LRW) for electron microscopic imaging. Additional details on the experimental methods are provided in the online supplement. For immuno-TEM, samples were labeled overnight with an anti-NTHi monoclonal antibody, 6E4, followed by goat anti-mouse IgG ultra-small gold secondary antibody labeling (Electron Microscopy Sciences, Hatfield, PA) and silver enhancement. The 6E4 antibody recognizes an NTHi lipooligosaccharide (LOS) epitope and is specific for H. influenzae LOS (18). Controls for immuno-TEM included the following: no primary antibody on LRW-embedded CF BALF; LRW-embedded NTHi 2019 for a positive control; and LRW-embedded 107 cfu NTHi 2019 with 105 primary human airway epithelial cells for a centrifugation artifact control.
Bacterial Strains and Culture Conditions
NTHi strain 2019 is a clinical isolate obtained from a patient with chronic obstructive pulmonary disease (COPD) (19). The 10 clinical isolates of NTHi from patients with CF were obtained from the Clinical Microbiology Laboratory at the University of Iowa. We also studied NTHi strains 2019, 2019siaA (20), 2019siaB (21), and 2019wecA (7) in our NTHi–epithelial coculture model. Plasmid pRSM2211 expressing green fluorescent protein (GFP) mut3 was kindly provided by Lauren Bakaletz (22) and was electroporated into NTHi strain 2019 and NTHi strain 2019wecA by methods previously described (23). All strains were reconstituted from frozen glycerol stock cultures and propagated on brain heart infusion (BHI) agar or broth (Difco, Detroit, MI) supplemented with 10 μg of hemin (Sigma Chemical Co., St. Louis, MO) per milliliter and 10 μg of nicotinamide-adenine nucleotide (Sigma) per milliliter at 37°C. For pRSM2211 plasmid selection, kanamycin (Sigma) 20 μg/ml was added to BHI broth or agar.
Microtiter Biofilm Formation Assay
Biofilm formation by NTHi clinical isolates was assessed using a 96-well microplate as previously described (8, 24). Additional details on the experimental methods are provided in the online supplement.
Airway Epithelial–NTHi Cocultures
Calu-3 cells (ATCC No. HTB-55) were prepared and grown at the air– liquid interface with media containing Ultroser-G (Biosepra, Cergy-Saint-Christophe, France) as previously described for primary cultures of human airway epithelia (25). Penicillin, streptomycin, gentamicin, and fluconazole used in initial establishment of epithelial cultures were removed by repeated apical and basolateral washings, and antibiotic-free media changes over 4 to 5 d. To establish bacterial colonization of the epithelial surface, 20 multiplicity of infection (MOI) (2 × 107 cfu) of NTHi bacteria were suspended in 50 μl of phosphate-buffered saline (PBS) and added to the apical surface. The antibiotic-free basolateral media were changed daily throughout all coculture experiments. Transepithelial resistance was measured using an EVOM-G ohm meter (World Precision Intruments, Sarasota, FL) after the addition of 200 μl of Dulbecco's modified Eagle medium (DMEM)/Hams F-12 (Gibco, Carlsbad, CA) on the apical surface.
Confocal Microscopy
All confocal microscopy experiments used NTHi strain 2019 or 2019wecA containing pRSM2211 expressing GFPmut3 imaged on a Bio-Rad 1024 confocal microscope (Bio-Rad, Hercules, CA). Kanamycin was used for plasmid selection pressure until suspension in PBS before inoculation. In separate experiments, we found that pRSM2211 was highly stable in NTHi strain 2019 and showed no decrease in fluorescence until passaged 10 to 15 times without antibiotic selection. For each time point, five stacked images at 400× magnification (270 × 270 μm) were obtained and average values were used for statistical analyses. Each of the five stacked images were selected from an apical surface area at random using the following method: (1) the z-focal plane was brought above the point where bacteria could be visualized; (2) the sample was moved on the x and/or y axis; (3) the z-focal plane was then lowered; (4) if the cell surface was obscured by sloughed cells or debris, the process was restarted; (5) the first unobstructed cell surface area was imaged regardless of epithelial cell contour or bacterial content; (6) the z axis was again brought above the focal plane and the process repeated. The first five stacked images were used for further analyses. Three-dimensional rendering was performed using Imaris 4.2 (Bitplane, St. Paul, MN), and quantification of biofilm mass was performed using MatLab (The MathWorks, Inc., Natick, MA) and COMSTAT analysis (BioCentrum-DTU, Lungby, Denmark) (26).
NTHi Biofilm Adherence
NTHi–airway epithelial cocultures were grown as described above. To assess NTHi biofilm adherence to epithelia, apical surfaces of the coculture underwent daily washes to remove nonadherent bacteria. Washes consisted of 200 μl of 1:1 DMEM:Hams F12 media pipetted onto the cell culture insert interior wall and then aspirated off the contralateral wall vigorously five times on a daily basis before sample processing for transepithelial resistance, colony counts, and electron microscopy.
