Abstract
Plastid transformation has become an attractive tool in biotechnology. Because of the prokaryotic nature of the plastid's gene expression machinery, expression elements (promoters and untranslated regions) that trigger high-level foreign protein accumulation in plastids usually also confer high expression in bacterial cloning hosts. This can cause problems, for example, when production of antimicrobial compounds is attempted. Their bactericidal activity can make the cloning of the corresponding genes in plastid transformation vectors impossible. Here, we report a general solution to this problem. We have designed a strategy (referred to as toxin shuttle) that allows the expression in plastids of proteins that are toxic to Escherichia coli. The strategy is based on blocking transcription in E. coli by bacterial transcription terminators upstream of the gene of interest, which subsequently are excised in planta by site-specific recombination. We demonstrate the applicability of the strategy by the high-level expression in plastids (to up to 30% of the plant's total soluble protein) of 2 phage-derived protein antibiotics that are toxic to E. coli. We also show that the plastid-produced antibiotics efficiently kill pathogenic strains of Streptococcus pneumoniae, the causative agent of pneumonia, thus providing a promising strategy for the production of next-generation antibiotics in plants.
Keywords: chloroplast, molecular farming, plastid transformation, phage lysin, site-specific recombination
Plants provide a safe, easily scalable, and cheap production system for proteinaceous pharmaceuticals (1). Transgene expression from the plastid genome represents a particularly promising strategy in molecular farming (2) because of the plastids' potential to accumulate foreign proteins to very high levels (up to >70% of the plant's total soluble protein; ref. 3) and the increased biosafety provided by the maternal mode of plastid inheritance, which greatly reduces transgene transmission via pollen.
In view of rapidly spreading antibiotic resistances and the stagnating discovery of new antibiotics, the development of novel inexpensive antibiotics represents one of the biggest challenges in pharmaceutical research. Because of their enormous diversity and high host specificity, bacteriophages represent a huge source of potent antibacterial agents. Phage therapy is therefore considered as one of the most promising alternatives to conventional antibiotics (4). At the end of their life cycle, phages kill their bacterial host by hydrolyzing its cell wall. Cell lysis is mediated by the action of a single enzyme encoded in the phage genome, a cell wall hydrolase also referred to as lysin. Purified phage lysin proteins have been demonstrated to rapidly kill pathogenic bacteria and effectively prevent bacterial colonization of mucosal surfaces (5, 6), thus qualifying as excellent candidates for next-generation antibiotics. We have recently shown that a lysin (PlyGBS) targeted against group A and group B streptococci can accumulate to high levels in plant cells if encapsulated inside the chloroplast (3).
The future therapeutic application of lysins as novel antibiotics will require efficient and inexpensive expression platforms that are suitable for producing large protein amounts for topical or systemic treatments. Synthesis of lysins to high levels in bacterial expression hosts is often difficult because of the cell wall-degrading bacteriolytic properties of these proteins. Indirectly, this also poses a serious problem to lysin production in chloroplasts. Because of the prokaryotic nature of the plastid's gene expression machinery, expression cassettes that confer high levels of foreign protein accumulation in plastids usually also do so in bacterial cloning hosts. This can make genes encoding pharmaceutical proteins with antibacterial activities unclonable and, in this way, prohibit the use of these transgenes for plastid transformation. Here, we present a solution to this problem. We have developed a strategy that effectively prevents transgene expression in bacteria, but still permits high-level expression in plastids. We demonstrate the applicability of the strategy by producing 2 lysin-type protein antibiotics against Streptococcus pneumoniae, the causative agent of pneumonia.
Results
Toxicity of Pal and Cpl-1 and Design of a Toxin Shuttle Vector.
Pneumococcal pneumonia represents a leading cause of death among the elderly and immunocompromized patients, who are chronically or terminally ill. Over the past decades, numerous antibiotic-resistant strains of S. pneumoniae have emerged and created a pressing need to develop alternative antibiotics against pneumonia (7). Two highly effective lysins have been identified from phages infecting S. pneumoniae: Cpl-1, a lysozyme hydrolyzing the polysaccharide chains in the bacterial cell wall (8) and Pal, an amidase cleaving the linker peptides in the peptidoglycan network (7). Because of their different mode of action, the 2 lysins act synergistically in that their combined application kills S. pneumoniae even more effectively (8).
