Abstract
We describe a simple proteomic approach to investigate the differential protein expression patterns and identify the physiologically relevant angiogenic and anti-angiogenic factors involved in the hyaloid vascular system regression. Differentially-expressed proteins were identified using two-dimensional gel electrophoresis followed by nano-flow chromatography coupled to tandem mass spectrometry. These proteins are expected to provide insight as to their function in the early maintenance and eventual regression of the hyaloid vascular system.
Keywords: hyaloid vascular system, vasa hyaloidea propria, tunica vasculosa lentis, proteomic analysis, kininogen
INTRODUCTION
The hyaloid vascular system is a transiently existing network of capillaries that nourishes the immature lens and primary vitreous of the developing eye.1 The hyaloid artery runs from the back of the eye to the embryonic lens, giving rise to a capillary plexus that surrounds the lens consisting of the vasa hyaloidea propria, the tunica vasculosa lentis, and the pupillary membrane.2 In humans, the first elements of the hyaloid vasculature to undergo regression are the vasa hyaloidea propria, followed by tunica vasculosa lentis, and the pupillary membrane and lastly, the main hyaloid trunk; this process commences at 12 weeks of gestation and culminates in the involution of the entire hyaloid by 35–36 weeks of gestation.3 Failure of the hyaloid vascular system to regress in humans can lead to a condition known as persistent hyperplastic primary vitreous, which, if untreated, can result in permanent blindness.
In the mouse, the hyaloid vascular system forms at embryonic day 10.5 (E10.5) and is completed by E13.5.4 The pupillary membrane is completely regressed on post gestational day 16 (P16), the vasa hyaloidea propria disappears between P12 and P16, and the vessels of the tunica vasculosa lentis and the hyaloid artery remaining at P16 undergo gradual regression from P14 to P30.5–8
The precise mechanism of the regression of the hyaloid vascular system is still unknown, but several hypotheses have been postulated to explain this process. Mechanisms that are involved in the hyaloid vascular system regression include programmed degradation of the pupillary membrane and the hyaloid vascular system by macrophages,9–11 anti-angiogenic molecules in the vitreous humor,12–17 apoptosis factors (bcl-2, bax, and bak)2, 18 and the production of transforming growth factor-β (TGF- β), a potent inhibitor of vascular endothelial cell proliferation by the hyalocytes.5, 19, 20, 21
In addition, a reduction of endogenous “survival factors” below a critical threshold may induce apoptosis; these include fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), and vascular endothelial growth factor (VEGF).22 Mitchell et al.4 hypothesized that, during lens growth, the progressive physical separation of the lens epithelial cells from the endothelial cells may deprive the endothelium of an essential survival factor, such as VEGF, and induce hyaloid vascular system endothelial cell apoptosis.1 However, Feenay et al. demonstrated that the tunica vasculosa lentis degeneration was not affected by the addition of a VEGF-A antibody, suggesting that programmed regression may be independent of VEGF-A.23
In an attempt to investigate the potential role of other factors that may be involved in hyaloid vascular system regression, we employed a proteomic approach to identify specific qualitative and quantitative protein changes in the early (maintenance) and later (regression) phases of the hyaloid vascular system. We initially examined the gradual regression of the hyaloid vascular system of the mouse eye to identify the optimal time points for our analyses. We used two-dimensional gel electrophoresis (2-DE) maps and liquid chromatography coupled to tandem mass spectrometry to study the modifications of the protein expression profiles occurring in the mouse lens and vitreous at various stages of the regression of the hyaloid vascular system. Screening and sub-classification of the differentially expressed proteins allowed us to identify proteins that were down-regulated or up-regulated during hyaloid vascular system regression.
EXPERIMENTAL SECTION
Animals
Ten pregnant C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). Fifty newborn litters were used for our experiments. The newborn mice were sacrificed on postnatal days 1, 4, 8, and 16. Four eyes per time point were used for the histological studies. For the protein extraction studies, 20 eyes were used for P1 and P4 samples, and 12 eyes for P8 and P16 samples. Experiments were conducted in accordance with the Animal Care and Use Committee guidelines of the Massachusetts Eye and Ear Infirmary and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Morphometric Analysis of Hyaloid Vascular System Regression
Newborn mice were sacrificed and fixed with paraformaldehyde in PBS (phosphate-buffered saline). Then eyes were removed, immersed in buffered neutral formalin solution (100 ml 40% formalin, 900 ml distilled water, 4.0 gm monobasic sodium phosphate, 6.5 gm dibasic sodium phosphate), and embedded in paraffin. Tissue sections, 5 microns in thickness, were obtained using a microkeratome and were mounted on slides. The slides were stained with hematoxylin and eosin. 10 serial images located at the sagittal axis of each of the 4 eyeballs were analyzed per P1, P4, P8 and P16. Two masked observers counted the number of vessels of the pupillary membrane and the tunica vasculosa lentis in each field at 20X magnification. T-test was performed to analyze the difference among each time period.
Tissue Preparation for 2-DE Gels
Using a dry ice bed, the frozen eyes were scraped with a blade (Beaver) to remove the cornea, aqueous humor, conjunctiva, sclera, iris, ciliary body, uvea, and retina, leaving the lens surrounded by the pupillary membrane, tunica vasculosa lentis, and primary vitreous (containing vasa hyaloidea propria). The dry ice bed was used to prevent warming of the specimen and subsequent protein denaturation, tissue melting, and contamination by eye components other than the lens and primary vitreous.
Twenty specimens of lens with pupillary membrane, tunica vasculosa lentis, and vasa hyaloidea propria of P1, 20 of P4, 12 of P8, and 12 of P16 were pooled and processed as described before.5 The specimens were independently solubilized in 250 μl of total protein extraction buffer (7 M urea, 2 M thiourea, 1% (w/v) ASB-14 detergent, 40 mM Tris base, 0.001% bromophenol blue, 20% carrier ampholyte) and 2 μl of 200 mM tributylphosphine by mechanically homogenizing with an electrical tissue homogenizer for 5 min on ice. The homogenate was transferred into 1.5 ml microcentrifuge tubes. The protein extracts were cleared by centrifugation at 14,000 rpm for 20 min at 4°C to remove particulates. Protein concentration of the cleared supernatants was determined by using a compatible protein assay (Bio-Rad Laboratories). 125 μg total protein was loaded onto each gel.
Three samples per post-gestational day were loaded onto nonlinear immobilized pH gradient (IPG) gel strips (7 cm, pH 3–10) and rehydrated overnight with rehydration/sample buffer (Bio-Rad Laboratories). Isoelectric focusing (IEF) and 2D Electrophoresis were performed as described.5 Briefly, IEF was performed with a programmed voltage gradient at 8,000 V for 5 hours. IPG strips were prepared for the second dimension by two sequential 10-min incubations in 6 M urea, 50 mM Tris (pH 8.8), 30% glycerol, 2% SDS, and 0.001% bromophenol blue containing, alternately, 2% DTT and 2.5% iodoacetamide. Following equilibration, second-dimension separation was then performed on 4–20% SDS-PAGE gels (Bio-Rad Laboratories) with the first-dimension IPG strip embedded in 0.5% agarose at the top. After electrophoresis, proteins on gel were fixed in 10% acetic acid and 20% methanol for 1 hour and then stained using SYPRO Ruby protein gel stain (Bio-Rad Laboratories).
Image Analysis
Images of the 2-DE gels were captured with Molecular Imager FX Pro Plus multi-imager system (Bio-Rad, 532 nm excitation, 555 nm emission filter, 1064 nm excitation filter). The gel images were warped and protein expression profiles at each time point were analyzed in triplicate using the Phoretix 2D image analysis software (Nonlinear Dynamics, UK). The mean spot volume was calculated per each time point and using those of corresponding spot in P1 as reference.
