Abstract
The histopathological diagnosis of melanoma can be challenging. No currently used molecular markers accurately distinguish between nevus and melanoma. Recent transcriptome analyses have shown the differential expression of several genes in melanoma progression. Here, we describe a multi-marker diagnostic assay using 5 markers (ARPC2, FN1, RGS1, SPP1, and WNT2) overexpressed in melanomas. Immunohistochemical marker expression was analyzed in 693 melanocytic neoplasms comprising a training set (tissue microarray of 534 melanomas and nevi), and 4 independent validation sets: tissue sections of melanoma arising in a nevus; dysplastic nevi; Spitz nevi; and misdiagnosed melanocytic neoplasms. Both intensity and pattern of expression were scored for each marker. Based on the differential expression of these 5 markers between nevi and melanomas in the training set, a diagnostic algorithm was obtained. Using this algorithm, the lesions in the validation sets were diagnosed as nevus or melanoma, and the results were compared with the known histological diagnoses. Both the intensity and pattern of expression of each marker were significantly different in melanomas compared to nevi. The diagnostic algorithm exploiting these differences achieved a specificity of 95% and a sensitivity of 91% in the training set. In the validation sets, the multi-marker assay correctly diagnosed a high percentage of melanomas arising in a nevus, Spitz nevi, dysplastic nevi, and misdiagnosed lesions. The multi-marker assay described here can aid in the diagnosis of melanoma.
Keywords: biomarkers, diagnosis, microarray analysis
Melanoma was estimated to be the 7th most common cancer in the United States in 2008 (1). The diagnosis of melanoma is rendered following pathological evaluation of the biopsied tissue. Numerous histopathological features of melanoma have been described (2). However, the diagnosis of melanoma remains problematic in a subset of cases (3–7), as the relative weights assigned to these criteria differ among pathologists. Moreover, many of these criteria overlap with atypical but otherwise benign nevi such as dysplastic or Spitz nevi. Thus, the level of discordance in the diagnosis of certain melanocytic neoplasms can be high, even when examined by a panel of experienced melanoma pathologists (4). In addition, no molecular assays are routinely performed to assist in this differential diagnosis, given that immunohistochemical markers (e.g., S100, HMB-45, MART-1, and MITF) have little utility in distinguishing nevi from melanomas (8–12).
Gene expression profiles have proven useful in refining approaches to cancer classification (13) and prognosis (13–15) and in determining predictive markers for therapy (16). Profiles developed using microarray analyses have been translated into smaller gene sets using more routinely available methodology (17, 18). However, few studies have reported the validation of gene signatures using immunohistochemical methods to resolve a differential diagnosis.
In a recent study, we demonstrated distinct gene expression profiles for nevi and melanomas (12). Here, we aimed to confirm the differential protein expression of a subset of the previously identified transcripts and to examine the utility of a multi-marker diagnostic assay for melanoma.
Results
Five markers (ARPC2, FN1, RGS1, SPP1, and WNT2) were selected for confirmation and incorporation into the multi-marker assay based on statistically significant differences in gene expression and the availability of commercially available antibodies. Expression of the selected markers was uniformly assessed both in the training set and in 4 validation sets. Each marker was scored on a 4-point scale for staining intensity.
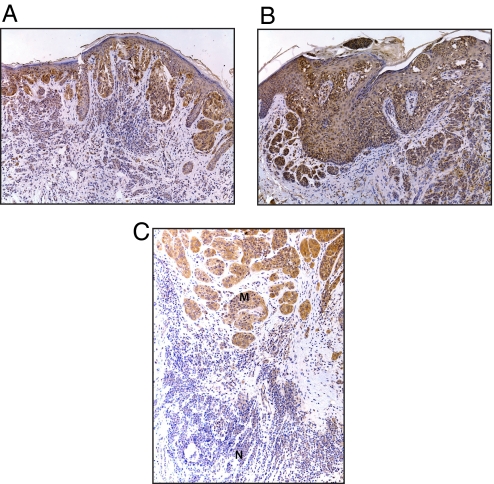
In the process of scoring expression intensity for the first marker in the training set, an intriguing expression pattern in benign nevi was observed, with a systematically stronger staining in the junctional zone of the nevus, along with a loss of expression at the nevus base (Fig. 1). In contrast, expression of the markers was more uniform in melanomas in the comparison between the lesional junctional zone and base (Fig. 1). As a result, we scored the tissue microarrays (TMAs) for intensity of expression both at the junctional zone (“top”), as well as at the base (“bottom”) of each melanocytic neoplasm in which the orientation of the lesion was clearly demonstrable in the specimen core present on the TMA section.