Biofilm Susceptibility to Antibiotics
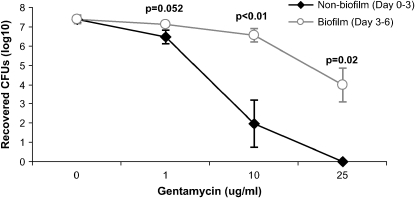
To determine if NTHi in biofilms exhibited decreased susceptibility to killing by antibiotics, epithelia were inoculated with NTHi 2019 as described above. Bacteria in coculture were then either (1) exposed to antibiotics from Days 0–3, starting at the time of NTHi inoculation (nonbiofilm condition) and bacteria collected on Day 3 or (2) allowed to form a biofilm over 3 d in antibiotic-free conditions before exposure to antibiotics on Days 3–6 (biofilm condition) and bacteria harvested on Day 6. For the nonbiofilm condition, we added gentamicin (Celgro, Warren, NJ) to the Day 0–3 basolateral media in varying concentrations (0, 1, 10, or 25 μg/ml). For the biofilm condition, antibiotic-free media was used on Days 0–3 before adding the same concentrations of gentamicin to the basolateral media on Days 3–6. Media were changed on a daily basis in both conditions. To collect bacteria from the coculture, 200 μl of 1:1 DMEM/Hams F12 were added and vigorously pipetted directly onto the apical cell culture surfaces. Bacteria were then vortexed to disrupt biofilm structures, diluted in PBS, plated on supplemented BHI plates, incubated overnight at 37°C, and manually counted to determine surviving colony-forming units (n = 3–6). Because gentamicin poorly penetrates cells, gentamicin concentrations were measured in the apical washings with a CEDIA assay (Diagnostix, Mississauga, ON, Canada) using a Hitachi P-module (Roche Modular Instruments, Indianapolis, IN) in the Clinical Laboratory at the University of Iowa. To determine if the recovered bacteria changed their gentamicin susceptibility during coculture, recovered NTHi bacteria from the apical washes on Day 3 or 6 were also plated on supplemented BHI plates with 1 μg/ml gentamicin. We additionally plated recovered bacteria on unsupplemented BHI plates with factor X and factor V discs (Remel, Lenexa, KS) to ensure that the recovered bacteria were H. influenzae and not a contaminant. Gentamicin invasion assays were performed as previously described (27, 28) using the standard 50-μg/ml and a higher 100-μg/ml gentamicin concentration because of the concern of decreased susceptibility of NTHi to gentamicin in a biofilm state.
Airway Epithelial Nuclear Factor-κB Signaling
Airway epithelial cultures were initially prepared as described above. Cell cultures were basolaterally infected with a replication incompetent adenoviral vector (MOI 10) containing nuclear factor (NF)-κB response elements driving a luciferase reporter as previously described (29). One day after transduction, epithelia were treated with 100 ng/ml recombinant human interleukin (IL)-1β (Sigma) on the apical and basolateral surfaces, or were apically inoculated with NTHi 2019, as described above. As a negative control, epithelia were incubated with 100 μl of PBS apically and 500 μl of media basolaterally. One day after stimulation with IL-1β or NTHi, cells were lysed and luciferase activity measured (Luciferase Assay System; Promega, Madison, WI) per the manufacturer's recommendations using a Monolight 3010 luminometer (Pharmogen, San Diego, CA).
Basolateral Chemokine and Cytokine Abundance
We measured IL-8 and tumor necrosis factor (TNF)-α protein concentrations in the basolateral media using IL-8 (Human IL-8 OptEIASet; BD Biosciences, San Jose, CA) and TNF-α (DuoSet DY210; R&D Systems, Minneapolis, MN) ELISA kits per the manufacturers' instructions. We also measured CCL20 abundance by ELISA using anti-human MIP-3 α/CCL20 monoclonal antibody and biotinylated affinity purified anti-human macrophage inflammatory protein (MIP)-3α/CCL20 polyclonal antibody (MAB360 and BAF360, respectively; R&D Systems). Additional detail on the method for the CCL20 ELISA is provided in the online supplement. All three ELISAs used the same samples stored at –80°C (n = 5) and were run in duplicate to ensure reproducibility.
Statistical Analysis
All analyses of statistical significance were performed with one-tailed Student's t tests using Microsoft Excel (Microsoft Corp., Redmond, WA). p values less than 0.05 were considered statistically significant. For quantification of biofilm mass, each datapoint used was the mean biomass of five randomly imaged areas from a single coculture. Mean biomass values from different coculture experiments were used for further statistical analyses.
RESULTS
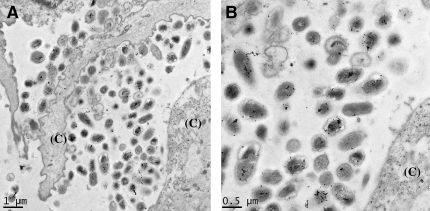
Using electron microscopy, we found evidence of structures consistent with NTHi biofilms in airway lavage samples from young, asymptomatic patients with CF. NTHi often colonizes the airway early in CF (12, 14, 30), but no H. influenzae biofilm structures have been previously observed in lung samples. As part of their routine care, BALF from patients with CF was obtained to monitor bacterial colonization and inflammation in the lung. In 10 young patients with CF never colonized or not currently colonized with P. aeruginosa in their lower airways, we obtained and fixed BALF material. In the 7 of the 10 samples that either grew no bacteria on culture or had less than 50% of neutrophils in their BALF (see Table 1), we found no bacteria in a biofilm morphology. Of these seven patients, only two (Patients 6 and 9) had bacteria detected on epithelial surfaces (data not shown). We found structures consistent with biofilms in two of the three patients with both greater than 105 cfu/ml of pathogenic bacteria and 50% or more neutrophils in their BALF (see Figure 1). The overall area and numbers of bacteria involved in these structures are more extensive than would be expected with a microcolony, and are suggestive of more mature biofilms. Of note, due to the nature of sample processing and TEM imaging, the matrix of the biofilm is not apparent. To visualize the matrix in subsequent in vitro experiments, we used scanning electron microscopy (SEM) with osmium–perfluorocarbon fixation to preserve the hydration of structures like biofilms. Both of these patients were 17 mo of age and had no pulmonary symptoms other than occasional cough. To further verify the bacteria as NTHi, we used immunolabeling with an antibody specific to an H. influenzae LOS antigen on both of these patients' samples. As shown in Figure 1, the bacteria in the biofilm from Patient 3 were labeled by the H. influenzae LOS-specific monoclonal antibody. This is the first in vivo evidence of NTHi biofilm formation in human lung disease. In contrast, the bacteria in the biofilm in Patient Sample 4, whose culture predominantly grew S. aureus, had no significant labeling with the NTHi-specific monoclonal antibody (data not shown). The “no primary antibody” control showed no significant labeling, and LRW-embedded NTHi 2019, which was immunostained in an identical manner, displayed the same pattern of bacterial labeling as seen in the CF BALF (see Figure E1 of the online supplement). To verify that the bacterial structures seen were not products of centrifugation artifact, we processed and immunolabeled 107 cfu of NTHi 2019 and 105 freshly dissociated primary human airway epithelial cells in the same manner as the BALF. We found no similar bacterial structures associated with epithelial cells (data not shown).
