Abstract
Rationale: Patients with severe chronic obstructive pulmonary disease (COPD) may have varying levels of disability despite similar levels of lung function. This variation may reflect different COPD subtypes, which may have different genetic predispositions.
Objectives: To identify genetic associations for COPD-related phenotypes, including measures of exercise capacity, pulmonary function, and respiratory symptoms.
Methods: In 304 subjects from the National Emphysema Treatment Trial, we genotyped 80 markers in 22 positional and/or biologically plausible candidate genes. Regression models were used to test for association, using a test–replication approach to guard against false-positive results. For significant associations, effect estimates were recalculated using the entire cohort. Positive associations with dyspnea were confirmed in families from the Boston Early-Onset COPD Study.
Results: The test–replication approach identified four genes—microsomal epoxide hydrolase (EPHX1), latent transforming growth factor-β binding protein-4 (LTBP4), surfactant protein B (SFTPB), and transforming growth factor-β1 (TGFB1)—that were associated with COPD-related phenotypes. In all subjects, single-nucleotide polymorphisms (SNPs) in EPHX1 (p ⩽ 0.03) and in LTBP4 (p ⩽ 0.03) were associated with maximal output on cardiopulmonary exercise testing. Markers in LTBP4 (p ⩽ 0.05) and SFTPB (p = 0.005) were associated with 6-min walk test distance. SNPs in EPHX1 were associated with carbon monoxide diffusing capacity (p ⩽ 0.04). Three SNPs in TGFB1 were associated with dyspnea (p ⩽ 0.002), one of which replicated in the family study (p = 0.02).
Conclusions: Polymorphisms in several genes seem to be associated with COPD-related traits other than FEV1. These associations may identify genes in pathways important for COPD pathogenesis.
Keywords: dyspnea, emphysema, exercise tolerance, genetic association, pulmonary function tests
Chronic obstructive pulmonary disease (COPD) is an inherently heterogeneous disorder. Within a given individual, there may be varying contributions of emphysema, chronic bronchitis, and small airway disease. However, the majority of studies of COPD genetics have focused only on the presence or absence of COPD diagnosis, which may or may not have been based on spirometry (1). Few studies have analyzed quantitative COPD-related traits, and most of these have examined only spirometric measures, such as FEV1 and the ratio of FEV1 to FVC (2).
The National Emphysema Treatment Trial (NETT) is a multicenter, randomized, clinical trial comparing lung volume reduction surgery (LVRS) with medical management for severe COPD (3). One of the important findings of NETT was that two COPD-related phenotypes, radiographic distribution of emphysema and exercise capacity, could be used to identify subgroups of patients with varying responses to LVRS (4). These subgroups identify patients with different treatment responses and might also be used to identify patients with different disease mechanisms. If so, using such subgroups may aid in the discovery of genes that may predict LVRS outcome or genes that may predispose to the development of COPD.
We hypothesized that different genetic factors influence different COPD-related functional capacity phenotypes, including pulmonary function measures, exercise capacity, and respiratory symptoms. To test this hypothesis, we examined genetic associations for these phenotypes in individuals participating in the NETT Genetics Ancillary Study. Some of the genotype data used in this study have been included in case-control studies for COPD susceptibility (5–7). Results from the current study have been previously reported as an abstract (8).
METHODS
Study Subjects
Subject enrollment and data collection in NETT have been described (3, 4). The current analysis included 304 non-Hispanic white subjects in the NETT Genetics Ancillary Study. After providing written informed consent, these NETT participants provided a blood sample for DNA extraction for genetic studies of COPD. Phenotypes were measured prior to randomization but after pulmonary rehabilitation. The study was approved by the institutional review boards at participating NETT centers. Additional details can be found in the online supplement.
Candidate Genes and Genotyping
Table 1 lists the 22 candidate genes, selected by one of three criteria: (1) positional candidate genes in chromosomal regions linked to COPD-related traits (2), (2) candidate genes based on presumed COPD pathophysiology, or (3) genes with published COPD associations. These candidate gene variants have been analyzed previously in genetic association studies where the NETT subjects (cases) were compared with smoking control subjects (5–7).
TABLE 1.