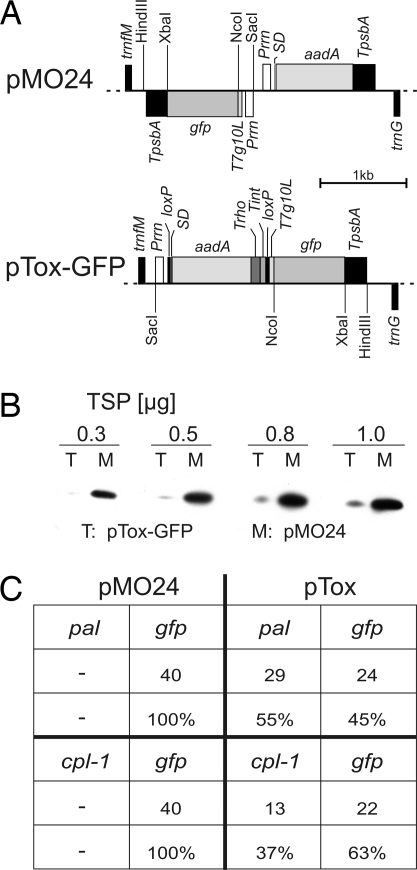
To facilitate high-level expression of the Pal and Cpl-1 protein antibiotics in plastids, the 2 coding regions were optimized by adjusting their sequences to the codon usage preferred by higher plant plastids (3, 9). However, when cloning of the synthetic genes into standard plastid expression cassettes (refs. 3, 9, and 10 and Fig. 1A) was attempted, no correct clones were obtained in Escherichia coli cloning hosts. The few clones that were obtained all contained mutations that either disrupted the reading frame or prevented expression of the genes. These mutations included frameshift mutations and deletions removing the Shine-Dalgarno sequence. We therefore suspected that expression of the genes from the (prokaryotic-type) plastid expression signals results in high levels of Pal and Cpl-1 accumulation in bacteria, which is lethal, most likely caused by the cell wall-hydrolyzing properties of the enzymes. As currently-available inducible expression systems for plastids suffer from either low efficiency (11) or problems with undesired side effects on plastid gene expression (12), we sought an alternative solution by developing a strategy that would prevent expression of the potentially toxic proteins in E. coli without impairing expression in plastids. One of the few differences in gene expression between plastids and eubacteria lies in the mechanism of transcription termination. Whereas bacterial transcription terminates at specific sequences (in either a rho-dependent or a rho-independent manner), plastid transcription does not normally terminate at specific sequences, but instead, mRNA 3′ ends are generated by posttranscriptional processing (13). We therefore designed an expression strategy for toxic gene products that is based on premature transcription termination in E. coli. To this end, we constructed a toxin shuttle vector (pTox; Fig. 1A), in which the transgene of interest represents the second cistron of an operon formed together with the upstream selectable marker gene aadA. To prevent cotranscription of the transgene of interest in E. coli, 2 strong transcriptional terminator sequences were cloned downstream of the aadA: the rho-dependent terminator λtR1 from the cro gene of phage λ (Trho) and the intrinsic (rho-independent) terminator rrnB T1 from E. coli (Tint). Trho terminates 97.1% of the nascent mRNAs (14) and Tint terminates > 90% (15). To facilitate in planta excision of both the selectable marker gene and the transcription terminators, loxP sites were integrated (i) between the promoter driving the operon and the aadA and (ii) between the transcription terminators and the translation initiation signal of the transgene of interest (Fig. 1A). Earlier work had shown that Cre-mediated site-specific recombination on loxP sites represents an effective method to induce sequence-specific deletions in transgenic plastid genomes (16, 17). We used the reporter gene gfp to assess the efficiency at which pTox reduces transgene expression in E. coli. When transcript and protein accumulation in the presence versus the absence of the transcription block (consisting of the 2 terminators) were compared, pTox turned out to strongly reduce GFP expression. Both mRNA and protein accumulation were reduced to approximately one-tenth of the expression levels obtained in the absence of the terminators (Fig. 1 A and B and Fig. S1).
Fig. 1.
The toxin shuttle. (A) Map of the vectors used to test the efficiency of the toxin shuttle strategy in E. coli. Vector pMO24 harbors the selectable marker gene aadA and a gfp gene driven by the ribosomal RNA operon promoter (Prrn) and the 5′ UTR derived from the strong translation initiation signals of the gene 10 transcript of bacteriophage T7 (T7G10L; ref. 10). The aadA gene is under the control of the ribosomal RNA operon promoter, an rbcL-derived Shine-Dalgarno sequence (SD; ref. 25), and the 3′UTR from the plastid psbA gene (TpsbA). Relevant restriction sites are indicated. Genes above the line are transcribed from left to right, genes below the line are transcribed in the opposite direction. In vector pTox-GFP, the aadA and gfp transgenes are cotranscribed from Prrn and separated by 2 bacterial transcription terminator sequences: the rho-dependent terminator λtR1 from the cro gene of the coliphage λ (Trho; ref. 14) and the intrinsic terminator rrnB T1 from E. coli (Tint; ref. 15). The aadA gene including 3′UTR and bacterial transcription terminators is flanked by directly orientated loxP sites. (B) GFP accumulation conferred by pMO24 and pTox-GFP in E. coli. The amount of TSP loaded is indicated above each pair of lanes. (C) Confirmation of the toxicity of the Pal and Cpl-1 proteins by cocloning experiments with gfp. Equal amounts of the gfp and pal genes (as NcoI/XbaI restriction fragments) were mixed and ligated into the similarly-cut vectors pMO24 and pTox. Whereas cocloning into pMO24 yielded only gfp clones, gfp and pal-containing clones were obtained in a ratio of ≈1:1 with pTox. Similarly, cpl-1 could be cocloned with gfp into pTox, but not into pMO24. Absolute clone numbers and percentages of analyzed clones are given.