TABLE 1.
ANALYSIS OF CYSTIC FIBROSIS BRONCHOALVEOLAR LAVAGE FLUID
| Pt | Age (yr) | H. flu | S. a. | WBC | PMNL | Adherence | Communal |
|---|---|---|---|---|---|---|---|
| 1 | 8.8 | ND | ND | 35 | 28 (80%) | ND | ND |
| 2 | 4.4 | ND | 640k | 1,145 | 882 (77%) | ND | ND |
| 3 | 1.4 | 800k | ND | 1,540 | 1,155 (75%) | + | +† |
| 4 | 1.4 | 50k | 10M | 4,050 | 2,957 (73%) | + | + |
| 5 | 7.4 | ND | ND | 1,255 | 678 (54%) | ND | ND |
| 6 | 3.1 | ND | ND | 1,705 | 887 (52%) | Rare | ND |
| 7 | 12.3 | ND | ND | 448 | 193 (43%) | ND | ND |
| 8 | 4.2 | 700k | ND | 409 | 86 (21%) | ND* | ND |
| 9 | 1.4 | 1M | ND | 547 | 33 (6%) | + | ND |
| 10 | 3.4 | ND | ND | 155 | 8 (5%) | ND | ND |
Definition of abbreviations:H. flu = Haemophilus influenzae cfu/ml; k = thousand; M = million; ND = not detected; PMNL = polymorphonuclear lymphocyte count per cubic milliliter of bronchoaleolar lavage fluid; Pt = patient; S. a. = Staphylococcus aureus cfu/ml; WBC = white blood cell count per cubic milliliter of bronchoalveolar lavage fluid.
Bronchoalveolar lavage fluid from 10 young patients with cystic fibrosis was analyzed by quantitative culture and cell count. Ages shown are in years at the time of bronchoscopy. Adherence and Communal refer to individual or groups (microcolony or biofilm) of bacteria adherent to airway epithelia, respectively.
No epithelia on sample.
H. influenzae by labeling using monoclonal antibody 6E4.
Figure 1.
Evidence for nontypeable Haemophilus influenzae (NTHi) biofilm formation early in cystic fibrosis (CF). Bronchoalveolar lavage fluid (BALF) from an asymptomatic, previously uncolonized 17-mo-old patient with CF revealed bacteria adherent to cell surfaces (CF BALF Patient 3 from Table 1). (A) Immuno–transmission electron microscopy (-TEM) of LR White–embedded CF BALF with a monoclonal antibody against an H. influenzae–specific lipooligosaccharide antigen with small immuno-gold and -silver enhancement, labeled bacteria in the biofilm. (B) Higher power magnification of immunolabeling. “(C)” indicates host cells.
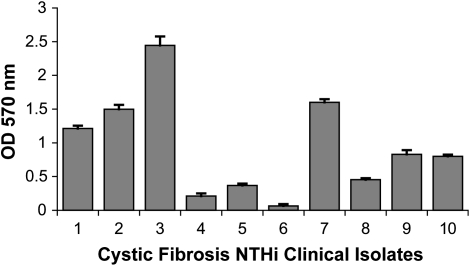
After finding evidence suggesting that NTHi formed structures consistent with biofilms in vivo, we sought to determine if clinical isolates of NTHi formed biofilms. Using an in vitro batch biofilm assay on plastic surfaces, all CF NTHi isolates formed biofilms to varying degrees (see Figure 2). Planktonic growth curves showed no significant differences between isolates (data not shown). Some isolates (Nos. 1, 2, 3, and 7) formed significantly greater biofilm mass than other isolates (Nos. 4, 5, 6, and 8; p < 0.0001). This finding demonstrates that the ability to form biofilms is common in CF NTHi isolates.
Figure 2.
Clinical isolates of H. influenzae from patients with CF form biofilms on plastic surfaces. Ten clinical strains of H. influenzae from patients with CF were grown in supplemented brain heart infusion (BHI) broth overnight. Biofilm density was quantified by crystal violet staining as described in Methods, with error bars indicating the standard error of the mean (n = 6). OD = optical density.
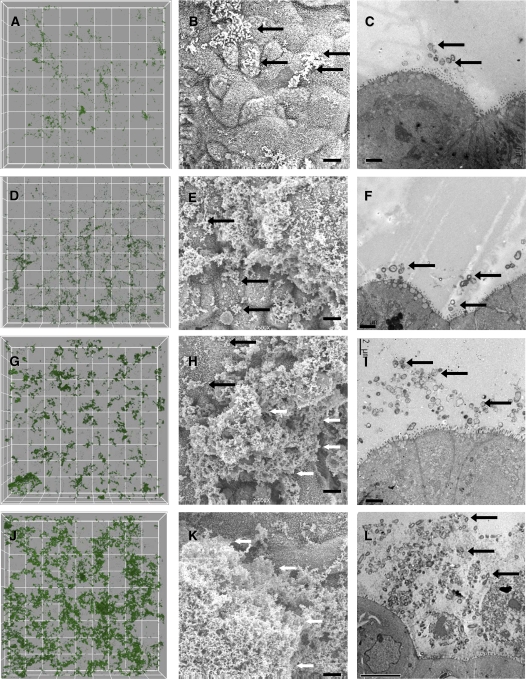
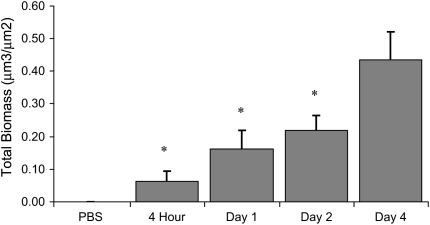
To further investigate NTHi biofilm formation, we developed a model of NTHi cocultured on polarized human airway epithelia. Importantly, no medium was apically supplied to the bacteria in this model, requiring the bacteria to obtain all nutrients and growth factors from or through the polarized airway epithelial layer, mimicking conditions in an in vivo lung infection. On the basis of several morphologic criteria, NTHi formed structures consistent with biofilms over 4 d of coculture on air–liquid interface cultured airway epithelia. Structurally, biofilms are a community of microorganisms encased in a matrix, attached to a surface. As shown in Figure 3, NTHi bacteria were initially scattered over the cell surface with little associated matrix formation. Over subsequent days of coculture, matrix formation increased, resulting in large biofilm structures measuring over 20 μm in depth. By Day 4, communities of NTHi bacteria were observed within a matrix on the apical surface of epithelia. Additional images of the NTHi–epithelial coculture model are shown in Figure E2. To quantitate bacterial structures, GFP-labeled bacteria were cocultured and imaged using confocal microscopy. Three-dimensional rendering and biomass analyses showed significantly (p < 0.01) increasing biofilm mass through Day 4 (see Figure 4) and were visually similar to the qualitative SEM and TEM images in Figure 3.