GENETIC POLYMORPHISMS ANALYZED IN THE NETT GENETICS ANCILLARY STUDY
| Gene Name | HUGO Symbol | Selection Criteria* | Markers | Comment |
|---|---|---|---|---|
| α1-Antichymotrypsin | SERPINA3 | Assoc. | 2 | |
| Elastin | ELN | Biology | 1 | |
| Microsomal epoxide hydrolase | EPHX1 | Assoc. | 8 | |
| Vitamin D binding protein | GC | Assoc. | 2 | |
| G-protein–coupled receptor C5A | GPCR5A | Position | 1 | |
| Glutathione S-transferase M1 | GSTM1 | Assoc. | 1 | Null deletion |
| Glutathione S-transferase P1 | GSTP1 | Assoc. | 2 | |
| Heme oxygenase-1 | HMOX1 | Assoc. | 1 | STR |
| Interleukin-8 receptor α | IL8RA | Position | 9 | |
| Interleukin-8 receptor β | IL8RB | Position | 6 | |
| Latent transforming growth factor β binding protein-4 | LTBP4 | Position | 4 | |
| Matrix Gla Protein | MGP | Position | 3 | |
| Microsomal glutathione S-transferase-1 | MGST1 | Position | 4 | |
| Matrix metalloprotease-1 | MMP1 | Assoc. | 1 | 1-bp insertion/deletion |
| Matrix metalloprotease-9 | MMP9 | Assoc. | 1 | STR |
| Matrix metalloprotease-12 | MMP12 | Biology | 3 | |
| Serine protease inhibitor E2 | SERPINE2 | Position | 17 | |
| Surfactant protein B | SFTPB | Assoc. | 2 | 1 SNP, 1 STR |
| Surfactant protein D | SFTPD | Assoc. | 2 | |
| Transforming growth factor β1 | TGFB1 | Assoc., Position | 5 | |
| Tissue inhibitor of metalloproteases-2 | TIMP2 | Assoc. | 1 | |
| Tumor necrosis factor α | TNF | Assoc. | 4 | |
| Totals: 22 genes, 80 polymorphisms |
Definition of abbreviations: SNP = single-nucleotide polymorphism; STR = short tandem repeat.
All markers are SNPs unless otherwise noted. See Table E1 of the online supplement for references.
Assoc., significant associations of variant(s) within this gene with chronic obstructive pulmonary disease (COPD) in the published literature (6); Biology, biologically plausible candidate gene for COPD; Position, positional candidate gene in regions of COPD linkage (2).
Details of genotyping methods have been reported previously (5–7). Single-nucleotide polymorphisms (SNPs) were genotyped using the 5′ to 3′ exonuclease assay in TaqMan (Applied Biosystems, Foster City, CA) (9) or with unlabeled minisequencing reactions and mass spectrometry in Sequenom (San Diego, CA) (10). For three short tandem repeat (STR) markers, polymerase chain reaction was performed using fluorescence-labeled and unlabeled primers, and product sizes were assessed by capillary electrophoresis on an ABI 3100 machine (Applied Biosystems). A 1-bp insertion–deletion in MMP1 and the GSTM1 null deletion were genotyped with TaqMan assays.
Statistical Analysis
To reduce the risk of false-positive results, the 304 subjects were randomly divided into a test set of 150 subjects and a replication set of the remaining 154 subjects. In the test sample, each genotype–phenotype association was analyzed using linear regression, assuming an additive genetic model, adjusted for relevant covariates. Markers with a p value less than 0.1, selected to increase sensitivity to detect valid associations, were analyzed in the replication sample. A p value less than 0.05 in the replication set was used to define a statistically significant result. All variants in genes that had at least one significant marker in the test–replication process were reanalyzed in all 304 subjects to improve precision of the effect estimates. Low exercise tolerance was defined using sex-specific thresholds (4) and was analyzed using logistic regression. STR markers were analyzed by comparing each allele (frequency > 5%) to all other alleles, assuming additive genetic models. Data were analyzed using SAS (SAS Institute, Cary, NC). More details regarding the genetic association analysis methods can be found in the online supplement.
Linkage disequilibrium (pairwise r2) between SNPs in the same gene was calculated in Haploview (11). Haplotype analysis was performed using the expectation-maximization algorithm and score tests, implemented in haplo.stats (12). Global haplotype p values were derived from a minimum of 1,000 simulations.
Dyspnea Replication Analysis
Genes associated with dyspnea in the test–replication analysis in the NETT subjects were examined in participants in the Boston Early-Onset COPD Study. Details of subject recruitment and phenotyping in the Boston Early-Onset COPD Study have been described previously (13); extended pedigrees were ascertained through a proband younger than 53 yr with FEV1 of less than 40% predicted. The Boston Early-Onset COPD Study questionnaire included a modified version of the Medical Research Council (MRC) dyspnea scale (14). Family-based association analysis was performed using the pedigree family-based association test, implemented in the PBAT software package (15), assuming additive genetic models, adjusting for age, sex, pack-years of smoking, and post-bronchodilator FEV1 (% predicted).
RESULTS
Study Subjects
Characteristics of the 304 non-Hispanic white participants in the NETT Genetics Ancillary Study are shown in Table 2. There was a predominance of men (63.8%) and a mean age of 67.3 yr. These findings are similar to the entire cohort of 1,218 NETT participants (4). On average, subjects had severe impairment on tests of pulmonary function and exercise capacity, although there was more variability in the measurements of the latter. Phenotype distributions were not significantly different in the test and replication samples, with the exception of the University of California, San Diego, Shortness of Breath Questionnaire (UCSD SOBQ) score (t test, p = 0.02) (16). This difference did not remain significant when adjusted for multiple testing.
TABLE 2.