We next wanted to see whether pTox allows the cloning of pal and cpl-1 and, at the same time, ultimately confirm the suspected toxicity of Pal and Cpl-1 to bacteria. We therefore performed cocloning experiments, in which we ligated 1:1 mixes of gfp and either pal or cpl-1 into pTox and the control vector lacking the transcription block. Whereas in the control vector only gfp-containing clones were recovered, plasmids with gfp and pal or cpl-1 were obtained at about the same frequencies with pTox (Fig. 1C). These results unambiguously demonstrate that insertion of pal and cpl-1 into a strong plastid expression cassette is lethal to E. coli.
Integration of pal and cpl-1 into the Plastid Genome with the Toxin Shuttle.
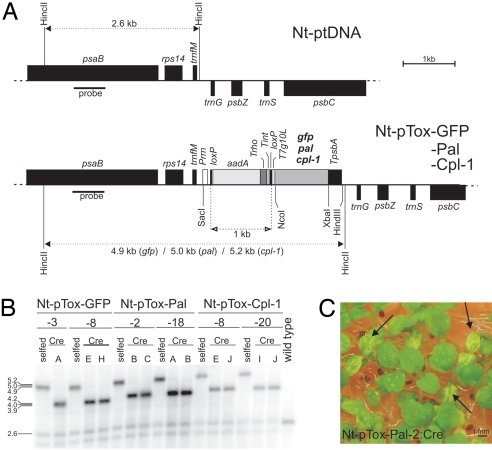
To test whether transgenes inserted into pTox can be expressed to high levels in plastids, we performed chloroplast transformation experiments with pTox vectors carrying 3 different transgenes: pal (vector pTox-Pal), cpl-1 (pTox-Cpl), and, as a control, gfp (pTox-GFP; Fig. 2A). pTox targets the transgene to the intergenic spacer between the trnfM and trnG genes by homologous recombination (ref. 18 and Fig. 2A). Biolistic chloroplast transformation of tobacco leaves followed by selection for spectinomycin resistance produced several plastid-transformed (transplastomic) lines. Two lines per construct were characterized in detail (Nt-pTox-GFP-3, Nt-pTox-GFP-8, Nt-pTox-Pal-2, Nt-pTox-Pal-18, Nt-pTox-Cpl-8, and Nt-pTox-Cpl-20; Fig. 2B). Homoplasmic transplastomic lines were isolated by conducting additional regeneration rounds under continuous antibiotic selection. Southern blot analyses confirmed successful transgene integration into the plastid genome (Fig. 2B). Inheritance assays demonstrated that, after 3 regeneration rounds, homoplasmic lines had been obtained and all wild-type copies of the plastid genome had been eliminated (Fig. S2).
Fig. 2.
Plastid transformation with pTox vectors. (A) Physical maps of the targeting region in the wild-type plastid genome and the transgenic plastid genomes in transplastomic Nt-pTox-GFP, Nt-pTox-Pal, and Nt-pTox-Cpl-1 lines. Relevant restriction sites used for cloning and RFLP analysis of transplastomic lines are marked. Sizes of expected restriction fragments in RFLP analyses are indicated. The location of the RFLP probe is shown as black bar. (B) RFLP analysis of transplastomic lines. Total DNA was digested with HincII and hybridized to a probe detecting the region of the plastid genome that flanks the transgene insertion site. Fragment sizes are indicated in kb. Transplastomic lines before (selfed) and after (Cre) elimination of the aadA marker and the transcription block are compared. Numbers of independently-generated transplastomic lines and individually-regenerated plants (capital letters) are indicated above the blot. Faint wild-type-like bands in all transplastomic lines are caused by promiscuous DNA in the nucleus, as shown (30). (C) Identification of leaf sectors lacking the aadA marker by germination of seedlings on spectinomycin-containing medium. Removal of the aadA by site-specific recombination is evidenced by sectorial bleaching (arrows).