Figure 3.
Airway epithelial cultures infected with NTHi show progressive biofilm formation over time. Samples were inoculated with NTHi and imaged at 4 h, 1 d, 2 d, and 4–5 d. Representative confocal (A, D, G, J), scanning electron microscopic (SEM; B, E, H, K), and TEM images (C, F, I, L) are shown. Four hours (A–C): scattered bacteria on the apical surface with virtually no matrix; however, some microcolonies are starting to form; Day 1 (D–F): thin matrix production and areas of larger microcolonies; Day 2 (G–I): more extensive matrix and larger biofilm structures have formed; Days 4–5 (J–L): thick matrix entirely obscuring the apical surface in some areas. Grid boxes on confocal images represent 30 μm2. Scale bars on all SEM images indicate 5 μm. TEM scale bars indicate 2 μm on 4-h–Day 2 images and 10 μm on Days 4–5. Black arrows indicate areas of individual bacteria, and white arrows indicate larger biofilm structures.
Figure 4.
Quantification of NTHi biofilms in coculture. Quantification of green fluorescent protein–labeled NTHi biofilms grown in coculture on Calu-3 cells as shown in Figure 3. Each datapoint represents the mean biomass of five randomly imaged areas from a single coculture at the times shown. Each graphed value represents the average of the mean biomass values from different coculture experiments, with error bars indicating the standard error of the mean (n = 6–8, except phosphate-buffered saline [PBS], n = 3). *p < 0.001 versus Day 4.
Because NTHi biofilm formation on plastic surfaces may be less clinically relevant than biofilm formation on epithelial surfaces or in vivo, we selected a robust biofilm-forming CF NTHi isolate (No. 3) and a poor biofilm-forming isolate (No. 4) as identified in Figure 2, and determined their capacity to form biofilms in the epithelial coculture model. Isolate 3 formed larger and more extensive biofilms than Isolate 4 (see Figure E3). This was especially apparent at lower magnifications. Thus, CF NTHi isolates form biofilms on airway epithelia and the quantitative biofilm formation on plastic surfaces correlates with the qualitative biofilm formation in coculture.
After showing morphologic evidence of NTHi biofilms, we next investigated if these structures displayed functional characteristics of biofilms. To test if NTHi biofilms displayed decreased susceptibility to antibiotics, we exposed NTHi–epithelial cocultures to gentamicin for 3 d, either starting on Day 0 at the time of NTHi inoculation (nonbiofilm condition) or starting on Day 3 after coculturing the bacteria in antibiotic-free conditions to establish biofilms (biofilm condition). In the NTHi biofilm condition, we recovered significantly more bacteria from the apical epithelial surface in the 10- and 25-μg/ml gentamicin concentrations (p < 0.05; see Figure 5). To ascertain if the bacteria had become intrinsically less susceptible to killing by antibiotics or if growth in the biofilm state conferred decreased antibiotic susceptibility, the recovered bacteria were additionally inoculated on supplemented BHI agar plates with 1 μg/ml gentamicin. All apically recovered NTHi remained susceptible to 1 μg/ml gentamicin. The minimal inhibitory concentration of gentamicin for NTHi 2019 by microbroth dilution and radial diffusion assay was 0.25 μg/ml or less. Because gentamicin poorly penetrates cells and apical abundance of antibiotic may have been lower in the basolateral media, we measured concentrations of gentamicin in the recovered apical bacterial washings. Gentamicin concentrations averaged 1.4, 6.7, and 17.5 μg/ml for the 1-, 10-, and 25-μg/ml basolateral conditions, respectively, indicating a 93% penetration of gentamicin to the apical surface on average. We also determined the relative proportion of intracellular, invasive bacteria versus extracellular bacteria in Day 3 cocultures using established methods of apical gentamicin applied for 1.5 h to kill extracellular bacteria, and then lysing the cells to release intracellular bacteria. We found that an average of 1.4 and 0.18% (2.6 and 0.32% SD) of bacteria invaded intracellularly in the 50- and 100-μg/ml apical gentamicin concentrations, respectively (n = 4). However, because NTHi bacteria in biofilms display increased survival with gentamicin exposure, as shown above, this may complicate this invasion assay. Therefore, we cannot exclude the possibility of NTHi surviving for reasons other than intracellular invasion. We also found decreased NTHi susceptibility to penicillin in similar studies (data not shown). To assess bacterial adherence to epithelia, we vigorously washed the apical surface of the cell cultures on a daily basis through the coculture experiment. Recovered bacteria were not statistically different in the unwashed control and washed samples, except on Day 4 of the experiment, which averaged 3.6 × 106 cfu for the washed sample, versus 2.2 × 107 cfu in the unwashed control (p = 0.04, n = 3). Although the washings moderately disrupted the biofilms, as shown by decreased biofilm size and less extensive coverage of the epithelial surface, biofilm structures persisted throughout the 4-d experiment and were qualitatively similar to Day 2 unwashed biofilms (see Figure E4). Therefore, despite vigorous daily washing of the apical surface, bacterial counts and biofilm formation persisted. Although in both the antibiotic susceptibility and adherence experiments, the bacteria in biofilms were disrupted by extensive vortexing, we cannot exclude the possibility that residual clumping of the bacteria, or the bacteria being in a viable but nonculturable biofilm state (31) may have underestimated colony counts. Thus, NTHi bacteria in this coculture model exhibit two functional characteristics of biofilms: adherence and decreased susceptibility to killing by antibiotics.