Fig. 1.
Photomicrographs of WNT2 immunostaining in a benign nevus (A), melanoma (B), and melanoma arising in association with a nevus (C), where M represents the melanoma and N represents the nevus.
Initially, each of the 5 markers was evaluated individually for its ability to diagnose melanoma versus nevus, using the 4-point intensity scale applied to the base of each lesion. The optimal way to partition each marker's 4-point scale was identified using univariate logistic regression analysis. Each of the 5 molecular markers was shown to be significantly overexpressed in melanomas when compared with nevi (Table 1). This provided directional confirmation of the results of the previous cDNA microarray analysis (12).
Table 1.
Discrimination of melanoma from nevus in the training data with the use of single-marker expression scores alone via logistic regression analysis
| Marker | Optimal scale partitioning | Chi-square | P value |
|---|---|---|---|
| ARPC2 | 0 vs. 1, 2 vs. 3 | 24.2 | <0.001 |
| FN1 | 0 vs. 1, 2 vs. 3 | 4.75 | 0.029 |
| RGS1 | 0, 1, 2 vs. 3 | 8.98 | 0.0027 |
| SPP1 | 0, 1 vs. 2, 3 | 11.1 | <0.001 |
| WNT2 | 0, 1, 2, 3 (entire scale) | 86.6 | <0.001 |
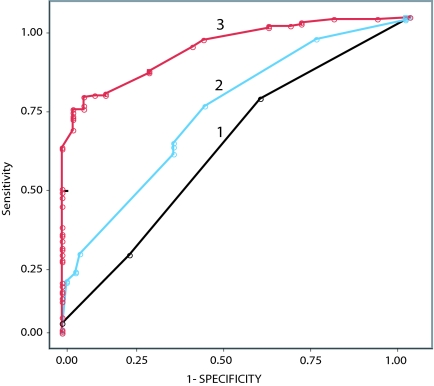
Several analyses were performed to examine whether the combination of markers was useful in its ability to diagnose melanoma. First, we examined the intensity of bottom expression scores for the 5 markers, taken together. Optimally partitioned marker expression scores were combined via multiple logistic regression, and a probability of being malignant was assigned to each lesion. Assigned probabilities were then partitioned at their optimal point of separation, achieving a diagnostic specificity of 94% and sensitivity of 76%. In addition, receiver operating characteristic (ROC) curves were constructed for various combinations of the 5 markers (Fig. 2), revealing an area under the ROC curve (AUC) of 0.911 for all 5 markers combined.
Fig. 2.
ROC plots in the diagnosis of melanoma using marker intensity scores for 1 marker alone (FN1, curve 1), 3 markers (FN1, ARPC2, SPP1, curve 2), and all 5 markers (curve 3).
Second, we performed an analysis using the differences in “top-to-bottom” marker expression for each of the 5 markers. This analysis showed that nevi consistently lost expression for each of the 5 markers (i.e., the top score exceeded the bottom score) when compared with melanomas, in which the marker immunostaining was much more uniform. This pattern of different top-to-bottom marker expression between nevi and melanomas was replicated significantly for all 5 markers, with sensitivities and specificities for each marker shown in Table 2.
Table 2.