CHARACTERISTICS OF THE PARTICIPANTS IN THE NETT GENETICS ANCILLARY STUDY
| Male sex, n (%) | 194 (63.8) |
| Age, yr | 67.3 ± 6.0 |
| Pack-years of smoking (n = 300) | 67.4 ± 31.6 |
| Post-bronchodilator FEV1, % predicted | 27.9 ± 7.4 |
| DlCO, % predicted (n = 302) | 29.9 ± 10.0 |
| Maximum work, watts | 42.8 ± 22.2 |
| 6-min walk test distance, ft | 1,240.5 ± 301.1 |
| Body mass index, kg/m2 | 25.0 ± 3.5 |
| UCSD SOBQ score* | 59.4 ± 17.8 |
| BODE index (modified)† | 4.7 ± 1.6 |
Definition of abbreviations: BODE = Body mass index, airflow Obstruction, Dyspnea, Exercise capacity; UCSD SOBQ = University of California, San Diego, Shortness of Breath Questionnaire.
Values are mean ± SD unless otherwise indicated; n = 304, except as noted.
Higher scores indicate more severe dyspnea (16).
The BODE index is a 10-point composite emphysema severity score; higher scores predict poorer outcomes (17). We modified the original BODE by using quartiles of the UCSD SOBQ to calculate the dyspnea measure instead of the Medical Research Council dyspnea scale. UCSD SOBQ score ⩽ 52 contributed 0 points, 53–62 contributed 1 point, 63–77 contributed 2 points, and > 77 contributed 3 points.
Each of the quantitative phenotypes tested was significantly correlated with each of the other phenotypes (p < 0.01, Pearson correlation), although most of the correlations were weak (|r| < 0.4). The strongest correlations were between maximum work achieved during the exercise test and 6-min walk test distance (r = 0.59, p < 0.0001) and between modified BODE (Body mass index, airflow Obstruction, Dyspnea, Exercise tolerance) score (17) and each of its components (FEV1: r = −0.43, p < 0.0001; UCSD SOBQ score: r = 0.79, p < 0.0001; 6-min walk distance: r = −0.63, p < 0.0001). BODE score was also correlated with maximum work (r = −0.51, p < 0.0001).
Test–Replication Association Analysis
The results of the test–replication analysis in the NETT Genetics Ancillary Study are shown in Table 3. In the test set, 20 markers in eight genes showed nominal evidence for association (p < 0.1) with maximum work capacity as a continuous trait (data not shown). Only two SNPs were significant in the replication analysis, including coding SNPs in microsomal epoxide hydrolase (EPHX1 rs1051740) and latent transforming growth factor-β binding protein 4 (LTBP4 rs2077407). Eleven markers in six genes were associated (p < 0.1) with low exercise capacity as a binary outcome in the screening analysis; none was replicated. In the analysis of 6-min walk test distance, 14 markers in nine genes were associated in the test set. Only one marker, the 263-bp allele of the surfactant protein B (SFTPB) STR, was significant on replication. Another allele of the SFTPB STR, length 259 bp, was not associated in the test set but was marginally associated (p = 0.05) in the replication analysis.
TABLE 3.
RESULTS OF THE TEST–REPLICATION ANALYSIS OF FUNCTIONAL CAPACITY TRAITS IN THE NETT GENETICS ANCILLARY STUDY
| p Value
|
||||
|---|---|---|---|---|
| Phenotype* | Gene | Marker | Test Set | Replication Set |
| Maximum work, watts | EPHX1 | rs1051740 | 0.04 | 0.001 |
| LTBP4 | rs2077407 | 0.02 | 0.003 | |
| Low exercise capacity† | — | — | — | n.s. |
| 6MWT distance, ft | SFTPB | D2S388, 263bp allele | 0.07 | 0.03 |
| FEV1, post-BD, liters | — | — | — | n.s. |
| DlCO, % predicted | EPHX1 | rs868966 | 0.06 | 0.02 |
| UCSD SOBQ score | EPHX1 | rs2292566 | 0.02 | 0.005 |
| SFTPB | D2S388, 259bp allele | 0.02 | 0.03 | |
| TGFB1 | rs1800469 | 0.008 | 0.02 | |
| rs2241712 | 0.007 | 0.04 | ||
| BODE index | — | — | — | n.s. |
Definition of abbreviations: BODE = Body mass index, airflow Obstruction, Dyspnea, Exercise tolerance; n.s. = not significant; UCSD SOBQ = University of California, San Diego, Shortness of Breath Questionnaire; 6MWT = 6-min walk test.
Associations with p < 0.1 in the test set and p < 0.05 in the replication set are shown.
Table 3a.
| *Phenotype | Covariates Included in regression models |
|---|---|
| Maximum work | Age, sex, pack-years, FEV1 (% predicted), height |
| Low exercise capacity | Age, pack-years, FEV1 (% predicted), height |
| 6MWT distance, ft | Age, sex, pack-years, FEV1 (% predicted) |
| FEV1, liters | ge, sex, pack-years, height |
| DlCO, % predicted | Age, sex, pack-years, FEV1 (% predicted) |
| UCSD SOBQ score | Age, sex, pack-years, FEV1 (% predicted) |
| BODE index (modified) | Age, sex, pack-years |
Low exercise capacity is defined as maximum work ⩽ 40 W for men or ⩽ 25 W for women (4).