Figure 5.
NTHi in a biofilm survives high concentrations of gentamicin. NTHi–airway epithelial cocultures were exposed to the indicated concentrations of gentamicin. For the nonbiofilm condition, gentamicin was added from Day 0 at time of NTHi inoculation, until Day 3, when the apical bacteria were recovered. For the biofilm condition, we allowed NTHi to form a biofilm over 3 d in antibiotic-free conditions before exposure to gentamicin on Days 3–6, after which the bacteria were recovered. Mean values for each condition with error bars indicating the standard error of the mean are shown. Significantly more bacteria survived in the biofilm/Day 3–6 biofilm condition compared with the nonbiofilm/Day 0–3 condition at the 10- and 25-mg/ml gentamicin concentrations (n = 3–6).
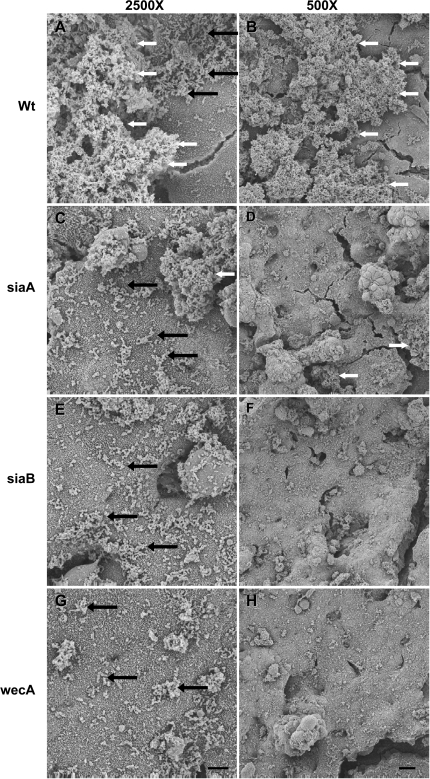
We hypothesized that NTHi biofilm formation on epithelia requires matrix formation and the ability to incorporate sialic acid into the biofilm, as suggested by previous studies (6, 7). To test this hypothesis, NTHi 2019siaA, 2019siaB, and 2019wecA were cocultured on airway epithelia. These three bacterial strains are deficient in producing the sialylated exopolysaccharide that forms NTHi's biofilm matrix. By Day 3, many bacteria were observed scattered above the microvilli on the apical surface, but matrix production was diminished and no substantial biofilm structures were observed (see Figure 6). In contrast, large biofilm structures with extensive matrix formed in the parent NTHi 2019 and clinical CF NTHi samples. The 2019wecA mutant generally had the thinnest biofilm formation; in contrast, both the siaA and siaB mutants varied more in biofilm thickness. Quantification of Day 3 cocultured NTHi biofilms using confocal microscopy showed similar results and revealed significantly decreased biofilm mass in the NTHi strain 2019wecA mutant (mean, 0.04 μm3/μm2; SD, 0.02 μm3/μm2) compared with the 2019 wild-type strain (mean, 0.21 μm3/μm2; SD, 0.09 μm3/μm2; p = 0.02, n = 4). For Imaris three-dimensional renderings of NTHi strain 2019 and 2019wecA biofilms, see Figure E5.
Figure 6.
NTHi bacteria deficient in producing a sialylated exopolysaccharide form biofilms poorly. Epithelial cultures were apically infected with NTHi and allowed to form biofilms for 3 d. (A, B) Wild-type NTHi 2019 (Wt) or clinical isolates form robust biofilms. NTHi 2019 with mutations in siaA2019 (siaA; C, D), 2019siaB (siaB; E, F), and 2019wecA (wecA; G, H) had many bacteria on the epithelial surface, but formed biofilms much more poorly. Scale bars on 2,500× and 500× magnifications indicate 5 and 20 μm, respectively. Black arrows indicate areas of individual bacteria, and white arrows indicate larger biofilm structures.
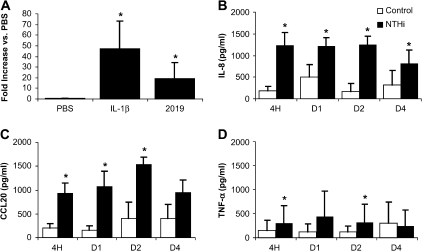
This bacterial–epithelial model allows for intimate contact between host and pathogen. We hypothesized that airway epithelia would recognize and respond to NTHi biofilm formation on their apical surface. When grown at the air–liquid interface, airway cells form a polarized epithelial monolayer with apical tight junctions, creating a barrier with high transepithelial resistance. The transepithelial resistance remained stable and unchanged from uninfected controls throughout the 4 d of coculture with NTHi (data not shown). However, while retaining barrier function, epithelia actively responded, as shown by NF-κB signaling (see Figure 7A), and significantly increased secretion of IL-8 at all time points, CCL20 (MIP-3α) at all time points except Day 4, and TNF-α at the 4-h and 2-d time points (see Figures 7B–7D). Thus, the airway epithelia responded to NTHi infection by increasing innate and adaptive immune responses.