Diagnostic accuracy for melanoma using lesion junctional zone versus lesion base (″top-to-bottom″) score in training data for each individual marker
| Marker | Specificity, % | Sensitivity, % | P value |
|---|---|---|---|
| ARPC2 | 60 | 96 | <0.001 |
| FN1 | 23 | 99 | 0.01 |
| RGS1 | 33 | 99 | <0.001 |
| SPP1 | 62 | 99 | <0.001 |
| WNT2 | 100 | 100 | <0.001 |
In the case of WNT2, there was perfect separation of the 2 distributions of top-to-bottom difference scores between nevi and melanomas in the lesions in which the orientation was clearly demonstrable. Thus, every nevus analyzed lost expression from its junctional zone to its base, whereas every melanoma had uniform expression from the junctional zone to its base. This perfect discrimination between nevus and melanoma rendered impossible further reliance on logistic regression as our sole analytical tool. The logistic regression estimation procedure could not produce precise P values for the maximum likelihood estimates of the regression coefficients in the face of such perfect discrimination between nevus and melanoma. Thus, a diagnostic algorithm was obtained that focused on the differences in vertical expression scores when all 5 markers were combined. Application of this diagnostic algorithm to the lesions in the training set yielded a sensitivity of 99% and a specificity of 85%.
We further examined the utility of marker expression scores and top-to-bottom differences using a digital imaging analysis. The WNT2-stained lesions in the training set were scanned digitally, and mean densitometric intensity was calculated for each lesion. The correlation between scores assigned by the study dermatopathologist and mean densitometric density was assessed, revealing a Spearman rank-order correlation coefficient of 0.84 (P < 0.001) between corresponding WNT2 bottom scores. We also evaluated the concordance in differences in the top-to-bottom expression scores, and observed a 96.8% agreement between the marker expression scores and mean densitometric intensity (P < 0.001, binomial sign test).
Finally, we explored the utility of the multi-marker assay in diagnosing melanoma combining both top and bottom marker intensity scores and top-to-bottom differences. Once again, we were unable to use multiple logistic regression. Thus, 2 different classification schemes were selected to analyze the combined diagnostic accuracy of the multi-marker assay. A classification tree generated by a freely available software program (rpart) identified 4 independent classifiers (i.e., WNT2 top-to-bottom difference score, FN1 top score, RGS1 bottom score, and WNT2 top score). Application of this classification scheme resulted in a sensitivity of 84% and a specificity of 86% in the diagnosis of melanoma in the training set.
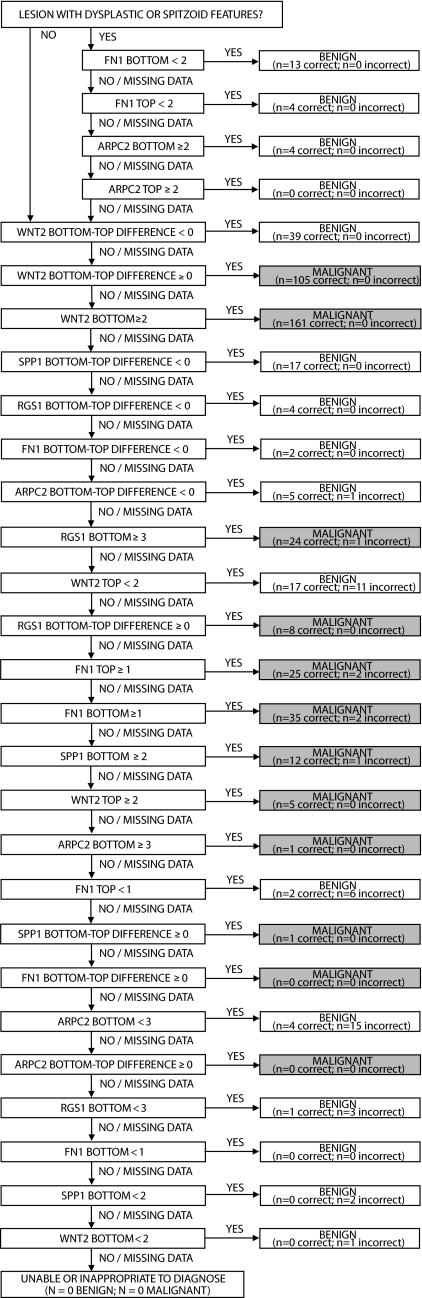
Next, a tailored diagnostic algorithm was obtained to exploit differential marker expression in distinct nevus subtypes. Intriguingly, all of the 21 dysplastic nevi included in the training set were confirmed to be dysplastic solely on the basis of their ARPC2 and FN1 intensity scores. This confirmatory algorithm for dysplastic nevi was then combined with a second, separate algorithm for all other lesions based on top-to-bottom differences and with a third algorithm for all other lesions based on marker intensity scores to form a composite diagnostic algorithm. Application of the composite algorithm (that incorporated the 4 classifiers generated by rpart) to the training set (Fig. 3) achieved a specificity of 95% and a sensitivity of 91% in the diagnosis of melanoma. All subsequent analyses were conducted using this composite algorithm.