We compared the volume of each spot presented in P4, P8 and P16 with the volume of the corresponding spot presented in P1. We divided the spots presented in each gel in three groups: (1) spots with a volume increased more than 2 folds; (2) spots with a volume decreased more than 2 folds and (3) spots with a volume between 2 folds increased and 2 folds decreased when compared to those of P1.
Protein Identification
Three additional samples for P1 and three additional samples for P16 were loaded onto nonlinear IPG gel strips (7 cm, with pH 4–7, pH 5–8, pH 7–10) and rehydrated overnight with rehydration/sample buffer. Two groups of proteins were identified; (a.) Proteins up-regulated in P1 (i.e. present in P1 and absent in P16 with a protein expression level in P1 two-fold greater than P16) and (b.) proteins up-regulated in P16 (i.e. present in P16 and absent in P1 with a protein expression level in P16 two-fold greater than P1).
Gel spots corresponding to differentially expressed proteins were excised from 2-DE gels and transferred to a multiwell plate using Xcise robotic station (Shimadzu, Japan). The excised gel plugs were washed and destained three times by adding 200 μl of 25 mM NH4HCO3 in 50% acetonitrile and incubating for 10 minutes. Then, 100 μL of acetonitrile was added into each well, incubated for 10 minutes after which the acetonitrile was discarded. The gel pieces were re-swelled in 50–100 μL for 45 min in buffer containing 10–15 μg/mL trypsin (Promega, Madison, WI, USA) and 50 mM NH4HCO3, pH 8.0. The digestion was performed overnight at 37°C. 20–30 μL of deionized water was added to each well every 2.5 hours. After overnight digestion, the clear liquid was transferred into a new 96-well plate. The tryptic peptides were extracted by one change of 50–100 μL of 20 mM NH4HCO3, two changes of 50–100 μL of 5% formic acid in 50% acetonitrile, and one change of 50–100 μL of 5% formic acid in 50% isopropanol at room temperature. The collected digests were concentrated using Speedvac and resuspended in 300 μL of 2% aqueous acetonitrile with 1% formic acid solution. About 30% of the digest was subjected to nano-LC ESI IT MS/MS analysis.
An LCQ Deca XP Plus 3D ion trap ESI mass spectrometer (Thermo Electron, San Jose, CA) was used for all experiments. Protein digest (100 μL) was injected onto a C18 solid phase extraction column (300 μm i.d. × 5 mm, Dionex, CA) to concentrate and desalt the sample. A self-packed reversed-phase analytical fused silica nano-LC column (75 μm i.d. × 15 cm; stationary phase: Magic C18AQ, 3 μm, 100 Å (Michrom Bioresources, MA); column: PicoFrit, 15 μm i.d. pulled tip (New Objective, MA) was used in all LC-MS/MS experiments. The columns were connected using a 10-port Valco valve. A desalted and concentrated sample was eluted from the SPE column and separated on the nano-LC column applying a linear gradient of acetonitrile in 0.1% formic acid. The eluent was introduced into the LCQ Deca XP Plus mass spectrometer by nanoelectrospray. A forty five minute long gradient (2%B to 40%B in 45 min and 40%B to 95%B in 5 minutes; where solvent A was 2% acetonitrile 0.1% formic acid and solvent B was 5% isopropanol, 0.1% formic acid, 85% acetonitrile) was used for the reversed phase separation of each in-gel digested sample. A full MS scan between 400 and 1800 m/z followed by five full MS/MS scans for the five most intense ions from the MS scan were acquired in data-dependent MS/MS scanning mode. The scan time was set to 50 μs and up to three scans were accumulated. The temperature of a heated capillary was set to 180 °C. The spray voltage of 1.95 kV was used in all experiments.
The .raw files were converted to .dta files using Bioworks 3.1 software (ThermoElectron). The .dta files were submitted to a database search using SEQUEST algorithm. The search was performed against a combined forward (“normal”, downloaded from www.ncbi.nml.nih.gov) and reverse (“decoy”) mouse FASTA databases containing 273,531 protein entries. An in-house built database containing sequences of common contaminants was appended to the mouse concatenated database. Methionine, histidine and tryptophan oxidation (+15.9949 atomic mass units (amu)) and cysteine alkylation (+57.0215 amu) were set as differential modifications. Peptide mass tolerance equal 1.5 amu and fragment ion mass tolerance equal to 0.8 amu was used in all searches. Monoisotopic mass type, fully tryptic peptide termini and up to 2 missed cleavages were applied in all searches. The SEQUEST output (.out files) were analyzed using DTASelect software (Scripps Research Institute). The balance between reliability and sensitivity of the protein identification data was set by adjusting the estimated false positive peptide identification rate to 1%. Optimizations of thresholds for adjusted primary scores (XCorr), relative scores (ΔCn and Sp) and minimum ion proportion score was essential to accomplish the targeted error rate without compromising sensitivity significantly. Duplicate peptide matches were purged on the basis of Xcorr with use of DTASelect to eliminate redundancy caused by homological proteins and protein isoforms. Similar (indistinguishable) proteins were listed in a single protein group entry without adding redundancy into the protein IDs list. Functional classification was based on the classification provided in the TrEMBL (http://embl-heidelberg.de/ Molecular Biology Laboratory, Heidelberg, Germany) and SwissProt protein knowledge database (http://www.expasy.org/ Swiss Institute of Bioinformatics, Geneva, Switzerland).
Immunohistochemistry Staining
To validate the differential expression of one of the inflammatory response proteins, kininogen. P1 and P16 mouse eyes were harvested and embedded in OCT compound (Tissue-Tek, Miles Laboratories, Naperville, IL) at −20°C. Frozen sections were cut at a thickness of 8 um, placed on microscope slides, air dried, and then fixed in 4% paraformaldehyde for 5 min. After blocking with 3% BSA in PBS, P1 and P16 sections were immunostained with goat anti-kininogen antibody (R&D Systems, Inc., Minneapolis, MN). The secondary antibody used was fluorescein isothiocyanate-conjugated anti-goat IgG secondary antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Stained sections were observed by confocal microscopy (TCS 4D; Leica, Heidelberg, Germany). Propidium iodide (Vector Laboratories, Burlingame, CA) was used for nuclear staining.
Data analysis
Gene bank IDs derived from peptide information were analyzed using Protein knowledgebase uniprotkb program (http://beta.uniprot.org/) to determine their corresponding Swiss protein ID. Additionally, biological process of the proteins derived from the protein knowledgebase were analyzed and grouped with reference to the biological/chemical MeSH class and KEGG pathway programs (Table 1 and Figure 3). 24
Table 1.