Figure 7.
Airway epithelial responses to NTHi biofilms. (A) Epithelial cultures were basolaterally transfected with adenovirus containing nuclear factor-κB response elements driving luciferase. Cell lysates from adenovirus transduced cells were obtained 1 d after stimulation with PBS (negative control), 100 ng/ml interleukin (IL)-1β, (positive control), or apical infection with NTHi (n = 11). (B–D) ELISA analysis of the basolateral media from control uninfected and NTHi-infected airway epithelial cultures sampled at 4 h, 1 d, 2 d, and 4 d for the indicated chemokines and cytokines (n = 5). Mean values for each condition with error bars indicating the standard error of the mean are shown. *p < 0.05.
DISCUSSION
Lung lavage samples from very young patients with CF contained H. influenzae in structures consistent with biofilm formation. This is the first in vivo evidence of NTHi biofilm formation within the human lower airways. In support of this finding, clinical NTHi isolates from patients with CF formed biofilms in vitro. Furthermore, our in vitro coculture results suggest that NTHi bacteria adhere and form antibiotic-resistant biofilms on the apical surface of airway epithelia. The epithelia, in turn, respond by increased secretion of several innate and adaptive immune factors that mediate inflammation. Because inflammation is believed to be a key element initiating CF lung disease (for a review, see Reference 32), aggressively managing NTHi infections and the resultant proinflammatory products could potentially help to delay the onset of lung damage and/or chronic bacterial colonization. The observation of biofilms in BALF from very young patients with CF may have particular relevance to early CF lung disease pathogenesis and the treatment of asymptomatic children colonized with H. influenzae.
H. influenzae Forms Biofilms on Airway Epithelia
We show that NTHi forms biofilms on airway epithelia with both morphologic and functional data. These are the first data showing H. influenzae biofilms exhibit decreased susceptibility to killing by antibiotics. NTHi resistance to killing depended on growth in the biofilm state, as bacteria removed from the gentamicin- and penicillin-resistant biofilms remained susceptible to planktonic minimal inhibitory concentration antibiotic concentrations. Other bacteria that form biofilms exhibit similar patterns of antibiotic susceptibility (33, 34). Several other lines of evidence support NTHi biofilm formation in vivo. NTHi biofilms were directly visualized in middle ear aspirates from children with OM (10). Similarly, NTHi forms biofilms in the chinchilla model of OM (9). In addition, RNA from NTHi can be detected in sterile middle ear aspirates of children with OM (31), providing indirect evidence of biofilm formation. Furthermore, monthly sputum cultures from patients with COPD reveal intermittent negative cultures despite continuous colonization by the same isolate proven by molecular typing, suggesting that the organism is present despite negative sputum cultures (35). One hypothesis for the intermittently negative cultures is that culture methods designed to detect planktonic growth may be less sensitive for detecting slow-growing bacteria in biofilms.
Part of the definition of a biofilm is that its matrix is self-produced. Because Calu-3 cells produce mucus, it could be argued that the structures seen in the coculture model were bacteria covered in epithelial mucus. Definitive proof of bacterial matrix versus epithelial mucus is complicated by the fact that NTHi biofilm matrix consists of 2,6 linked sialic acid on a polysaccharide backbone that is virtually indistinguishable from human mucus (7). Although we did not definitively show that NTHi produced the matrix seen in the coculture model, there are three lines of evidence that strongly suggest that this is the case. First, NTHi can be seen throughout these structures on confocal microscopic and TEM imaging. Second, the structures seen in these studies are morphologically similar to NTHi biofilms in other models only containing bacteria (7). Third, and most compelling, the 2019wecA matrix mutant and 2019siaA/B sialylation mutants failed to produce robust biofilms with a prominent matrix in coculture (Figure 6). If the structures associated with the wild-type bacteria were epithelially derived mucus produced in response to bacterial stimulation, we would expect similar biofilm-like mucus production by the sialylation mutants and wild-type NTHi, since they had the same numbers of bacteria.
NTHi in Early CF Lung Disease
NTHi is a major pathogen in CF. Often disregarded as an oral contaminant, NTHi commonly colonizes the upper respiratory tract (36, 37). However, this common human upper respiratory tract commensal is also a significant pathogen in the lower respiratory tract of patients with CF (11–13, 15, 38–40). In BALF studies on infants with CF, H. influenzae is the bacteria most commonly cultured, and it is only later in childhood that P. aeruginosa becomes more prevalent (12). Approximately 82% of children with P. aeruginosa had previous isolates of S. aureus or H. influenzae (14). During exacerbations of CF lung disease in children, the isolation rate of H. influenzae was significantly greater than at other times (38).
NTHi Biofilms: A Possible Link to Early CF Lung Disease
For reasons that are not fully understood, NTHi causes persistent infections despite appropriate antibiotic therapy in diseases with abnormalities of mucociliary clearance, such as COPD (35, 41) and CF (11, 38–40). Because H. influenzae forms biofilms on airway epithelia that display decreased susceptibility to antimicrobials, this may help to explain why such infections persist despite antibiotic treatment in OM, COPD, and CF (40, 42, 43). In CF, some strains of drug-resistant H. influenzae can persist for up to 7 yr, and many patients are colonized with multiple strains of H. influenzae (40). Supporting the relevance and importance of these findings, several other lines of evidence suggest NTHi disease may be biofilm related (9, 10, 31, 35).
We also show that H. influenzae biofilms cause the release of proinflammatory cytokines and chemokines by airway epithelia. Previous studies reported that H. influenzae stimulates respiratory epithelial production of macrophage-inflammatory proteins, IL-8, and TNF-α in cell culture (44) and mouse models (45). These data support the idea that airway epithelia play key roles in innate immunity and may help to signal and coordinate both innate and adaptive immune responses. Because some of these factors, such as CCL20, possess antimicrobial activity (46), the epithelia may additionally have direct innate immune roles to help clear bacterial infections. It is currently unknown if any of these factors may influence bacterial biofilm formation, but other airway surface liquid components, such as lactoferrin, are known to possess such properties (47).