In the analysis of post-bronchodilator FEV1 (L), six markers in six genes were associated (p < 0.1) in the test set; none of these were replicated. Nine markers in seven genes were found in the screening analysis of carbon monoxide diffusing capacity (DlCO); one of these, a promoter SNP in EPHX1 (rs868966), was replicated. In the screening analysis of the UCSD SOBQ score, 23 markers in nine genes were nominally associated (p < 0.1); four of these associations were significant on replication. These replicated associations include an SNP in EPHX1 (rs2292566), the 259-bp allele of the SFTPB STR, and two SNPs in TGFB1 (rs1800469, rs2241712). In the analysis of the modified BODE index, 14 markers in seven genes had p values of less than 0.1 on screening, but none were replicated. One allele of the SFTPB STR, length 269 bp, was significantly associated with modified BODE index in the test set, but another allele, length 263 bp, had a p value of less than 0.05 in the replication set.
Comprehensive Association Analysis of Promising Candidate Genes
The test–replication process identified four genes for further analysis; at least one marker in each of these genes was associated with at least one trait. All markers in these genes were then tested for association with all seven phenotypes in the full set of 304 NETT subjects. The variants analyzed included eight SNPs in EPHX1, four SNPs in LTBP4, one SNP in SFTPB, one STR near SFTPB, and five SNPs in TGFB1. These regression models are shown in Table 4.
TABLE 4.
GENETIC ASSOCIATION RESULTS FOR FOUR SIGNIFICANT GENES (IDENTIFIED IN TEST–REPLICATION PROCEDURE) IN 304 PARTICIPANTS IN THE NETT GENETICS ANCILLARY STUDY
| A. Exercise Capacity Phenotypes
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maximum Work (watts)
|
Low Exercise Capacity
|
6MWT Distance (ft)
|
||||||||||||
| Gene | SNP or STR (allele) | β (SE) | p Value | OR (95% CI) | p Value | β (SE) | p Value | |||||||
| EPHX1 | rs1877724 intron | −3.7 (1.6) | 0.03 | n.s. | n.s. | |||||||||
| rs1051740 exon, coding | −5.6 (1.5) | 0.0002 | 1.75 (1.16–2.62) | 0.007 | n.s. | |||||||||
| LTBP4 | rs2303729 exon, coding | 5.1 (1.4) | 0.0003 | 0.43 (0.29–0.65) | < 0.0001 | 49.1 (23.3) | 0.04 | |||||||
| rs1131620 exon, coding | 3.0 (1.4) | 0.03 | 0.58 (0.39–0.85) | 0.006 | 44.8 (22.3) | 0.05 | ||||||||
| rs1051303 exon, coding | 3.7 (1.4) | 0.007 | 0.56 (0.38–0.83) | 0.004 | 46.6 (22.2) | 0.04 | ||||||||
| rs2077407 exon, silent | 8.9 (2.3) | 0.0001 | 0.46 (0.24–0.89) | 0.02 | n.s. | |||||||||
| SFTPB | D2S388, 259bp | n.s. | 1.58 (1.01–2.48) | 0.05 | n.s. | |||||||||
| D2S388, 263bp | n.s. | n.s. | 68.7 (24.5) | 0.005 | ||||||||||
| B. Pulmonary Function Phenotypes
|
||||||||||||||
| DlCO (% predicted)
|
Post-BD FEV1 (L)
|
|||||||||||||
| Gene | SNP or STR (allele) | β (SE) | p Value | β (SE) | p Value | |||||||||
| EPHX1 | rs868966 promoter | 2.2 (0.8) | 0.004 | n.s. | ||||||||||
| rs2292566 exon, silent | 3.8 (1.2) | 0.002 | n.s. | |||||||||||
| rs2234922 exon, coding | 2.2 (1.1) | 0.04 | n.s. | |||||||||||
| C. Symptom Severity Phenotypes
|
||||||||||||||
| UCSD SOBQ Score
|
BODE Index
|
|||||||||||||
| Gene | SNP or STR (allele) | β (SE) | p Value | β (SE) | p Value | |||||||||
| LTBP4 | rs1131620 exon, coding | n.s. | −0.26 (0.12) | 0.03 | ||||||||||
| rs1051303 exon, coding | n.s. | −0.26 (0.12) | 0.03 | |||||||||||
| SFTPB | D2S388, 263bp allele | n.s. | −0.34 (0.13) | 0.01 | ||||||||||
| TGFB1 | rs2241712 promoter | 5.4 (1.5) | 0.0005 | 0.42 (0.14) | 0.002 | |||||||||
| rs1800469 promoter | 5.7 (1.5) | 0.0002 | 0.46 (0.14) | 0.0008 | ||||||||||
| rs1982073 exon, coding | 4.7 (1.5) | 0.002 | 0.41 (0.13) | 0.002 | ||||||||||
Definition of abbreviations: BODE = Body mass index, airflow Obstruction, Dyspnea, Exercise tolerance; CI = confidence interval; n.s. = not significant; SNP = single-nucleotide polymorphism; STR = short tandem repeat; UCSD SOBQ = University of California, San Diego, Shortness of Breath Questionnaire; 6MWT = 6-min walk test.