List of differentially-expressed proteins from the lens and vitreous of P1 and P16 mice.
| Protein up-regulated in P1 (PI 4–7) | ||||
| Swisspro ID | NCBI GI # | Protein | Molecular weight (kDa) | Biological Process |
| Q8CGQ2 | 27923590 | A disintegrin and metalloprotease domain 4 | 84.4 | Protein metabolism |
| Q5XJY5 | 52789310 | Archain | 57.2 | Protein transport |
| P70327 | 6755722 | T-box transcription factor TBX6 | 58.6 | Transcription regulation |
| Q60803 | U33840 | Tnf receptor-associated factor 3 | 64.2 | Apoptosis |
| Q8WLM9 | AAH21760 | H2-T10 protein | 43 | Antigen processing and presentation |
| P15092 | 14714795 | Ifi204 protein | 71.6 | Transcription regulation |
| O08760 | 2197093 | N-glycosylase/DNA lyase | 38.8 | DNA repair |
| Q9JIL4 | AAH13512 | PDZ domain containing 1 | 56 | Protein binding |
| A2ANU5 | NP_075755 | Zinc finger protein 120 | 15 | Regulation of transcription |
| Q91WU6 | NP_598728 | Palmitoyltransferase ZDHHC7 | 35 | Protein palmitoylation mitochondrial |
| Q99L13 | 21704140 | 3-hydroxyisobutyrate dehydrogenase, | 35 | |
| Q9R1P4 | AAH05762 | Proteasome subunit alpha type-1 | 29.5 | Protein metabolism |
| Q8R1C9 | NP_666197 | Amyloid beta (A4) precursor protein-binding, family B, member 3 | 52.6 | Negative regulation of apoptosis |
| Q5D0F3 | AAH24897 | Tap1 protein | 63.4 | Defense response |
| Q8BTW3 | NP_082550 | Exosome complex exonuclease MTR3 | 28.3 | rRNA processing |
| P63158 | CAA56631 | High mobility group protein B1 | 28.3 | Transport |
| Q8VED5 | 17512384 | Keratin, type II cytoskeletal 79 | 57.5 | Structural molecule activity |
| Q6P904 | 38173917 | D430015B01Rik protein | 72 | Unclassified |
| P21126 | XP_356994 | Ubiquitin-like protein 4A | 17.8 | Protein metabolism |
| Q60872 | AF026481 | Eukaryotic translation initiation factor 1A | Protein synthesis | |
| O88569 | AAH59107 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 37.4 | mRNA processing |
| B1AZ15 | NP_795999 | Cobl-related 1 | 133 | neural tube formation |
| Q923J1 | AY032951 | Transient receptor potential cation channel subfamily M member 7 Also known as: | 212 | Cation transport |
| Q91YM8 | NP_033129 | Ribonucleoside-diphosphate reductase | 90 | DNA replication |
| Q8CIE6 | AAH47429 | Coatomer subunit alpha | 138 | Protein transport |
| Q8BNA6 | XP_146821 | Similar to fat3; fat3 protein | 502 | Cell adhesion |
| Q811W1 | NP_780712 | Alanine and arginine rich domain containing protein | 18.9 | Unclassified |
| Q8BW70 | AAH57122 | Ubiquitin carboxyl-terminal hydrolase 38 | 116 | Ubl conjugation pathway |
| Q2KHS8 | NP_473422 | Mta1 protein | 79.4 | Cancer progression |
| Q60668 | AAH49098 | Heterogeneous nuclear ribonucleoprotein D0 | 38.3 | DNA binding |
| P16858 | AK002273 | Glyceraldehyde-3-phosphate dehydrogenase | 35.8 | Glucose metabolism |
| Q80ZS8 | AAH48503 | DiGeorge syndrome critical region gene 6 | 22.8 | Development |
| P15105 | NP_032157 | Glutamine synthetase | 42 | Glutamate metabolism |
| Q02789 | L06234 | Voltage-dependent L-type calcium channel subunit alpha-1S | 213 | Ion transport |
| Q6PB52 | AAH59887 | Lrpap1 protein | 35.4 | Chaperone |
| Q80XR6 | AAH43069 | Hnrpab protein (Heterogeneous nuclear ribonucleoprotein A/B) | 33.8 | DNA binding |
| O08749 | NP_031887 | Dihydrolipoyl dehydrogenase, mitochondrial precursor | 54.2 | mitochondrial electron transport |
| Q61414 | AAH57934 | Keratin, type I cytoskeletal 15 | 49 | Cytoskeletal |
| Q02257 | AAH40757 | Junction plakoglobin | 81.8 | Cell adhesion |
| O35098 | NP_036123 | Dihydropyrimidinase-related protein 4 | 61.9 | axon growth |
| P80317 | NP_033968 | T-complex protein 1 subunit zeta | 58 | Chaperone |
| Q8R081 | 20072624 | Heterogeneous nuclear ribonucleoprotein L | 60 | RNA synthesis/degradation |
| Q8VEI1 | AAH18459 | Cct6a protein | 42.9 | Protein metabolism |
| Q62420 | 10720274 | Endophilin-A1 | 40 | Unclassified |
| Q61183 | NP_035242 | Poly (A) polymerase alpha | 82.3 | RNA synthesis |
| Q6PGC1 | AAH57112 | Putative ATP-dependent RNA helicase DHX29 | 153.9 | RNA helicase |
| Q6A085 | BC035547 | Zinc finger protein 629 | 96 | DNA binding |
| Q9CWJ9 | AAH39925 | Bifunctional purine biosynthesis protein PURH | 64.2 | Purine nucleotide biosynthesis |
| Q60864 | NP_058017 | Stress-induced phosphoprotein 1 | 62.5 | Chaperone |
| Q8BUU7 | NP_780363 | N-Acetylgalactosamine kinase | 49.3 | Galactose metabolism |
| Q9QXL1 | AF202893 | Kinesin-like protein KIF21B | 186 | Motor protein |
| Q9Z0T6 | AF116459 | Polycystic kidney disease and receptor for egg jelly-like protein | 241 | Ion transport |
| Q3TNZ7 | NP_031756 | Procollagen, type XII, alpha 1 | 70 | Extracellualr matrix |
| Q07E15 | DP000183 | Cortactin-binding protein | 178 | Cytoskeleton organization |
| Q9WU62 | AF117610 | Inner centromere protein | 101 | Cell cycle |
| Q7SIG6 | BC096022 | Development and differentiation-enhancing factor 2 | 106 | GTPase activation |
| Q8VCW4 | AAH18388 | Unc93 homolog B | 67 | Antigen presentation |
| Protein up-regulated in P16 (PI 4–7) | ||||
| Swisspro ID | NCBI GI # | Protein | Molecular weight (kDa) | Biological Process |
| Q9ESS2 | AB036765 | Fibroblast growth factor 22 | 18.9 | Growth factor |
| Q6PAC1 | AAH60377 | Gsn protein | 80.7 | Vesicle-mediated transport |
| Q3UGX3 | AK147307 | N-acetyltransferase 8-like protein | 32.7 | Acyltransferase |
| Q9R1S3 | AAH21148 | GPI ethanolamine phosphate transferase 1 | 105 | phosphotransferase |