Site-specific recombination was used to remove the aadA and the transcription block. To this end, all homoplasmic transplastomic lines were crossed to nuclear-transgenic plants expressing the Cre recombinase fused to a plastid transit peptide (16). Deletion of the aadA marker was evidenced by appearance of pale leaf sectors upon seed germination on spectinomycin-containing medium (Fig. 2C). The sectors were excised and regenerated into plants. Southern blot analysis confirmed that marker-free transplastomic plants had been obtained for all constructs (Fig. 2B). Seed tests and RNA gel blot analysis demonstrated that the aadA had been eliminated from all copies of the plastid genome (Fig. S2 and Fig. 4B).
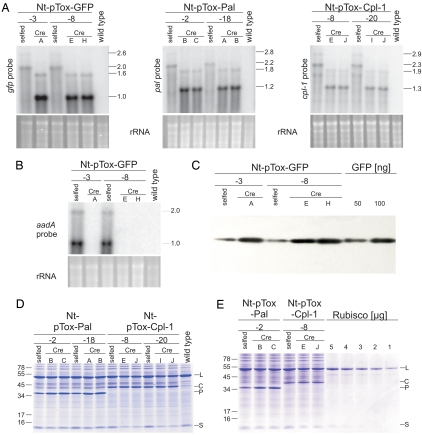
Fig. 4.
mRNA and foreign protein accumulation in transplastomic plants. (A) Analysis of gfp, pal, and cpl-1 mRNA accumulation in plants before (selfed) and after (Cre) excision of the transcription block. The ≈1-kb size difference between the major transcripts in the selfed lanes and the corresponding Cre lanes corresponds to the size of the aadA marker and the transcription block. To confirm equal loading, the ethidium bromide-stained agarose gels are also shown. (B) Confirmation of complete removal of the marker gene by Northern blot analysis with an aadA-specific probe. (C) Increase in GFP accumulation by removal of the transcription block. Three micrograms TSP was loaded per lane. For quantitation, 2 samples of purified GFP protein were included. (D) Detection of Pal and Cpl-1 by Coomassie staining of total soluble protein. The large (L) and small (S) subunits of Rubisco and the Pal (P) and Cpl-1 (C) proteins are indicated. Band sizes of the molecular mass marker are given in kDa. (E) Quantitation of Pal and Cpl-1 protein accumulation levels by comparison with a dilution series of purified Rubisco.
High-Level Expression of Pal and Cpl-1 in Plastids.
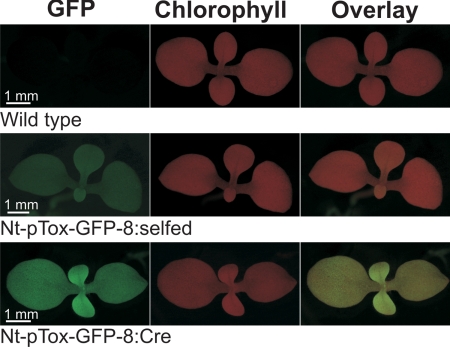
To visualize transgene expression from pTox vectors in vivo, we comparatively analyzed GFP accumulation before (Nt-pTox-GFP-8:selfed) and after (Nt-pTox-GFP-8:Cre) marker excision by UV microscopy. Although GFP was readily detectable in both lines (suggesting that it can also be expressed from the dicistronic aadA-gfp transcript), fluorescence was significantly stronger after marker excision (Fig. 3) suggesting that removal of the aadA and the transcription block enhance GFP expression. We next analyzed gene expression at the RNA and protein levels more quantitatively for all 3 transgenes. RNA gel blot analyses revealed the presence of dicistronic transcripts in all lines carrying the aadA marker, confirming that the transcription block does not terminate transcription in plastids (Fig. 4A). Elimination of the bacterial terminators and the aadA by Cre-mediated recombination resulted in appearance of abundant monocistronic mRNAs. For gfp and pal, removal of the aadA resulted in increased mRNA abundance (Fig. 4A), possibly suggesting that the monocistronic transcripts are more stable than the (considerably longer) dicistronic ones. This effect, however, was less pronounced for cpl-1. Consistent with their higher RNA accumulation levels, GFP accumulation in the Nt-pTox-GFP:Cre plants was 4-to 5-fold higher than in the Nt-pTox-GFP:selfed plants and was estimated to represent 5–10% of the total soluble protein (TSP; Fig. 4C).
Fig. 3.
Excision of the bacterial transcription block in plastids. Removal of the transcription block (Nt-pTox-GFP-8:Cre plants) results in strongly-enhanced GFP fluorescence.