It is hypothesized that inflammation from H. influenzae causes damage to the airway and paves the way for colonization with P. aeruginosa (13). Supporting a role for H. influenzae in causing inflammation and airway damage, single-pathogen infections with H. influenzae, S. aureus, or P. aeruginosa have no significant differences in BALF IL-8 or neutrophil counts, two hallmark inflammatory markers in CF lung disease (15). Many CF centers currently do not treat H. influenzae detected on throat or BALF cultures. The data presented here provide a rationale for considering treatment of early, asymptomatic colonization with H. influenzae. A recent European consensus report for early intervention in CF recommended 2- to 4-wk antibiotic courses for eradication of H. influenzae when it is detected (48).
Conclusions
To our knowledge, this is the first in vivo evidence of NTHi biofilm formation within the human lower airways. Furthermore, this is the first in vitro model for bacterial biofilm formation on polarized human airway epithelia. Over 4 d of coculture on airway epithelial cells, NTHi formed large, adherent biofilm structures with decreased antibiotic susceptibility. These data also suggest that the airway epithelium plays an active role in innate immune responses to NTHi infections, as evidenced by cytokine and chemokine release, and NF-κB signaling. Most significantly, in addition to NTHi isolates forming biofilms in vitro, these data provide preliminary in vivo evidence that H. influenzae forms structures consistent with biofilms very early in the pathogenesis of CF, even before the onset of clinical signs or symptoms of lung disease.
Supplementary Material
Acknowledgments
The authors thank the Cell Culture Core at the University of Iowa for support in preparing airway epithelial cultures. They thank Jian Q. Shao for his technical assistance on microscopic sample preparation and imaging, Meg Ketterer for her help with the 6E4 antibody preparation, and Dwight Look for providing critical commentary on the manuscript. They also thank Lauren Bakaletz for graciously providing plasmid pRSM2211.
Supported primarily by grants from the National Institutes of Health (HD27748 and HL67992, T.D.S; AI30040 and AI24616 M.A.A.; HL61234, P.B.M.), with additional support from the Cell Culture Core, funded by the Center for Gene Therapy for Cystic Fibrosis (NIH P30 DK-54759, John Englehardt) and the Cystic Fibrosis Foundation, and the American Lung Association (RG-11408-N, T.D).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200509-1459OC on May 4, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Lam J, Chan R, Lam K, Costerton JW. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun 1980;28:546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000;407:762–764. [DOI] [PubMed] [Google Scholar]
- 3.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 2003;57:677–701. [DOI] [PubMed] [Google Scholar]
- 4.Nickel JC, Ruseska I, Wright JB, Costerton JW. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 1985;27:619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol 2003;171:4329–4339. [DOI] [PubMed] [Google Scholar]
- 6.Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun 2004;72:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greiner LL, Watanabe H, Phillips NJ, Shao J, Morgan A, Zaleski A, Gibson BW, Apicella MA. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect Immun 2004;72:4249–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol 2002;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, Daigle BJ, Ehrlich MD, Post JC. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2002;287:1710–1715. [DOI] [PubMed] [Google Scholar]
- 10.Post JC. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 2001;111:2083–2094. [DOI] [PubMed] [Google Scholar]
- 11.Bilton D, Pye A, Johnson MM, Mitchell JL, Dodd M, Webb AK, Stockley RA, Hill SL. The isolation and characterization of non-typeable Haemophilus influenzae from the sputum of adult cystic fibrosis patients. Eur Respir J 1995;8:948–953. [PubMed] [Google Scholar]
- 12.Rosenfeld M, Gibson RL, McNamara S, Emerson J, Burns JL, Castile R, Hiatt P, McCoy K, Wilson CB, Inglis A, et al. Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr Pulmonol 2001;32:356–366. [DOI] [PubMed] [Google Scholar]
- 13.Smith A. Pathogenesis of bacterial bronchitis in cystic fibrosis. Pediatr Infect Dis J 1997;16:91–95. [Discussion 95–96, 123–126.] [DOI] [PubMed] [Google Scholar]
- 14.Abman SH, Ogle JW, Harbeck RJ, Butler-Simon N, Hammond KB, Accurso FJ. Early bacteriologic, immunologic, and clinical courses of young infants with cystic fibrosis identified by neonatal screening. J Pediatr 1991;119:211–217. [DOI] [PubMed] [Google Scholar]
- 15.Muhlebach MS, Stewart PW, Leigh MW, Noah TL. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am J Respir Crit Care Med 1999;160:186–191. [DOI] [PubMed] [Google Scholar]
- 16.Starner TD, Zhang N, Shao J, Apicella MA, McCray PB Jr. Haemophilus influenzae forms biofilms on airway epithelia: implications for early cystic fibrosis lung disease. Pediatr Pulmonol Suppl 2005;28:300. [Google Scholar]
- 17.Starner TD, Zhang N, Shao J, Apicella MA, McCray PB Jr. Nontypeable Haemophilus influenzae forms biofilms on human airway epithelia. Pediatr Pulmonol Suppl 2004;27:278. [Google Scholar]
- 18.Campagnari AA, Spinola SM, Lesse AJ, Kwaik YA, Mandrell RE, Apicella MA. Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb Pathog 1990;8:353–362. [DOI] [PubMed] [Google Scholar]
- 19.Campagnari AA, Gupta MR, Dudas KC, Murphy TF, Apicella MA. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect Immun 1987;55:882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PA, Samuels NM, Phillips NJ, Munson RS Jr, Bozue JA, Arseneau JA, Nichols WA, Zaleski A, Gibson BW, Apicella MA. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J Biol Chem 2002;277:14598–14611. [DOI] [PubMed] [Google Scholar]