Fig. 3.
Diagnostic algorithm obtained from the training data and applied to the validation sets based on marker intensity scores as well as top-to-bottom differences. For each diagnostic statement in the algorithm the number of correct and incorrect diagnoses achieved in the training set are indicated.
To validate the multi-marker assay developed from the training set, we examined both the intensity and pattern of expression of the 5 markers in 4 distinct and independent validation sets with greater relevance to the histological distinction between nevus and melanoma. Thus, we amassed a validation set of 38 cases of primary melanoma in association with a nevus, resulting in 75 evaluable melanocytic neoplasms, for which many potential confounding factors (age, sex, anatomical location, and potentially irrelevant histologic subtypes of nevus and melanoma, among others) are automatically controlled. In addition, we collected 2 additional validation sets directly relevant to this histological differential diagnosis: 21 cases of Spitz nevus and 39 cases of dysplastic nevus. These are the 2 most commonly problematic nevus subtypes in the differential diagnosis of nevus versus melanoma. Finally, we examined marker expression in a prospectively collected data set of 24 initially misdiagnosed lesions, including 6 lesions initially diagnosed as melanoma that were subsequently diagnosed as nevus by a consensus dermatopathology review, and 18 lesions initially diagnosed as benign or ambiguous that subsequently recurred locally and/or metastasized and were diagnosed as melanoma.
Using the composite diagnostic algorithm generated from the training data, the multi-marker assay achieved 95% specificity and 97% sensitivity in the melanomas arising in a nevus (P < 0.001, Fisher exact test). In addition, this algorithm correctly identified 95% (37/39) of dysplastic nevus sections and 95% (20/21) of Spitz nevi. Finally, the multi-marker assay correctly diagnosed 18/24 (75%) of the previously misdiagnosed lesions. A comparison of the sensitivity, specificity, and AUC of the multi-marker assay using various diagnostic algorithms is presented in Table 3.
Table 3.
Comparison of sensitivity, specificity, and AUC of multi-marker assay for melanoma diagnosis using different diagnostic algorithms
| Diagnostic algorithm | Specificity, % | Sensitivity, % | AUC |
|---|---|---|---|
| Marker intensity alone | 94 | 76 | 0.911 |
| Vertical expression alone | 85 | 99 | 0.917 |
| Combined expression scores | 95 | 91 | 0.928 |
Discussion
Our results demonstrate that a multi-marker assay composed of ARPC2, FN1, RGS1, SPP1, and WNT2 protein expression levels can be useful in the differential diagnosis of nevus versus melanoma. Our study is the largest to date analyzing the utility of molecular markers in melanoma diagnosis, and unique in using a comprehensive set of tissues necessitated by the heterogeneity of both nevi and melanomas. The multi-marker assay corrected three-quarters of the cases in which incorrect pathological diagnoses had been rendered, including melanomas initially misdiagnosed as nevi, in which the clinical behavior of the lesion had initiated review of the prior biopsy. The multi-marker assay described here could be used to assist in the histological diagnosis of melanoma, thereby providing important information to pathologists and other clinicians responsible for caring for patients with melanocytic neoplasms that are difficult to classify.