To test whether the Pal and Cpl-1 proteins accumulate in transplastomic plants to very high levels, we analyzed electrophoretically-separated TSP samples by Coomassie staining (which usually allows foreign protein detection if accumulation levels are >1–2% of TSP). Interestingly, all transplastomic lines showed strong additional bands, the molecular mass of which corresponded to the theoretical molecular masses of Pal and Cpl-1, respectively (Fig. 4D). Mass spectrometric protein identification confirmed that the massively-accumulating novel proteins were indeed Pal and Cpl-1. Consistent with the RNA accumulation data (Fig. 4A), removal of the aadA resulted in substantially-increased protein accumulation in the Pal-expressing lines, but gave no strong increase in Cpl-1 accumulation (Fig. 4D). Quantification of protein accumulation against a purified Rubisco standard revealed that Pal accumulates to ≈30% of TSP (in Nt-pTox-Pal:Cre plants) and Cpl-1 accumulated to ≈10% of TSP (Fig. 4E). These expression levels are equivalent to ≈0.5 g (Cpl-1) to 2 g (Pal) of protein product per kg of fresh weight (19).
Bacteriolytic Activity of Plastid-Produced Pal and Cpl-1.
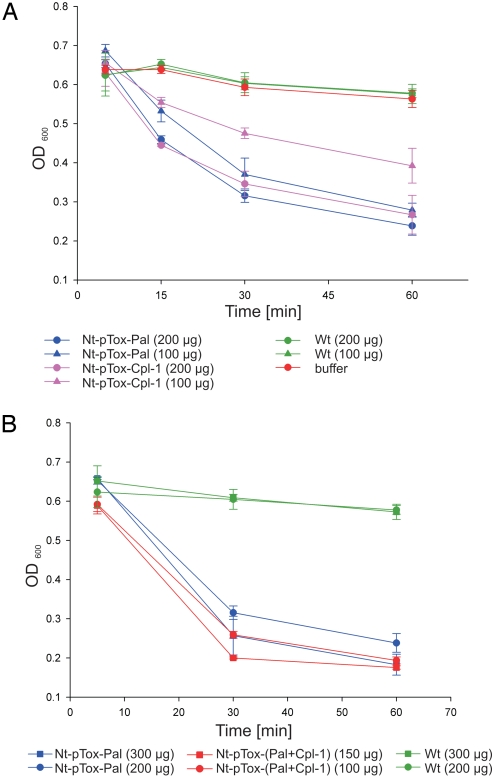
We next wanted to test whether the chloroplast-produced Pal and Cpl-1 lysins were biologically active in that they would effectively kill S. pneumoniae, the causative agent of pneumonia. The extremely high accumulation levels made it unnecessary to purify the lysins and allowed the use of a crude total protein extract (including treatments with wild-type extract as a control). Proteins from wild-type plants did not exhibit endogenous bacteriolytic activity (Fig. 5) as had been shown before (3). In contrast, protein extracts from transplastomic Pal- and Cpl-1-expressing plants effectively killed the streptococci (Fig. 5A). As little as 100 μg of TSP from Pal-expressing plants and 200 μg of TSP from Cpl-1-expressing plants was sufficient to kill the bacteria in a dense 1-mL culture in a time-dependent manner (Fig. 5A). Essentially complete killing within 1 h was achieved with the residual low optical density (OD) of the culture reflecting the OD of the bacterial lysate. Combined addition of Pal and Cpl-1 exhibited a synergistic effect and enhanced bacterial killing (Fig. 5B) as had been demonstrated with purified phage proteins (8). Effective killing of S. pneumoniae by the chloroplast-expressed lysins was confirmed by live/dead fluorescence staining, which stains live bacteria green and dead ones red. After 30-min exposure to 100 μg of protein extract from Pal-expressing transplastomic plants, most of the remaining (i.e., not yet lyzed) bacteria stained red, indicating that they were already dead (Fig. S3). Finally, the killing of the bacteria was also followed by determining the colony-forming units (CFU) after addition of the plant extracts. These assays revealed that, 30 min after addition of 300 μg of protein extract from Pal-expressing plants, the CFU had decreased to 23% and decreased further to 7% after 60 min.
Fig. 5.
Bactericidal activity of the chloroplast-produced Pal and Cpl-1 proteins on S. pneumoniae cultures. (A) Time course of bacterial cell death as monitored by clearing of bacterial cultures. The amounts of total soluble plant protein added to the cultures at time point 0 are indicated. The OD of the cultures was measured at 600 nm (OD600). Data are from 2 sets of experiments with 3 replicas each. The standard deviation is indicated. (B) Synergistic action of Pal and Cpl-1 in bacterial killing assays is shown.
The observed toxicity of Pal and Cpl-1 to E. coli (Fig. 1C) seemed to challenge the idea that phage-derived lysins usually act quite specifically on the host bacteria of the phage (4). To test whether Pal and Cpl-1 indeed act as lysins on E. coli, we exposed E. coli cultures to protein extracts from transplastomic plants. Interestingly, no significant decline in cell density was observed, indicating that Pal and Cpl-1 are only toxic when expressed inside E. coli cells, but do not kill the cells from the outside. This finding confirms that both lysins are specific to pneumococci (7, 8) and suggests that treatments with Pal and Cpl-1 should not have undesired nontarget effects.