| P06804 | NP_038721 | Tumor necrosis factor alpha | 25.8 | cytokine |
| Q3UHA3 | BAC98263 | Spatacsin | 273.9 | Axon transport |
| Q6PD36 | NP_109614 | Nptxr protein | 34.9 | Synapse formation and remodeling |
| Q9Z148 | AAH58357 | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | 138 | Germ cell development |
| Q8VBW1 | NP_598748 | Sodium- and chloride-dependent creatine transporter 1 | 70.9 | Ion transport |
| Q8BM14 | AK037106 | Lipase member K | 45.2 | Lipid metabolism |
| Q6P216 | BC064769 | Sytl2 protein | 59.5 | vesicular trafficking transport |
| Q99KD7 | AAH04714 | Heat shock protein 2 | 69.6 | Stress response |
| Q9DCH4 | NP_079620 | eukaryotic translation initiation factor 3, | 38 | Protein biosynthesis |
| Q91YS8 | BC014825 | Calcium/calmodulin-dependent protein kinase type 1 | 41.6 | Signal transduction |
| O08677 | AAH18158 | Kininogen-1 precursor | 73.1 | Inflammatory response |
| P28656 | NP_056596 | Nucleosome assembly protein 1-like 1 | 45.3 | modulating chromatin formation |
| P14211 | X14926 | Calreticulin | 47.9 | Cytoskeleton organization |
| Q61937 | M33212 | Nucleophosmin | 32.5 | DNA binding |
| P51859 | AAH05713 | Hepatoma-derived growth factor | 26.2 | Cellular process |
| Q5SXA9 | AAH06733 | Protein WWC1 | 124 | Protein transport |
| Q80XK0 | AAH46617 | Serine/threonine protein phosphatase | 61.4 | Hydrolase |
| Q6DD96 | NP_291068 | Protocadherin gamma subfamily A, 7 | 101.5 | Cell adhesion |
| Q8K0Z5 | AAH29186 | Tropomyosin 3, gamma | 33 | Regulation of muscle contraction |
| Q68EF4 | BC080284 | glutamate receptor, metabotropic 4 | 101.8 | Synaptic transmission |
| Q62209 | NP_035646 | Synaptonemal complex protein 1 | 115.9 | Cell cycle |
| Q9CQV8 | AF058797 | 14-3-3 protein beta/alpha | 28 | Protein targeting |
| Q8CGT7 | AAN75582 | Argonaute 5 protein | 72.1 | Enzyme |
| P97479 | U81453 | Myosin-VIIa | 254.8 | Biogenesis |
| Q792Y6 | NP_033456 | Trypsin 2 | 26 | Protease |
| Q91Z49 | NP_081502 | Putative 40-2-3 protein | 35.8 | Unclassified |
| Protein up-regulated in P1 (PI 5–8) | ||||
| Swisspro ID | NCBI GI # | Protein | Molecular weight (kDa) | Biological Process |
| Q9CQV8 | AF058797 | 14-3-3 protein beta/alpha | 28 | Protein targeting |
| P24638 | NP_031413 | Acid phosphatase 2, lysosomal | 48.5 | Biogenesis |
| Q8BGB7 | AAH21429 | Enolase-phosphatase E1 | 28.6 | Biosynthesis |
| Q810U4 | AJ543321 | Neuronal cell adhesion molecule | 138 | Cell adhesion |
| O54879 | AF022465 | High mobility group protein B3 | 23 | Regulate cell differentiation |
| Q9JIF7 | AAH30837 | Coatomer subunit beta | 107 | Vesicle-mediated transport |
| O89020 | NP_660128 | Afamin; alpha albumin; | 69.3 | transport |
| Q9CUU3 | DQ103262 | Synaptonemal complex protein 2 | 172 | Cell cycle |
| O88968 | AAH03720 | Transcobalamin 2 | 47.5 | Transport |
| Protein up-regulated in P16 (PI 5–8) | ||||
| Swisspro ID | NCBI GI # | Protein | Molecular weight (kDa) | Biological Process |
| Q6P4T0 | AAH56482 | Autophagy-related protein 2 homolog A | 210 | Unclassified |
| O88623 | NP_932760 | Ubiquitin carboxyl-terminal hydrolase 2 | 68.8 | Ubl conjugation pathway |
| Q8BK06 | AAH20074 | Fbxo9 protein | 50.7 | Ubl conjugation pathway |
| Q8CBY3 | AK034310 | leukocyte receptor cluster (LRC) member 8 | 86.7 | Unclassified |
| Q78ZA7 | NP_032698 | Nucleosome assembly protein 1-like 4 | 42.6 | Nucleosome assembly |
| Q6NVD9 | CAC83162 | Phakinin, CP49 | 45.7 | Cytoskeleton organization and biogenesis |
| O70566 | AK088648 | Diaphanous homolog 2 | 124.8 | Actin filament polymerization |
| Q6PD28 | AAH58977 | Protein phosphatase 2, regulatory subunit B (B56) | 57.3 | Signal transduction |
| O70572 | AAH10978 | Sphingomyelin phosphodiesterase 2 | 47.4 | Hydrolase |
| P49442 | AAH25072 | Inositol polyphosphate-1-phosphatase | 43.3 | Signal transduction |
| Q924W5 | NP_079971 | Structural maintenance of chromosomes protein 6 | 127.1 | DNA repair |
| Q8R2K7 | NP_899139 | snRNA-activating protein complex subunit 5 | 11.3 | Transcription |
| Q922M0 | NP_034826 | lymphocyte cytosolic protein 2 | 60.2 | Cytokine secretion |
| Q9Z0S1 | AAH11036 | Bisphosphate 3′-nucleotidase 1 | 33.1 | DNA binding |
| Q9D6R2 | AAH49956 | Isocitrate dehydrogenase 3 (NAD+) alpha | 39.6 | Tricarboxylic acid |
| P04247 | AAH25172 | Myoglobin | 17 | Transport |
| Q69ZB0 | AAH61202 | Leucine-rich repeat and coiled-coil domain-containing protein 1 | 119.1 | Unclassified |
| Protein up-regulated in P1 (PI 7–10) | ||||
| Swisspro ID | Mouse GI # | Protein | Molecular weight (kDa) | Biological Process |
| P47754 | NP_031630 | F-actin-capping protein subunit alpha-2 | 32.9 | Cytoskeleton organization |
| Q9CWS0 | NP_081269 | N(G), N(G)-dimethylarginine dimethylaminohydrolase 1 | 31.3 | hydrolase |
| O09114 | AAH38083 | Prostaglandin-H2 D-isomerase precursor | 21 | Prostaglandin biosynthesis/transport |
| Q04365 | AAA39399 | L1Md-9 repetitive sequence | 21.5 | Unclassified |
| Q9QZ09 | AJ242864 | Putative homeodomain transcription factor 1 | 86.7 | Regulation of transcription |
| Protein up-regulated in P16 (PI 7–10) | ||||
| Swisspro ID | Mouse GI # | Protein | Molecular weight (kDa) | Biological Process |
| Q9JJV0 | AAH56444 | Crystallin, beta A4 | 22.4 | Structural protein |
| P13705 | NP_034959 | MutS homolog 3 | 123 | DNA repair |
| Q8VEH2 | AAH18483 | CDKN1A interacting zinc finger protein 1 | 93.5 | mRNA splicing |
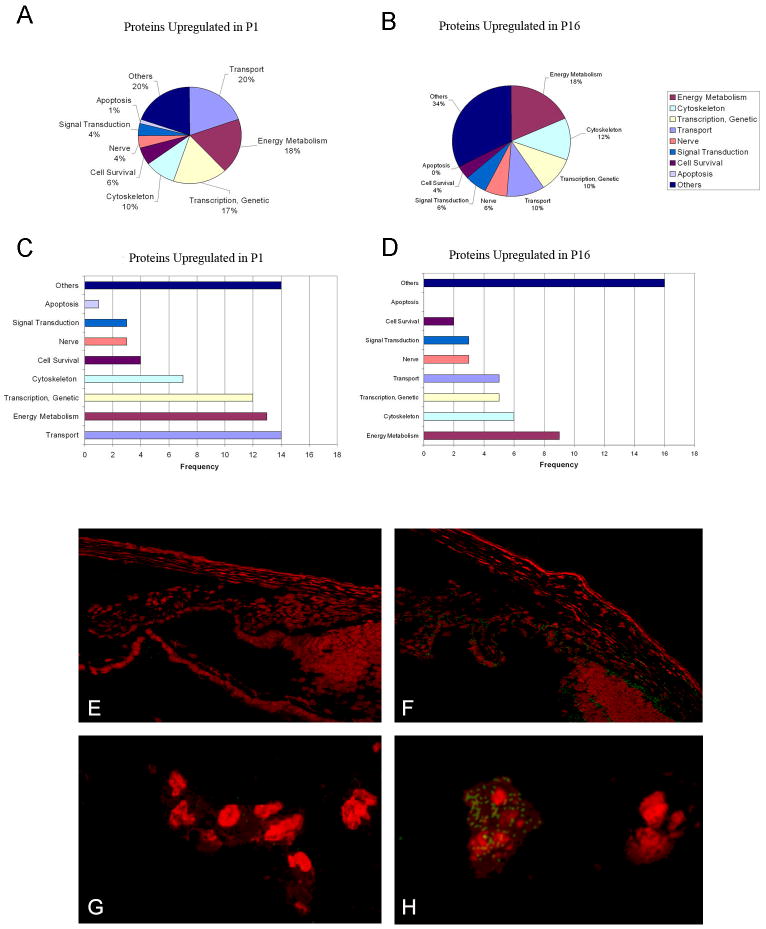
Figure 3.