Effect estimates (β or odds ratio [OR]) are adjusted for covariates listed in footnote to Table 3. Additive genetic models are assumed. Significant associations (p ⩽ 0.05) are shown.
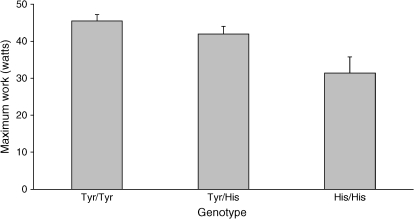
Two SNPs in EPHX1, including a coding variant (rs1051740, Tyr113His, also known as the “slow” allele), were associated with a reduction in maximal work output (Table 4A, Figure 1); the “slow” variant also increased the odds of being classified in the low-exercise subgroup (odds ratio, 1.75; 95% confidence interval, 1.16–2.62). Four SNPs in LTBP4 were significantly associated with an increase in work capacity and reduced odds of low exercise tolerance; three of these also led to a greater 6-min walk test distance. Presence of one allele of the SFTPB STR (259 bp) increased the odds of low exercise tolerance; individuals with another allele (263 bp) had greater 6-min walk test distance. SNPs in TGFB1 were not associated with exercise capacity traits.
Figure 1.
Effect of EPHX1 rs1051740 coding SNP (Tyr113His) on exercise capacity. Tyr113 is the wild-type allele, and His113 is the “slow” variant. Mean values (+ SEM) for maximum work (watts) for each genotype are shown. p = 0.0002 for an additive genetic model, adjusted for age, sex, pack-years of smoking, FEV1 (% predicted), and height.
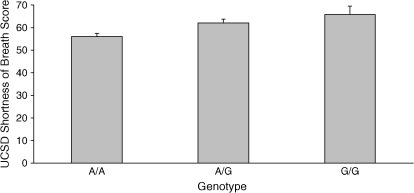
Three SNPs in EPHX1, including a coding variant (rs2234922, His139Arg, the “fast” allele), were associated with increased values of DlCO (Table 4B), but they were not associated with post-bronchodilator FEV1. None of the markers in LTBP4, SFTPB, and TGFB1 were associated with these pulmonary function phenotypes. Three SNPs in TGFB1, including two promoter SNPs (rs1800469, rs2241712) and one coding SNP (rs1982073, Leu10Pro), were associated with a higher UCSD SOBQ score, indicating more severe dyspnea (Table 4C, Figure 2). These three SNPs also led to an increase in modified BODE index (more severe disease). Two SNPs in LTBP4 and the 259-bp allele of the SFTPB STR were associated with a lower BODE index. EPHX1 was not associated with either symptom severity phenotype.
Figure 2.
Effect of TGFB1 rs2241712 promoter single-nucleotide polymorphism on dyspnea. Mean (+ SEM) of the University of California, San Diego (UCSD), Shortness of Breath Questionnaire scores for each genotype are shown. p = 0.0005 for an additive genetic model, adjusted for age, sex, pack-years of smoking, and FEV1 (% predicted).
Linkage Disequilibrium
In three of the four significant genes (EPHX1, LTBP4, and TGFB1), pairwise linkage disequilibrium (LD) between SNPs was determined by r2 (Table 5). Pairwise LD was not calculated for the fourth gene (SFTPB) because one of the two markers was a multiallelic STR. There was low LD between the eight SNPs in EPHX1; the highest r2 was 0.47 for two exonic SNPs (Table 5A, SNPs 6–7). Three SNPs in LTBP4 (Table 5B, SNPs 1–3) formed a block of LD. The two promoter SNPs in TGFB1 (Table 5C, SNPs 1–2) were in tight LD; these were both in LD with the Leu10Pro coding SNP (SNP 3). The two 3′ SNPs in TGFB1 (SNPs 4–5) were in strong LD. Haplotype analyses were performed as a secondary analysis to corroborate the results of the single SNP analyses. The haplotype results are available in the online supplement.
TABLE 5.