High Stability of Pal and Cpl-1 in Chloroplasts.
As the chloroplast protein biosynthesis capacity declines with leaf age, the age-dependent decrease in foreign protein accumulation (9, 20) provides a good proxy for protein stability. To determine the stability of Pal and Cpl-1 in planta, we analyzed a developmental series of 9 leaves from a mature transplastomic plant. Remarkably, no age-dependent decline in Pal and Cpl-1 accumulation was detectable and protein amounts in old leaves seemed to be even slightly higher than in young leaves (Fig. S4). This result suggests that the enormous accumulation levels of the lysins are, at least to a large extent, caused by their high stability inside chloroplasts.
Discussion
In the course of this work, we have developed a strategy facilitating the high-level expression of plastid transgenes that are unclonable into conventional expression cassettes. Taking advantage of one of the few mechanistic differences in plastid versus bacterial gene expression, we have effectively prevented lethal transgene expression in E. coli by inducing premature transcription termination upstream of the transgene. The bacterial terminators did not inhibit expression in plastids and, moreover, could be easily deleted from the plastid genome along with the selectable marker gene by using site-specific recombination. We have provided proof of principle for the applicability of our strategy by successfully expressing 2 phage-derived lysins, Pal and Cpl-1, that both proved unclonable into plastid expression cassettes. Lysin protein accumulation in chloroplasts to up to 30% of the plant's TSP demonstrates the effectiveness of our strategy and suggests that the same approach should be applicable also to the production of other biopharmaceuticals that exhibit antimicrobial activity (21).
The enormous lysin expression levels obtained here and in a previous study (3) are caused by high protein stability inside the chloroplast, which is consistent with the idea that phage lysins have evolved considerable resistance to the proteases of their host bacteria. Because plastids possess a prokaryotic-type proteolytic machinery (22), it seems conceivable that lysins are poor substrates also for chloroplast proteases. Thus, chloroplasts seem to provide an ideal expression platform for lysin-type protein antibiotics. In addition to low costs caused by inexpensive plant production and high expression levels, chloroplasts have the advantage that they lack a bacterial-type cell wall, and therefore high-level lysin expression does not interfere with the metabolism of the plastid. Also, the therapeutic use of plant protein extracts is safer than the use of lysates from phage-infected bacteria and avoids expensive purification steps to eliminate bacterial endotoxins. Finally, the transplastomic technology provides increased transgene containment caused by maternal plastid inheritance in most crops, which greatly reduces unwanted pollen transmission of transgenes (23).
Lysins from phages infecting pneumococci have been shown to kill all common capsular serotypes of S. pneumoniae, including highly antibiotic-resistant strains (7, 8). In addition to potentially helping to control or eliminate nasopharyngeal colonization by penumococci, efficient synthesis in plastids of the highly active protein antibiotics Pal and Cpl-1 therefore provides a promising step forward toward coping with the worldwide increase of resistance to multiple antibiotics in pneumococci.
Materials and Methods
Plant Material.
Sterile tobacco (Nicotiana tabacum cv. Petite Havana) plants were grown on agar-solidified Murashige and Skoog (MS) medium (24) with sucrose (20 g/L). Regenerated shoots from homoplasmic transplastomic lines were rooted and propagated on the same medium, followed by transfer to soil and growth to maturity under standard glasshouse conditions. For inheritance assays, seeds were surface-sterilized and sown on medium with or without spectinomycin (500 mg/L).
Cloning Procedures.
The plastid transformation vector pTox (GenBank accession no. EU450674) is based on the previously described plasmid pRB94 (18). A loxP site was inserted behind the Prrn promoter (3 bp upstream of the SD sequence; ref. 25) by PCR with the primers PTox-profwd (5′-AAAGAGCTCGCTCCCCCGCCGTCGTTCAA-3′) and PTox-prorev (5′-TTCTGCCATGAATCCCTCCCTAATAACTTCGTATAGCATACATTATACGAAGTTATCAACTGTATCCAA-3′) by using Deep VentR DNA polymerase (NEB). The amplified fragment was cloned as a SacI/FatI fragment into a progenitor clone of pZS197 (25) cut with SacI and NcoI, resulting in plasmid pMO22. A synthesized DNA fragment (GENEART) covering the rho-dependent terminator λtR1 from the cro gene of phage λ (Trho; ref. 14), the intrinsic terminator rrnB T1 from E. coli (Tint; ref. 15), a loxP site and the leader sequence from the gene 10 transcript of phage T7 (T7G10L; ref. 10; 5′-ataaccccgctcttacacattccagccctgaaaaagggcatcaaattaaaccacacctatggtgtatgcatttatttgcatacattcaatcaattgttagctttcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttaatctgataacttcgtataatgtatgctatacgaagttatggatcctagaaataattttgtttaactttaagaaggagatatacccatggaaatctaga-3′) was cloned as a SpeI–XbaI fragment into pMO22 (cut with XbaI and dephosphorylated with antarctic phosphatise; NEB), resulting in pMO23. The complete expression cassette (Fig. 1A) was cloned as a SacI/DraI fragment into a pRB94 derivative lacking the aadA marker and cut with SacI and SmaI, resulting in the transformation vector pTox. Synthetic genes for pal (GenBank accession no. EU450672) and cpl-1 (GenBank accession no. EU450673) were produced by DNA synthesis (GENEART) using the codon usage preferred by tobacco plastids and introducing suitable restriction sites (NcoI and XbaI) for cloning into the expression cassettes (Fig. 1A). To assess the toxicity of Pal and Cpl-1 to E. coli, the genes were cocloned in a 1:1 ratio with gfp into the pRB95-derived vector pMO24 (3, 18). Plasmid DNA isolated from bacterial clones was digested with NcoI and XbaI to determine the identity of the insert.