A–D: Pie-chart/frequency bar graphs showing the summary of protein classes that were upregulated in P1 (A, C) and P16 (B, D). Differential expression of one of the inflammatory response proteins, kininogen, is shown in E–H, using goat anti kininogen primary antibodies and FITC donkey anti-goat IgG secondary antibodies (with PI counterstaining). Proteome analysis showed greater than two fold increase of kininogen on P16 as compared to P1. Immunoconfocal microscopy demonstrates localization of kininogen on P16 in the iris and ciliary body (3F) and tunica vasculosa lentis (3H); immunolocalization was undetectable on P1 (3 E, G).
RESULTS
Morphometric Analysis of Hyaloid Vascular System Regression
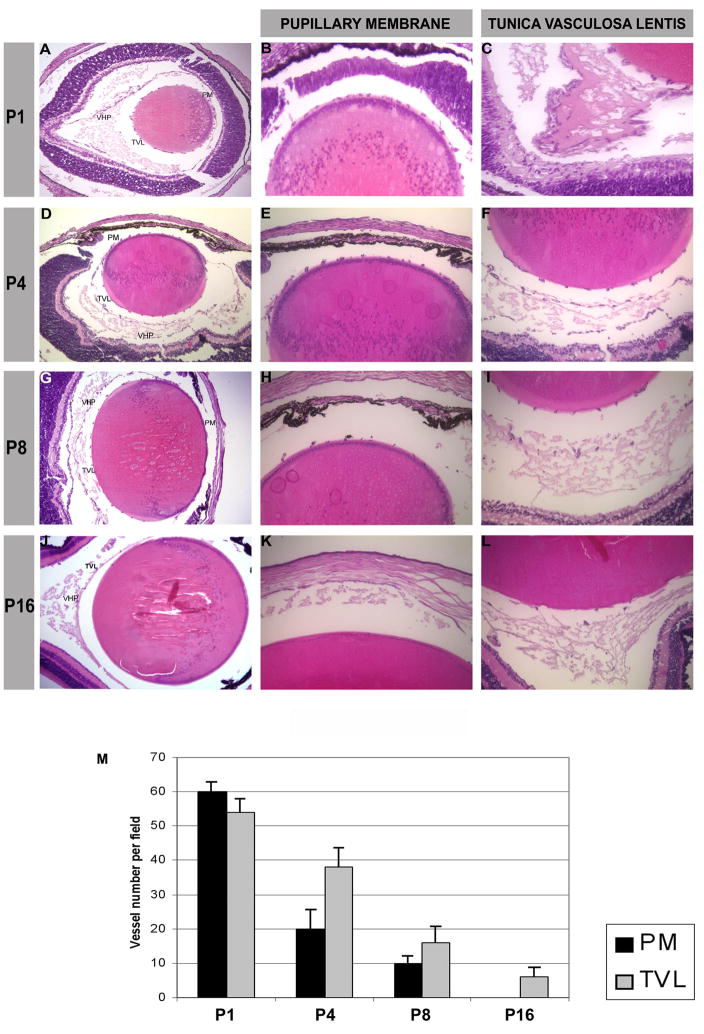
From embryonic day 14 to birth, the blood vessels that occupy most of the vitreous cavity in earlier stages become less prominent except around the lens and along the inner retinal surface. The pupillary membrane and the tunica vasculosa lentis remain prominent.8 The pupillary membrane of the newborn mice was a dense plexus of capillaries that began to regress on post-gestation day four and became vestigial on day eight. The pupillary membrane disappeared completely in all eyes by day sixteen (Fig. 1).
Figure 1.
Hematoxylin-eosin staining of mouse eyes at different stages of development: P1 (A), P4 (D), P8 (G), and P16 (J), magnification (10x). Enlargement of the anterior part of the mouse lens with pupillary membrane (PM) vessels, magnification (20x) (B, E, H, K). Posterior part of the lens with the tunica vasculosa lentis (TVL) and the vasa hyaloidea propria (VHP), magnification (20x) (C, F, I, L).
(M) Graph showing the vessel number per field at magnification (20x).
(N) Graph showing the mean spot volume in pixels.
(O) Graph showing the fold increase/decrease of each time point over P1.
The radial branches of the tunica vasculosa lentis that lie on the lens surface were prominent until P4, particularly posterior to the lens equator. Later there was a gradual disappearance of the tunica vasculosa lentis (Fig. 1); it had completely regressed in about one-third of the eyes examined on day 24 and in all eyes on day 30. The number of vessel(s) forming the papillary membrane and the tunica vasculosa lentis showed a statistically significant difference among P1, P4, P8 and P16 (p<0.005) (Fig. 1M).
The hyaloid artery arose from the optic disc as a single, thick proximal trunk and entered the vitreous, subsequently branching in several vessels of the vasa hyaloidea propria running from the anterior to the posterior pole of the lens. As the whole eye developed in size and the volume of the vitreous increased, the vasa hyaloidea propria ran toward the lens joining the tunica vasculosa lentis and became less prominent (Fig. 1). Involution of hyaloid artery and vasa hyaloidea propria began by day one after gestation, and a gradual disappearance of those vessels occurred from day twelve to day thirty.
We designed a time course experiment to examine the changes in protein expression at the critical time points throughout the hyaloid vascular system regression process. The lens and vitreous at P1, P4, P8, and P16 were selected because the histological analysis showed that on post-gestation day 1 the hyaloid network was still prominent, and by post-gestation day 16, the hyaloid network was already vestigial. We generated protein expression maps in triplicate for each time point, obtaining a reproducible separation of the protein spots on the 2-DE gels as described before.5 Phoretix 2D image analysis software could detect 770 spots in P1, 768 spots in P4, 742 spots in P8 and 714 spots in P16. This software allowed us to warp different gel images, improving the quality of the protein spot matching between the gels and allowing an easy detection of the differences among the gels. The mean spot volume progressively decreased from P1 to P16 (Fig. 1N). The comparison of the volume of each spot present in P4, P8 and P16 with the volume of the same spot present in P1 showed that 113 spots in P4, 78 spots in P8, and 76 spots in P16 had a volume increase more than two fold when compared with the corresponding spots in P1. 104 spots in P4, 186 spots in P8 and 209 spots in P16 had a volume reduced more than two folds when compared with the correspondent spots in P1. The analysis showed that the highest difference in terms of increased or decreased volume was between P1 and P16 (Fig. 1O).
This evidence suggested an active protein expression in the earlier post-gestation days coexisting with a prominent hyaloid vascular system and a presence of factors in P16 whose synthesis or degradation is related to the regression of the hyaloid vascular system itself.