- 21.Hood DW, Cox AD, Gilbert M, Makepeace K, Walsh S, Deadman ME, Cody A, Martin A, Mansson M, Schweda EK, et al. Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol Microbiol 2001;39:341–350. [DOI] [PubMed] [Google Scholar]
- 22.Mason KM, Munson RS Jr, Bakaletz LO. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect Immun 2003;71:3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell MA, Skowronek K, Kauc L, Goodgal SH. Electroporation of Haemophilus influenzae is effective for transformation of plasmid but not chromosomal DNA. Nucleic Acids Res 1991;19:3625–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 1998;28:449–461. [DOI] [PubMed] [Google Scholar]
- 25.Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, Welsh MJ. An in vitro model of differentiated human airway epithelia: methods for establishing primary cultures. Methods Mol Biol 2002;188:115–137. [DOI] [PubMed] [Google Scholar]
- 26.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000;146:2395–2407. [DOI] [PubMed] [Google Scholar]
- 27.Isberg RR, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 1985;317:262–264. [DOI] [PubMed] [Google Scholar]
- 28.Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol 2000;37:13–27. [DOI] [PubMed] [Google Scholar]
- 29.Sanlioglu S, Williams CM, Samavati L, Butler NS, Wang G, McCray PB Jr, Ritchie TC, Hunninghake GW, Zandi E, Engelhardt JF. Lipopolysaccharide induces Rac1-dependent reactive oxygen species formation and coordinates tumor necrosis factor-alpha secretion through IKK regulation of NF-kappa B. J Biol Chem 2001;276:30188–30198. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Olinsky A, Phelan PD. Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr Pulmonol 1996;21:267–275. [DOI] [PubMed] [Google Scholar]
- 31.Rayner MG, Zhang Y, Gorry MC, Chen Y, Post JC, Ehrlich GD. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 1998;279:296–299. [DOI] [PubMed] [Google Scholar]
- 32.Chmiel JF, Berger M, Konstan MW. The role of inflammation in the pathophysiology of CF lung disease. Clin Rev Allergy Immunol 2002;23:5–27. [DOI] [PubMed] [Google Scholar]
- 33.Anwar H, van Biesen T, Dasgupta M, Lam K, Costerton JW. Interaction of biofilm bacteria with antibiotics in a novel in vitro chemostat system. Antimicrob Agents Chemother 1989;33:1824–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams I, Venables WA, Lloyd D, Paul F, Critchley I. The effects of adherence to silicone surfaces on antibiotic susceptibility in Staphylococcus aureus. Microbiology 1997;143:2407–2413. [DOI] [PubMed] [Google Scholar]
- 35.Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:266–272. [DOI] [PubMed] [Google Scholar]
- 36.Kuklinska D, Kilian M. Relative proportions of Haemophilus species in the throat of healthy children and adults. Eur J Clin Microbiol 1984;3:249–252. [DOI] [PubMed] [Google Scholar]
- 37.Farjo RS, Foxman B, Patel MJ, Zhang L, Pettigrew MM, McCoy SI, Marrs CF, Gilsdorf JR. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr Infect Dis J 2004;23:41–46. [DOI] [PubMed] [Google Scholar]
- 38.Rayner RJ, Hiller EJ, Ispahani P, Baker M. Haemophilus infection in cystic fibrosis. Arch Dis Child 1990;65:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moller LV, Timens W, van der Bij W, Kooi K, de Wever B, Dankert J, van Alphen L. Haemophilus influenzae in lung explants of patients with end-stage pulmonary disease. Am J Respir Crit Care Med 1998;157:950–956. [DOI] [PubMed] [Google Scholar]
- 40.Roman F, Canton R, Perez-Vazquez M, Baquero F, Campos J. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J Clin Microbiol 2004;42:1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miravitlles M, Espinosa C, Fernandez-Laso E, Martos JA, Maldonado JA, Gallego M. Relationship between bacterial flora in sputum and functional impairment in patients with acute exacerbations of COPD. Study Group of Bacterial Infection in COPD. Chest 1999;116:40–46. [DOI] [PubMed] [Google Scholar]
- 42.Dagan R, Abramson O, Leibovitz E, Greenberg D, Lang R, Goshen S, Yagupsky P, Leiberman A, Fliss DM. Bacteriologic response to oral cephalosporins: are established susceptibility breakpoints appropriate in the case of acute otitis media? J Infect Dis 1997;176:1253–1259. [DOI] [PubMed] [Google Scholar]
- 43.Groeneveld K, van Alphen L, Eijk PP, Visschers G, Jansen HM, Zanen HC. Endogenous and exogenous reinfections by Haemophilus influenzae in patients with chronic obstructive pulmonary disease: the effect of antibiotic treatment on persistence. J Infect Dis 1990;161:512–517. [DOI] [PubMed] [Google Scholar]
- 44.Frick AG, Joseph TD, Pang L, Rabe AM, St Geme JW III, Look DC. Haemophilus influenzae stimulates ICAM-1 expression on respiratory epithelial cells. J Immunol 2000;164:4185–4196. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Moser C, Louboutin JP, Lysenko ES, Weiner DJ, Weiser JN, Wilson JM. Toll-like receptor 4 mediates innate immune responses to Haemophilus influenzae infection in mouse lung. J Immunol 2002;168:810–815. [DOI] [PubMed] [Google Scholar]
- 46.Starner TD, Barker CK, Jia HP, Kang Y, McCray PB Jr. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol 2003;29:627–633. [DOI] [PubMed] [Google Scholar]
- 47.Singh P, Parsek M, Tack B, Costerton J, Greenberg E, Welsh M. The activity of airway surface liquid antimicrobial factors against P. aeruginosa biofilms [abstract]. Am J Respir Crit Care Med 2000;161:A128. [Google Scholar]
- 48.Doring G, Hoiby N. Early intervention and prevention of lung disease in cystic fibrosis: a European consensus. J Cyst Fibros 2004;3:67–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.