Plastid Transformation.
Plastid transformation was carried out by the biolistic protocol (25) using 0.6-μm gold particles and a helium-driven biolistic gun (PDS1000He; BioRad). Transplastomic lines were selected on regeneration medium containing 500 mg/L spectinomycin (26). Primary transplastomic lines were subjected to additional regeneration rounds on spectinomycin-containing medium to obtain homoplasmic tissue.
Isolation of Nucleic Acids.
DNA was extracted from frozen leaf material using a published protocol (27) with minor modifications (extraction buffer: 100 mM Tris·HCl, 1 M NaCl, 10 mM EDTA, pH 8.0; extraction temperature: 75 °C). Total cellular RNA was extracted by using the peqGOLD TriFast reagent (Peqlab).
Gel Blot Analyses.
For Southern blot analyses, samples of 2 μg of total cellular DNA were digested with HincII, separated by electrophoresis in 1% agarose gels, and transferred onto Hybond XL membranes (GE Healthcare) by capillary blotting. Samples of 3 μg of total cellular RNA were electrophoresed in formaldehyde-containing 1% agarose gels and blotted onto Hybond XL membranes. A 550-bp PCR product from the psaB-coding region (28) was used as an restriction fragment length polymorphism (RFLP) probe to verify plastid transformation and test for homoplasmy. For transcript detection, the complete coding regions of pal and cpl-1, respectively, were used as probes. A gfp-specific probe was obtained by PCR amplification of the coding region. Probes were labeled with α[32P]dCTP by random priming (GE Healthcare). Hybridizations were performed at 65 °C in Church buffer (29).
Protein Extraction, Western Blot Analysis, and Mass Spectrometric Protein Identification.
TSP was extracted, quantitated, and analyzed by electrophoresis in SDS-containing polyacrylamide gels as described (3). Pal and Cpl-1 accumulation levels were determined by comparison with purified Rubisco as described (3). For activity tests, protein extracts (in 10% glycerol) were purified by dialysis against 25 mM Tris·HCl, pH 7.0, at 4 °C (molecular mass cut-off: 10 kDa) overnight. For Western blot analysis, 3 μg of Nt-pTox-GFP protein extract and samples of 50 and 100 ng of purified GFP were electrophoretically separated and transferred to a Hybond P membrane (GE Healthcare) using a standard transfer buffer (25 mM Tris·HCl, 192 mM glycine, pH 8.3). Immunobiochemical detection was carried out with the ECL system (GE Healthcare). The Pal and Cpl-1 proteins were identified by standard mass spectrometry procedures. Tryptically-digested peptide mixtures were analyzed by LC/MS/MS using nanoflow HPLC (Proxeon Biosystems) and a linear ion trap instrument (LTQ; Thermo Electron) as mass analyzer.
Fluorescence Microscopy.
Wild-type, Nt-pTox-GFP-selfed, and Nt-pTox-GFP-Cre plants grown on synthetic medium were analyzed for GFP and chlorophyll fluorescence by using a Leica MZ16 FA fluorescence stereomicroscope using the filters GFP3 (excitation filter 470/40 nm; barrier filter 525/50 nm) and Texas red (excitation filter 560/40 nm; barrier filter 610 LP).
Bacterial Assays.