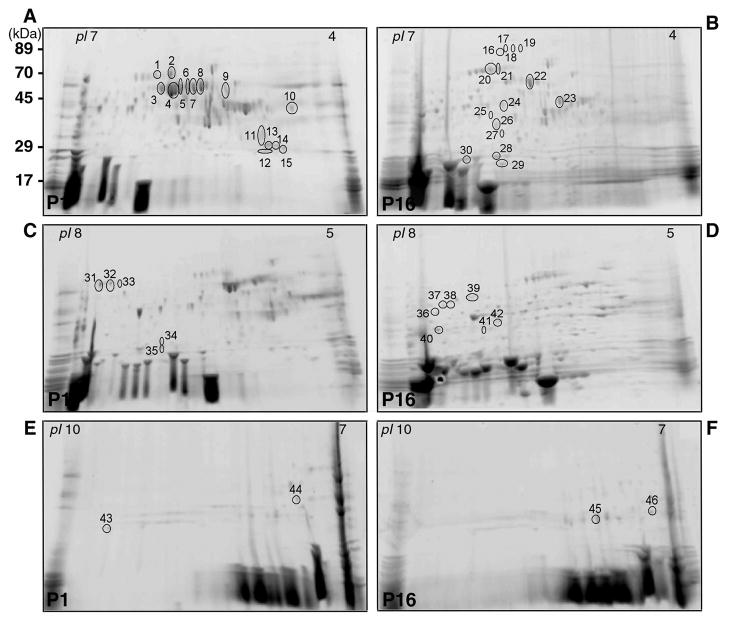
We analyzed the protein expression of P1 and P16 in greater detail using a narrow-range gradient strip (Figure 2). The use of broad-range strips allows the display of most proteins in a single gel. With narrow-range and micro-range overlapping gradient strips, resolution was increased by expanding a small pH range across the entire width of a gel, and more spots were able to be displayed for each sample. Because proteins outside of the pH range of the strip were excluded, more total protein mass could be loaded per strip, allowing more proteins to be detectable.
Figure 2.
Representative 2-DE gels of proteins obtained from the lens and vitreous of P1 mouse (A, C, E) and P16 mouse (B, D, F) using IPG strips with pH range 4–7 (A, B), 5–8 (C, D), 7–10 (E, F). The proteins excised for analysis and identification by MS are marked with numbers from 1 to 46.
Using the Phoretix 2D imaging analysis software, we could warp and match the P1 map with the P16 map and compare and identify the various protein expressions of these gels (Fig. 2A–2F). Comparing the P1 map pH 4–7 with the P16 map pH 4–7, we found fifteen spots up-regulated in P1 and fifteen other spots up-regulated in P16. Comparing the P1 map pH 5–8 with the P16 map pH 5–8, we found five spots up-regulated in P1 and seven other spots up-regulated in P16. Comparing the P1 map pH 7–10 with the P16 map pH 7–10, we found two spots up-regulated in P1 and two other spots up-regulated in P16. All of these spots were identified with mass spectrometry analysis. The results are shown in Table 1 and Figure 3.
DISCUSSION
Several hypothetical mechanisms of the hyaloid vascular regression have been proposed by different groups. 1, 5, 9–17, 20, 25–28 Given the complexity of the factors that may be involved in this system, we used a proteomic approach at various stages of development, focusing our attention on the different protein expressions existing between two different time points: postnatal day one and sixteen. Our results from the morphometric analysis of hyaloid vascular system are consistent with the studies of Ito and Smith:7, 8 the hyaloid vascular system progressively regresses between P1, in which pupillary membrane, tunica vasculosa lentis, and hyaloid vascular system are present, and P16, in which these mature capillary structures29 are almost completely regressed. Recently, several studies have attempted to analyze differential gene expression during eye development using cDNA-microarray technology.30, 31 Accordingly, proteomic analysis has been employed in retina development, macular degeneration and retinal and brain-derived progenitor cells.32, 33 However, there is no report using proteomic analysis to dissect mouse hyaloid vascular regression system. In this report, we were able to detect a series of striking functional differences between P1 and P16, including FGF-22, hepatocyte growth factor and kininogen. Their functions may be involved in hyaloid vascular system regression.
Proteins Up-regulated in P1
Energy metabolism
P1 samples exhibited a high abundance of glucose and galactose metabolic chain components (glyceraldehyde-3-phosphate dehydrogenase, dihydrolipoamide dehydrogenase). This may reflect the reduced energy requirement profile of the vitreous after the regression of the hyaloid vascular system.
Cell proliferation and development
Several proteins classified in the gene ontology database (http://www.geneontology.org/) as DNA-binding transcriptional factors (Ifi204 protein, zinc finger proteins), DNA replication proteins (ribonucleotide reductase M1), embryonic developmental proteins (T-box 6), and proteins involved in RNA synthesis and degradation are expressed far more often in P1 than in P16. They are involved, as transcriptional factors, in the embryonic developmental phase and in the processing of mRNAs, and they play a role in the regulation of cell proliferation and transformation.
Proteins Up-regulated in P16
Signal transduction
G-protein pathway and signal transduction proteins of the calcium/calmodulin pathways are involved in transcriptional regulation, hormone production, translational regulation, and regulation of cytoskeleton organization. These proteins are up-regulated in P16, suggesting an active role during the specialization and maturation of the intraocular structures.
Cancer-related and inflammatory proteins
In P1, we found the expression of two proteins related to cancer progression. Neuronal cell adhesion molecule precursor (CAM) is a member of the L1 subfamily of cell adhesion molecules that belongs to the immunoglobulin superfamily. Induction of N-CAM transcription by β-catenin or plakoglobin plays a role in melanoma and colon cancer tumorigenesis, probably by promoting cell growth and motility.34
In P16, we found the presence of tumor necrosis factor α . TNFα is mainly secreted by macrophages and can induce the cell death of certain tumor cell lines.35 It is a potent pyrogen that causes fever by direct action or stimulation of interleukin-1 secretion and is implicated in the induction of cachexia. Under certain conditions, it can stimulate cell proliferation and induce cell differentiation. Also in P16, we found an over-expression of two proteins that are known to facilitate angiogenesis: hepatoma-derived growth factor (HDGF) and fibroblast growth factor (FGF) 22. These two proteins may facilitate angiogenesis during the development of the hyaloid vascular system. Hepatoma-derived growth factor is involved in proliferative, angiogenic, and neurotrophic activity.36 It plays a putative role in the development and progression of cancer. When expressed in cells, HDGF’s mitogenic activity depends on its nuclear localization, but it also stimulates proliferation when added to the cell culture medium.37, 38, 39 FGF 22 is the homolog of human FGF 10, a factor required for embryonic epidermal morphogenesis and it also implicated as a primary factor in the wound-healing process.40 It also induces angiogenesis and stabilizes the endothelial barriers protecting the microvascular and epithelial tissues against mild injuries, and it speeds their repair after major damage.41
In P16, we also found the overexpression of kininogen, a well-known protein that can inhibit angiogenesis. It is a plasma protein that plays important roles in fibrinolysis, thrombosis, and inflammation.42, 43 Recent studies have revealed that high-molecular-weight kininogen, or HKa, inhibits angiogenesis, plays an important role in blood coagulation, and inhibits the thrombin- and plasmin-induced aggregation of thrombocytes.44–46 Our immunolocalization experiments confirm the differential expression of kininogen in P16 in the iris, ciliary body, and tunica vasculosa lentis (figure 3F,H).
Apoptosis-related proteins
In P1, we found the expression of aphamin. Aphamin enhances the survival of cortical neurons under apoptotic conditions.