LINKAGE DISEQUILIBRIUM IN 304 SUBJECTS IN THE NETT GENETICS ANCILLARY STUDY
| A. Microsomal Epoxide Hydrolase | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rs868966 | rs1877724 | rs1051740 | rs2292566 | rs2260863 | rs2234922 | rs1051741 | rs360063 | ||||||||
| 1. rs868966 promoter | 0.08 | 0.03 | 0.08 | 0 | 0 | 0 | 0.02 | |||||||||
| 2. rs1877724 intron | 0.03 | 0.01 | 0.1 | 0 | 0 | 0 | ||||||||||
| 3. rs1051740 exon, Tyr113His | 0.05 | 0.14 | 0 | 0.01 | 0.1 | |||||||||||
| 4. rs2292566 exon, silent | 0.05 | 0 | 0.01 | 0 | ||||||||||||
| 5. rs2260863 intron | 0.03 | 0.02 | 0.03 | |||||||||||||
| 6. rs2234922 exon, His139Arg | 0.47 | 0.15 | ||||||||||||||
| 7. rs1051741 exon, silent | 0.08 | |||||||||||||||
| 8. rs360063 3′ UTR | ||||||||||||||||
| B. Latent Transforming Growth Factor-β Binding Protein-4 | ||||||||||||||||
| SNP | rs2303729 | rs1131620 | rs1051303 | rs2077407 | ||||||||||||
| 1. rs2303729 exon, coding | 0.72 | 0.73 | 0.12 | |||||||||||||
| 2. rs1131620 exon, coding | 0.99 | 0.13 | ||||||||||||||
| 3. rs1051303 exon, coding | 0.12 | |||||||||||||||
| 4. rs2077407 exon, silent | ||||||||||||||||
| C. Transforming Growth Factor β1 | ||||||||||||||||
| SNP | rs2241712 | rs1800469 | rs1982073 | rs6957 | rs2241718 | |||||||||||
| 1. rs2241712 promoter | 0.95 | 0.67 | 0.01 | 0.02 | ||||||||||||
| 2. rs1800469 promoter | 0.68 | 0 | 0.01 | |||||||||||||
| 3. rs1982073 exon, Leu10Pro | 0 | 0 | ||||||||||||||
| 4. rs6957 3′ genomic | 0.9 | |||||||||||||||
| 5. rs2241718 3′ genomic | ||||||||||||||||
Definition of abbreviation: SNP = single-nucleotide polymorphism.
Pairwise r2 values are shown.
Family-based Analysis
Associations with dyspnea were tested in 949 individuals from 127 extended pedigrees in the Boston Early-Onset COPD Study. The modified MRC dyspnea scale included in the study questionnaire ranged in value from 0 (no shortness of breath) to 5 (too breathless to leave the house or breathless on dressing or undressing) (14). Probands reported severe dyspnea, with a mean MRC score of 4.4. Using the extended pedigree family-based association test, one promoter SNP in TGFB1 was significantly associated with MRC score (rs2241712, p = 0.02); another promoter SNP showed a trend toward association (rs1800469, p = 0.07). Both of these SNPs had replicated associations with USCD SOBQ score in the NETT Genetics Ancillary Study subjects (Table 4C). Three other SNPs in TGFB1, eight SNPs in EPHX1, four SNPs in LTBP4, and one SNP and one STR in SFTPB were not associated with MRC dyspnea score in the Boston Early-Onset COPD families.
DISCUSSION
Using a well-characterized group of patients from NETT, we were able to demonstrate significant genetic associations for several COPD-related phenotypes. Polymorphisms in four genes—EPHX1, LTBP4, SFTPB, and TGFB1—were significantly associated with measures of functional capacity, including exercise capacity, pulmonary function tests, and respiratory symptoms. Many of the single SNP associations were confirmed in haplotype analyses. Spirometric traits have been analyzed previously as intermediate phenotypes in COPD genetics (2, 5, 6, 18). However, COPD genetics studies using measures of exercise tolerance and symptom severity have not been previously published.
Variants in EPHX1 were associated with traits in all three categories, including maximal work capacity on a cardiopulmonary exercise test, DlCO, and UCSD SOBQ score. SNPs in two genes in the TGF-β pathway, TGFB1 and LTBP4, were associated with maximal work capacity and UCSD SOBQ score. The association between TGFB1 and dyspnea was replicated in an independent population, using a family-based study design. In addition, an STR near SFTPB was associated with 6-min walk test distance and dyspnea index.
Microsomal epoxide hydrolase is an enzyme important in the metabolism of reactive epoxide intermediates, such as those found in cigarette smoke. A coding variant in exon 3 (rs1051740, Tyr113His), termed the “slow” variant because of its effect on enzyme activity, has been associated with COPD in case-control studies (19, 20); in the Lung Health Study, a haplotype carrying the slow variant was associated with rapid decline in lung function (21). A coding variant in exon 4 (rs2234922, His139Arg), the “fast” variant, was associated with COPD diagnosis in a study comparing the subjects in the NETT Genetics Ancillary Study to community control subjects (6); the slow allele was not associated. However, other studies have failed to confirm the associations with either polymorphism (22, 23). The functional effects of the “fast” and “slow” variants and their haplotypes found in vitro (24) have not been confirmed in vivo (25). Therefore, it is possible that other variants in EPHX1 may affect COPD susceptibility, leading to the inconsistency in previous genetic association studies.
TGFB1 is located on chromosome 19q, a region of the genome linked to COPD-related phenotypes (5). Wu and colleagues demonstrated an association between the Leu10Pro polymorphism (rs1982073) and COPD (26). Celedón and colleagues showed that the rs2241712 promoter polymorphism was associated with COPD and related traits in the Boston Early-Onset COPD Study families and in an analysis comparing the 304 NETT subjects to community control subjects (5). The other promoter SNP (rs1800469) and the Leu10Pro polymorphism were also associated with COPD susceptibility in this case-control analysis.
LTBP4 is a component of the extracellular matrix and is involved in TGF-β signaling. LTBP4 is also located in the linked region on chromosome 19q. No human genetic association studies have been reported, but a mouse model suggests the importance of LTBP4 in the development of pulmonary emphysema (27).