S. pneumoniae (RKI 720, capsule type 6B; kindly provided by Sven Hammerschmidt, University of Greifswald, Greifswald, Germany) was grown at 37 °C overnight to an OD600 of ≈0.5 in THY medium (Todd-Hewitt broth; BD) supplemented with 0.5% (wt/vol) yeast extract. Cells were harvested by centrifugation (5 min, 4,000 × g), and the pellet was washed with PBS buffer (pH 7.0). The cells were then resuspended in PBS and adjusted to OD600 = 1.0. Protein samples of 200 μL [extracted from young leaves and diluted in 25 mM Tris·HCl (pH 7.0), 10% glycerol] were mixed with 800 μL of the bacterial suspension, and the decline in OD600 was measured over time (in triplicate). A 10-μL cell suspension was removed for CFU determination by plating a dilution series on THY plates. At time points 3, 15, 30, and 60 min after lysin addition, 50-μL samples were taken for viability assays and stained by using a live/dead fluorescence staining method (LIVE/DEAD BacLight Bacterial Viability Kit; Invitrogen) as described (3).
Supplementary Material
Acknowledgments.
We thank Dr. Stephanie Ruf and Yvonne Weber (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for plant transformation, Dr. Waltraud Schulze (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for help with mass spectrometric protein identification, and Cordula Lembke, Thomas Köller, and Kerstin Standar (University of Rostock) for help with bacterial assays. This work was supported by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU450672, EU450673, and EU450674).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813146106/DCSupplemental.
References
- 1.Ma JK-C, et al. Molecular farming for new drugs and vaccines. EMBO Rep. 2005;6:593–599. doi: 10.1038/sj.embor.7400470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock R. Plastid biotechnology: Prospects for herbicide and insect resistance, metabolic engineering, and molecular farming. Curr Opin Biotechnol. 2007;18:100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Oey M, Lohse M, Kreikemeyer B, Bock R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 2009;57:436–445. doi: 10.1111/j.1365-313X.2008.03702.x. [DOI] [PubMed] [Google Scholar]
- 4.Fischetti VA, Nelson D, Schuch R. Reinventing phage therapy: Are the parts greater than the sum? Nat Biotechnol. 2006;24:1508–1511. doi: 10.1038/nbt1206-1508. [DOI] [PubMed] [Google Scholar]
- 5.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Q, Nelson D, Zhu S, Fischetti VA. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob Agents Chemother. 2005;49:111–117. doi: 10.1128/AAC.49.1.111-117.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 8.Loeffler JM, Fischetti VA. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob Agents Chemother. 2003;47:375–377. doi: 10.1128/AAC.47.1.375-377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou F, et al. High-level expression of HIV antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J. 2008;6:897–913. doi: 10.1111/j.1467-7652.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda H, Maliga P. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 2001;29:970–975. doi: 10.1093/nar/29.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlbauer SK, Koop H-U. External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J. 2005;43:941–946. doi: 10.1111/j.1365-313X.2005.02495.x. [DOI] [PubMed] [Google Scholar]
- 12.Magee AM, MacLean D, Gray JC, Kavanagh TA. Disruption of essential plastid gene expression caused by T7 RNA polymerase-mediated transcription of plastid transgenes during early seedling development. Transgenic Res. 2007;16:415–428. doi: 10.1007/s11248-006-9045-z. [DOI] [PubMed] [Google Scholar]
- 13.Stern DB, Gruissem W. Control of plastid gene expression: 3′ Inverted repeats act as mRNA processing and stabilizing elements but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 14.Richardson LV, Richardson JP. Rho-dependent termination of transcription is governed primarily by the upstream Rho utilization (rut) sequences of a terminator. J Biol Chem. 1996;271:21597–21603. doi: 10.1074/jbc.271.35.21597. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds R, Bermúdez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 16.Corneille S, Lutz K, Svab Z, Maliga P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox-site-specific recombination system. Plant J. 2001;27:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- 17.Hajdukiewicz PTJ, Gilbertson L, Staub JM. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 2001;27:161–170. doi: 10.1046/j.1365-313x.2001.01067.x. [DOI] [PubMed] [Google Scholar]
- 18.Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- 19.Gleba Y, Marillonnet S, Klimyuk V. Engineering viral expression vectors for plants: The “full virus” and the “deconstructed virus” strategies. Curr Opin Plant Biol. 2004;7:182–188. doi: 10.1016/j.pbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Birch-Machin I, Newell CA, Hibberd JM, Gray JC. Accumulation of rotavirus VP6 protein in chloroplasts of transplastomic tobacco is limited by protein stability. Plant Biotechnol J. 2004;2:261–270. doi: 10.1111/j.1467-7652.2004.00072.x. [DOI] [PubMed] [Google Scholar]
- 21.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 22.Adam Z. Protein stability and degradation in plastids. Top Curr Genet. 2007;19:315–338. [Google Scholar]
- 23.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 25.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson D, Henry R. Single-step protocol for preparation of plant tissue for analysis by PCR. BioTechniques. 1995;19:394–400. [PubMed] [Google Scholar]
- 28.Zhou F, Karcher D, Bock R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wurbs D, Ruf S, Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.