In P16, we found the expression of heat shock cognate 71 kDa protein and heat shock protein 2. They are a group of proteins present in all cells under normal conditions and are overexpressed under various types of environmental stresses like heat, cold, and oxygen deprivation. They act like chaperones, making sure that the cell’s proteins are in the right shape and in the right place at the right time. Endogenous gelsolin has been demonstrated to have an anti-apoptotic property that correlates to its dynamic actions on the cytoskeleton.29
CONCLUSION
In both P1 and P16, during the regression of the hyaloid vascular system, we found numerous proteins that may be involved in the apoptotic pathway. This supports the findings of Lang et al.9–11 regarding the programmed mechanism responsible for the involution of the hyaloid vascular system. We also found upregulation of pro- and anti-angiogenic factors in P16. The active form of kininogen may be involved in the hyaloid vessel regression, while the presence of FGF-22 and hepatoma-derived growth factor may be important in supporting and maintaining the development of the retinal vascularization, which occurred at the same time as hyaloid vascular system regression. These results support previous findings12–17 regarding the anti-angiogenic ability of the vitreous humor and vitreous extracts in the regression of the tunica vasculosa lentis, and the presence of a substance inhibitory to neovascularization in the vitreous is also supported.
The identification and purification of the vitreous component responsible for the regression of hyaloid vessels holds great potential in our understanding of and in future control of ocular neovascularization. Further investigation of candidate proteins is needed to determine their in vitro- and in vivo-specific effects and their potential contribution to vascular regression.
Acknowledgments
The authors wish to thank Drs. Faisal Tobaigy and Ramon Ghanem for immunoconfocal microscopy.
Supported by S. Elizabeth O’Brien Trust (DTA), NIH EY10101 (DTA), P30-001792 (DTA), EY14048 (JHC), and an unrestricted departmental support from Research to Prevent Blindness (New York, NY)
Footnotes
Commercial relationship and financial interest: None
References
- 1.Cheong C, Sung YH, Lee J, Choi YS, Song J, Kee C, Lee HW. Role of INK4a locus in normal eye development and cataract genesis. Mech Ageing Dev. 2006;127(7):633–8. doi: 10.1016/j.mad.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Hahn P, Lindsten T, Tolentino M, Thompson CB, Bennett J, Dunaief JL. Persistent fetal ocular vasculature in mice deficient in bax and bak. Arch Ophthalmol. 2005;123(6):797–802. doi: 10.1001/archopht.123.6.797. [DOI] [PubMed] [Google Scholar]
- 3.Mann I. The Development of the Human Eye. New York: 1964. [Google Scholar]
- 4.Mitchell CA, Risau W, Drexler HC. Regression of vessels in the tunica vasculosa lentis is initiated by coordinated endothelial apoptosis: a role for vascular endothelial growth factor as a survival factor for endothelium. Dev Dyn. 1998;213(3):322–33. doi: 10.1002/(SICI)1097-0177(199811)213:3<322::AID-AJA8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 5.Albe E, Escalona E, Rajagopal R, Javier JA, Chang JH, Azar DT. Proteomic identification of activin receptor-like kinase-1 as a differentially expressed protein during hyaloid vascular system regression. FEBS Lett. 2005;579(25):5481–6. doi: 10.1016/j.febslet.2005.08.078. [DOI] [PubMed] [Google Scholar]
- 6.Brown AS, Zhang M, Cucevic V, Pavlin CJ, Foster FS. In vivo assessment of postnatal murine ocular development by ultrasound biomicroscopy. Curr Eye Res. 2005;30(1):45–51. doi: 10.1080/02713680490894315. [DOI] [PubMed] [Google Scholar]
- 7.Ito M, Yoshioka M. Regression of the hyaloid vessels and pupillary membrane of the mouse. Anat Embryol (Berl) 1999;200(4):403–11. doi: 10.1007/s004290050289. [DOI] [PubMed] [Google Scholar]
- 8.Smith RS, John SWM, Nishina PM, Sundberg JP. Systematic Evaluation of the Mouse Eye: Anatomy, Pathology, and Biomethods. CRC Press; Boca Raton: 2002. [Google Scholar]
- 9.Diez-Roux G, Lang RA. Macrophages induce apoptosis in normal cells in vivo. Development. 1997;124(18):3633–8. doi: 10.1242/dev.124.18.3633. [DOI] [PubMed] [Google Scholar]
- 10.Lang R, Lustig M, Francois F, Sellinger M, Plesken H. Apoptosis during macrophage-dependent ocular tissue remodelling. Development. 1994;120(12):3395–403. doi: 10.1242/dev.120.12.3395. [DOI] [PubMed] [Google Scholar]
- 11.Lang RA, Bishop JM. Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell. 1993;74(3):453–62. doi: 10.1016/0092-8674(93)80047-i. [DOI] [PubMed] [Google Scholar]
- 12.Felton SM, Brown GC, Felberg NT, Federman JL. Vitreous inhibition of tumor neovascularization. Arch Ophthalmol. 1979;97(9):1710–3. doi: 10.1001/archopht.1979.01020020278019. [DOI] [PubMed] [Google Scholar]
- 13.Lutty GA, Thompson DC, Gallup JY, Mello RJ, Patz A, Fenselau A. Vitreous: an inhibitor of retinal extract-induced neovascularization. Invest Ophthalmol Vis Sci. 1983;24(1):52–6. [PubMed] [Google Scholar]
- 14.Preis I, Langer R, Brem H, Folkman J. Inhibition of neovascularization by an extract derived from vitreous. Am J Ophthalmol. 1977;84(3):323–8. doi: 10.1016/0002-9394(77)90672-9. [DOI] [PubMed] [Google Scholar]
- 15.Ramesh S, Bonshek RE, Bishop PN. Immunolocalisation of opticin in the human eye. Br J Ophthalmol. 2004;88(5):697–702. doi: 10.1136/bjo.2003.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor CM, Weiss JB. Partial purification of a 5.7K glycoprotein from bovine vitreous which inhibits both angiogenesis and collagenase activity. Biochem Biophys Res Commun. 1985;133(3):911–6. doi: 10.1016/0006-291x(85)91222-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Penfold PL, Madigan MC, Billson FA. Effect of human vitreous hyalocyte-derived factors on vascular endothelial cell growth. Aust N Z J Ophthalmol. 1997;25 Suppl 1:S57–60. doi: 10.1111/j.1442-9071.1997.tb01758.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Sorenson CM, Sheibani N. Attenuation of retinal vascular development and neovascularization during oxygen-induced ischemic retinopathy in Bcl-2−/− mice. Dev Biol. 2005;279(1):205–19. doi: 10.1016/j.ydbio.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Lutty GA, Merges C, Threlkeld AB, Crone S, McLeod DS. Heterogeneity in localization of isoforms of TGF-beta in human retina, vitreous, and choroid. Invest Ophthalmol Vis Sci. 1993;34(3):477–87. [PubMed] [Google Scholar]
- 20.McMenamin PG, Djano J, Wealthall R, Griffin BJ. Characterization of the macrophages associated with the tunica vasculosa lentis of the rat eye. Invest Ophthalmol Vis Sci. 2002;43(7):2076–82. [PubMed] [Google Scholar]