SFTPB is a hydrophobic protein involved in regulating surface tension in the alveoli. Mutations in SFTPB have been implicated in respiratory failure in full-term neonates (28). Several studies have found variants in or near SFTPB to be associated with COPD in adults (29, 30). Our group has reported an association between a coding SNP (Thr131Ile) and airflow obstruction in the Boston Early-Onset COPD Study families (6); in a model that accounted for gene-by-environment interaction, this SNP was also associated in the case-control study that included the NETT subjects.
The genes that we have found to be associated with COPD phenotypes can be placed into pathways that may relate to COPD pathogenesis. Pathways such as xenobiotic metabolism (EPHX1) (31), extracellular matrix properties (LTBP4) (32), and inflammation and cellular signaling (TGFB1) (33) have been areas of active investigation in COPD. The importance of surface tension (SFTPB) in COPD has not been widely studied, but a recent article describes mathematical models relating surface properties to the development of emphysema (34). The different genes associated in our study may underscore the heterogeneity of COPD. It is possible that different genes and pathways contribute in varying combinations to various COPD-related phenotypes. For example, we have found that SNPs in TGFB1 are associated with dyspnea but not with other functional measures in COPD, such as exercise capacity, despite the correlations between these traits. Narrower phenotype definitions and rational subgroup analyses may help to successfully identify and replicate associations for COPD candidate genes.
The present study has several limitations. Replication of significant association results is an important step in complex trait genetics, but we only had measurements of one of the phenotypes, dyspnea, in a separate replication population. The instrument used to measure dyspnea in this replication study, the modified MRC scale, has a narrower range of possible results than the UCSD SOBQ score used in NETT. This may reduce the power for replication. Despite these limitations, we were able to replicate the association of TGFB1 with dyspnea. The other phenotypes analyzed, such as performance on a cardiopulmonary exercise test, are not routinely collected in studies of COPD genetics but may be collected in future clinical trials of COPD therapies, allowing for continued study of these functional impairment traits. Replication of these associations will be the strongest protection from spurious results due to multiple testing.
A variable number of markers in each gene were genotyped in this study, with some genes having only one or two markers tested. If the markers tested were not the true functional variants (or in linkage disequilibrium with the functional variants), then significant associations could be missed. This may explain why three of our four most significant genes (EPHX1, LTBP4, and TGFB1) were genes with multiple genotyped SNPs. However, no significant associations were found for SERPINE2, which had the largest number of SNPs tested.
Spurious results arising from multiple testing are a major concern in genetic epidemiology, especially in studies of multiple markers and phenotypes, such as our analysis. The optimal approach to adjust for multiple testing is not clear (35, 36). Many of the widely used methods are inappropriate for correlated data, such as multiple SNPs in a single gene or multiple related phenotypes. Therefore, we used a test–replication procedure within the NETT Genetics Ancillary Study cohort to reduce the possibility of false-positive results due to multiple testing, accepting that it may have reduced power to detect valid genetic associations. Based on power calculations in the online supplement, the power would be adequate to detect genes with moderate effects using the split dataset approach. However, we cannot exclude the possibility that some of the other candidate gene polymorphisms we studied may be associated with COPD-related phenotypes. Nevertheless, we were able to identify significant associations for four candidate genes with measures of functional impairment in COPD. Future studies using similarly specific COPD-related phenotypes may be able to identify additional genetic associations for COPD in general and for more precise subgroups in particular. In the future, narrowly defined subgroups, possibly based on genetic polymorphisms, may be better able to predict response to COPD therapies, including LVRS.
Supplementary Material
Acknowledgments
The authors thank Jody Sylvia, Salvatore Mazza, Michael Hager, Molly Brown, Alison Brown, and Maura Regan for their technical assistance with sample management and genotyping and Greg Foster for providing the SAS code to compute BODE index. Other coinvestigators in the NETT Genetics Ancillary Study include Marcia Katz, Rob McKenna, Malcolm DeCamp, Mark Ginsburg, Neil MacIntyre, Philip Diaz, Andrew Ries, Mark Krasna, Larry Kaiser, and Zab Mosenifar.
Supported by National Institutes of Health grants HL61575, HL71393, HL075478, K08-HL080242, and T32-HL07427 and by an American Lung Association Career Investigator Award. The National Emphysema Treatment Trial was supported by the U.S. National Heart, Lung, and Blood Institute (contracts N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119), the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality.
This article has an online supplement, which is accessible from the issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200509-1452OC on February 2, 2006
Conflict of Interest Statement: C.P.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.L.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.C.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.A.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.O.B. received $2,500 and $3,500 in 2004 for speaking at conferences sponsored by Boehringer Ingelheim. G.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. F.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.D.S. received research funding during 2001–2005 from Boehringer Ingelheim, Dey Pharmaceuticals, GlaxoSmithKline, LaRoche, and ONO Pharmaceuticals. F.C.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.P.U. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.J.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.K.S. received grant support, consulting fees, and honoraria from GlaxoSmithKline for studies of COPD genetics. He also received a speaker fee from Wyeth for a talk on COPD genetics and has received honoraria from Bayer.