- 21.Dimaio TA, Wang S, Huang Q, Scheef EA, Sorenson CM, Sheibani N. Attenuation of retinal vascular development and neovascularization in PECAM-1-deficient mice. Dev Biol. 2008;315(1):72–88. doi: 10.1016/j.ydbio.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eastman A. Survival factors, intracellular signal transduction, and the activation of endonucleases in apoptosis. Semin Cancer Biol. 1995;6(1):45–52. doi: 10.1006/scbi.1995.0006. [DOI] [PubMed] [Google Scholar]
- 23.Feeney SA, Simpson DA, Gardiner TA, Boyle C, Jamison P, Stitt AW. Role of vascular endothelial growth factor and placental growth factors during retinal vascular development and hyaloid regression. Invest Ophthalmol Vis Sci. 2003;44(2):839–47. doi: 10.1167/iovs.02-0040. [DOI] [PubMed] [Google Scholar]
- 24.Rolfs A, Hu Y, Ebert L, Hoffmann D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, Shen B, Baqui MM, Pearlberg J, Taycher E, DeLoughery C, Hoerlein A, Korn B, LaBaer J. A biomedically enriched collection of 7000 human ORF clones. PLoS ONE. 2008;3(1):e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischoff PM, Wajer SD, Flower RW. Scanning electron microscopic studies of the hyaloid vascular system in newborn mice exposed to O2 and CO2. Graefes Arch Clin Exp Ophthalmol. 1983;220(6):257–63. doi: 10.1007/BF00231352. [DOI] [PubMed] [Google Scholar]
- 26.Jack RL. Regression of the hyaloid vascular system. An ultrastructural analysis. Am J Ophthalmol. 1972;74(2):261–72. doi: 10.1016/0002-9394(72)90543-0. [DOI] [PubMed] [Google Scholar]
- 27.Latker CH, Kuwabara T. Regression of the tunica vasculosa lentis in the postnatal rat. Invest Ophthalmol Vis Sci. 1981;21(5):689–99. [PubMed] [Google Scholar]
- 28.Meeson A, Palmer M, Calfon M, Lang R. A relationship between apoptosis and flow during programmed capillary regression is revealed by vital analysis. Development. 1996;122(12):3929–38. doi: 10.1242/dev.122.12.3929. [DOI] [PubMed] [Google Scholar]
- 29.Harms C, Bosel J, Lautenschlager M, Harms U, Braun JS, Hortnagl H, Dirnagl U, Kwiatkowski DJ, Fink K, Endres M. Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol Cell Neurosci. 2004;25(1):69–82. doi: 10.1016/j.mcn.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Kesinger JW, Zhou Q, Wren JD, Martin G, Turner S, Tang Y, Frank MB, Centola M. Identification and characterization of zebrafish ocular formation genes. Genome. 2008;51(3):222–235. doi: 10.1139/G07-098. [DOI] [PubMed] [Google Scholar]
- 31.Popp MP, Liu L, Timmers A, Esson DW, Shiroma L, Meyers C, Berceli S, Tao M, Wistow G, Schultz GS, Sherwood MB. Development of a microarray chip for gene expression in rabbit ocular research. Mol Vis. 2007;13:164–73. [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn-Thomas TE, Dobbs DL, Sakaguchi DS, Young MJ, Honovar VG, Greenlee MH. Proteomic differentiation between murine retinal and brain-derived progenitor cells. Stem Cells Dev. 2008;17(1):119–31. doi: 10.1089/scd.2007.0051. [DOI] [PubMed] [Google Scholar]
- 33.Frost MR, Norton TT. Differential protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Mol Vis. 2007;13:1580–8. [PMC free article] [PubMed] [Google Scholar]
- 34.Hakansson J, Xian X, He L, Stahlberg A, Nelander S, Samuelsson T, Kubista M, Semb H. Neural cell adhesion molecule-deficient beta-cell tumorigenesis results in diminished extracellular matrix molecule expression and tumour cell-matrix adhesion. Tumour Biol. 2005;26(2):103–12. doi: 10.1159/000085817. [DOI] [PubMed] [Google Scholar]
- 35.Wolf AM, Wolf D, Rumpold H, Ludwiczek S, Enrich B, Gastl G, Weiss G, Tilg H. The kinase inhibitor imatinib mesylate inhibits TNF-{alpha} production in vitro and prevents TNF-dependent acute hepatic inflammation. Proc Natl Acad Sci U S A. 2005;102(38):13622–7. doi: 10.1073/pnas.0501758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abouzied MM, El-Tahir HM, Prenner L, Haberlein H, Gieselmann V, Franken S. Hepatoma-derived growth factor. Significance of amino acid residues 81–100 in cell surface interaction and proliferative activity. J Biol Chem. 2005;280(12):10945–54. doi: 10.1074/jbc.M414652200. [DOI] [PubMed] [Google Scholar]
- 37.Kishima Y, Yamamoto H, Izumoto Y, Yoshida K, Enomoto H, Yamamoto M, Kuroda T, Ito H, Yoshizaki K, Nakamura H. Hepatoma-derived growth factor stimulates cell growth after translocation to the nucleus by nuclear localization signals. J Biol Chem. 2002;277(12):10315–22. doi: 10.1074/jbc.M111122200. [DOI] [PubMed] [Google Scholar]
- 38.Thakar K, Niedenthal R, Okaz E, Franken S, Jakobs A, Gupta S, Kelm S, Dietz F. SUMOylation of the hepatoma-derived growth factor negatively influences its binding to chromatin. Febs J. 2008;275(7):1411–26. doi: 10.1111/j.1742-4658.2008.06303.x. [DOI] [PubMed] [Google Scholar]
- 39.Sue SC, Lee WT, Tien SC, Lee SC, Yu JG, Wu WJ, Wu WG, Huang TH. PWWP module of human hepatoma-derived growth factor forms a domain-swapped dimer with much higher affinity for heparin. J Mol Biol. 2007;367(2):456–72. doi: 10.1016/j.jmb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Beer HD, Bittner M, Niklaus G, Munding C, Max N, Goppelt A, Werner S. The fibroblast growth factor binding protein is a novel interaction partner of FGF-7, FGF-10 and FGF-22 and regulates FGF activity: implications for epithelial repair. Oncogene. 2005;24(34):5269–77. doi: 10.1038/sj.onc.1208560. [DOI] [PubMed] [Google Scholar]
- 41.Beyer TA, Werner S, Dickson C, Grose R. Fibroblast growth factor 22 and its potential role during skin development and repair. Exp Cell Res. 2003;287(2):228–36. doi: 10.1016/s0014-4827(03)00139-3. [DOI] [PubMed] [Google Scholar]
- 42.Barbasz A, Guevara-Lora I, Rapala-Kozik M, Kozik A. Kininogen binding to the surfaces of macrophages. Int Immunopharmacol. 2008;8(2):211–6. doi: 10.1016/j.intimp.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Sainz IM, Wu Y, Pixley R, Espinola RG, Hassan S, Khan MM, Colman RW. The inhibition of tube formation in a collagen-fibrinogen, three-dimensional gel by cleaved kininogen (HKa) and HK domain 5 (D5) is dependent on Src family kinases. Exp Cell Res. 2008;314(4):774–88. doi: 10.1016/j.yexcr.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song JS, Sainz IM, Cosenza SC, Isordia-Salas I, Bior A, Bradford HN, Guo YL, Pixley RA, Reddy EP, Colman RW. Inhibition of tumor angiogenesis in vivo by a monoclonal antibody targeted to domain 5 of high molecular weight kininogen. Blood. 2004;104(7):2065–72. doi: 10.1182/blood-2004-02-0449. [DOI] [PubMed] [Google Scholar]
- 45.Wu Y, Rizzo V, Liu Y, Sainz IM, Schmuckler NG, Colman RW. Kininostatin associates with membrane rafts and inhibits alpha(v)beta3 integrin activation in human umbilical vein endothelial cells. Arterioscler Thromb Vasc Biol. 2007;27(9):1968–75. doi: 10.1161/ATVBAHA.107.148759. [DOI] [PubMed] [Google Scholar]
- 46.Bior AD, Pixley RA, Colman RW. Domain 5 of kininogen inhibits proliferation of human colon cancer cell line (HCT-116) by interfering with G1/S in the cell cycle. J Thromb Haemost. 2007;5(2):403–11. doi: 10.1111/j.1538-7836.2007.02352.x. [DOI] [PubMed] [Google Scholar]