References
- 1.Sandford AJ, Silverman EK. Chronic obstructive pulmonary disease. 1: susceptibility factors for COPD the genotype-environment interaction. Thorax 2002;57:736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002;70:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The National Emphysema Treatment Trial Research Group. Rationale and design of The National Emphysema Treatment Trial: a prospective randomized trial of lung volume reduction surgery. Chest 1999;116:1750–1761. [DOI] [PubMed] [Google Scholar]
- 4.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 5.Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-β1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649–1656. [DOI] [PubMed] [Google Scholar]
- 6.Hersh CP, Demeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol 2005;33:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua A, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersh CP, DeMeo DL, Lazarus R, Celedon JC, Raby BA, Benditt J, Martinez F, Scanlon P, Sciurba F, Utz J, et al. Genetic determinants of functional impairment in the National Emphysema Treatment Trial [abstract]. Proc Am Thorac Soc 2005;2:A393. [Google Scholar]
- 9.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991;88:7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Ding H, Hung K, Guo B. A new MALDI-TOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Res 2000;28:E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 12.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 1998;157:1770–1778. [DOI] [PubMed] [Google Scholar]
- 14.Ferris BG. Epidemiology Standardization Project (American Thoracic Society). Am Rev Respir Dis 1978(Suppl);118:1–120. [PubMed] [Google Scholar]
- 15.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eakin EG, Resnikoff PM, Prewitt LM, Ries AL, Kaplan RM. Validation of a new dyspnea measure: the UCSD Shortness of Breath Questionnaire. University of California, San Diego. Chest 1998;113:619–624. [DOI] [PubMed] [Google Scholar]
- 17.Celli BR, Cote CG, Marin JM, Casanova C, Montes de Oca M, Mendez RA, Pinto Plata V, Cabral HJ. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med 2004;350:1005–1012. [DOI] [PubMed] [Google Scholar]
- 18.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 19.Xiao D, Wang C, Du MJ, Pang BS, Zhang HY, Xiao B, Liu JZ, Weng XZ, Su L, Christiani DC. Relationship between polymorphisms of genes encoding microsomal epoxide hydrolase and glutathione S-transferase P1 and chronic obstructive pulmonary disease. Chin Med J (Engl) 2004;117:661–667. [PubMed] [Google Scholar]
- 20.Smith CA, Harrison DJ. Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet 1997;350:630–633. [DOI] [PubMed] [Google Scholar]
- 21.Sandford AJ, Chagani T, Weir TD, Connett JE, Anthonisen NR, Pare PD. Susceptibility genes for rapid decline of lung function in the lung health study. Am J Respir Crit Care Med 2001;163:469–473. [DOI] [PubMed] [Google Scholar]
- 22.Takeyabu K, Yamaguchi E, Suzuki I, Nishimura M, Hizawa N, Kamakami Y. Gene polymorphism for microsomal epoxide hydrolase and susceptibility to emphysema in a Japanese population. Eur Respir J 2000;15:891–894. [DOI] [PubMed] [Google Scholar]
- 23.Yim JJ, Park GY, Lee CT, Kim YW, Han SK, Shim YS, Yoo CG. Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax 2000;55:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet 1994;3:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosagrahara VP, Rettie AE, Hassett C, Omiecinski CJ. Functional analysis of human microsomal epoxide hydrolase genetic variants. Chem Biol Interact 2004;150:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 2004;59:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterner-Kock A, Thorey IS, Koli K, Wempe F, Otte J, Bangsow T, Kuhlmeier K, Kirchner T, Jin S, Keski-Oja J, et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev 2002;16:2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nogee LM, Wert SE, Proffit SA, Hull WM, Whitsett JA. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am J Respir Crit Care Med 2000;161:973–981. [DOI] [PubMed] [Google Scholar]
- 29.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J 2001;18:482–490. [DOI] [PubMed] [Google Scholar]
- 30.Seifart C, Plagens A, Brodje D, Muller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers 2002;18:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama H, Geddes DM. Genes, oxidative stress, and the risk of chronic obstructive pulmonary disease. Thorax 1998;53:S10–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro SD. Evolving concepts in the pathogenesis of chronic obstructive pulmonary disease. Clin Chest Med 2000;21:621–632. [DOI] [PubMed] [Google Scholar]
- 33.Bartram U, Speer CP. The role of transforming growth factor beta in lung development and disease. Chest 2004;125:754–765. [DOI] [PubMed] [Google Scholar]
- 34.Ingenito EP, Tsai LW, Majumdar A, Suki B. On the role of surface tension in the pathophysiology of emphysema. Am J Respir Crit Care Med 2005;171:300–304. [DOI] [PubMed] [Google Scholar]
- 35.Curran-Everett D. Multiple comparisons: philosophies and illustrations. Am J Physiol Regul Integr Comp Physiol 2000;279:R1–R8. [DOI] [PubMed] [Google Scholar]
- 36.Kraft P. Multiple comparisons in studies of gene x gene and gene x environment interaction. Am J Hum Genet 2004;74:582–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.