This is especially important given that mistakes in melanoma diagnosis cause many patients to undergo inappropriate therapy. Furthermore, the misdiagnosis of melanoma is the second most common reason for cancer malpractice claims in the United States, second only to mistakes in breast cancer diagnosis (19). Patients mistakenly diagnosed with a melanoma are under permanent fear of relapse and may not be able to obtain life or health insurance, whereas patients mistakenly diagnosed with a nevus are deprived of appropriate therapy for their malignancy, potentially including sentinel lymph node biopsy and systemic adjuvant therapy. Our results indicate that the multi-marker assay could reverse (and therefore potentially prevent) a high percentage of the errors that can result from routine histological analysis of melanocytic neoplasms. It will be important to test the utility of this multi-marker assay further in tissue sets obtained from different cohorts than the ones tested here, and for general pathologists in addition to those focusing on pigmented lesions to replicate the findings reported here.
Intriguingly, our data demonstrate that more than 1 marker is likely necessary to accurately diagnose melanoma, given the heterogeneity of both nevi and melanomas. Although WNT2 was the best single marker in the diagnosis of melanoma when the training set was analyzed, the other four markers included in the analysis played a crucial role in achieving the high level of diagnostic accuracy observed. For example, ARPC2 and FN1 expression levels alone sufficed in the classification of dysplastic and Spitz nevi. Moreover, all 5 markers contributed to the molecular reclassification of the misdiagnosed melanocytic neoplasms. As a result, a tailored diagnostic algorithm that separately analyzed Spitz and dysplastic nevi provided a higher diagnostic accuracy than a classification tree that did not distinguish among these nevus categories. Although this tailored algorithm contains many diagnostic statements, it is important to note that the diagnostic output for any given lesion is a simple binary conclusion (i.e., benign or malignant), and that such an output would, in practice, be computer-generated. This molecular diagnosis would be achieved by scoring marker intensity for each of the markers on a 0–3 scale for the top and bottom of the lesion in question.
Several of the markers incorporated into our assay have been previously demonstrated to have a role in driving melanocytic tumor progression. Fibronectin has been shown to contribute to melanoma cell invasion and metastasis (20, 21). SPP1 has been implicated in melanoma cell-growth and invasion by virtue of activation of the nuclear factor κB signaling pathway (22, 23). Recent studies have also shown the importance of WNT2 expression to melanoma cell proliferation (24). Thus, in addition to serving as diagnostic markers for melanoma, these proteins may be directly involved in promoting melanoma progression.
The differential expression of the markers suggested by the cDNA microarray analysis was not uniform in the immunohistochemical analysis of the nevus, as the nevus junctional zone frequently expressed the markers at a higher level than the nevus base. This was in contrast to most melanomas, which showed uniformity in the top-to-bottom analysis of marker expression. As a result, the pattern of protein expression was a significant discriminator between nevus and melanoma and, in general, superior to absolute transcript expression scores.
The vertical decrease in immunostaining observed here has been observed with a few other markers, including S100A6 (9), Melan-A (8), and HMB-45 (11). However, these markers have no value in distinguishing between nevi and primary melanomas, because they were not differentially expressed in nevi versus melanomas (12). In addition, in some of these studies (9), intradermal nevi were the dominant nevus type examined. In our study, the down-regulation of marker immunostaining was observed at the base of intradermal, compound, as well as Spitz and dysplastic nevi. This indicates the more broad-based utility of these markers in the diagnosis of melanoma.
In conclusion, we describe a multi-marker immunohistochemical assay that can accurately diagnose melanocytic neoplasms, thereby aiding in this differential diagnosis.
Materials and Methods
Study Population.
Five data sets, containing a total of 693 melanocytic neoplasms, were independently collected for the analyses performed in this study: a training set, consisting of a TMA of 118 benign nevi and 416 primary melanomas, and 4 independent validation sets: tissue sections of 38 melanomas arising in a nevus (resulting in 75 evaluable melanocytic neoplasms); 39 dysplastic nevi; 21 Spitz nevi; and 24 misdiagnosed melanocytic neoplasms. The training set was collected both to confirm the differential expression of the markers derived from the previous cDNA microarray analysis and to examine the utility of multi-marker expression scores. The validation sets were independently collected, focusing specifically on neoplasms where the diagnosis of nevus versus melanoma is more relevant or problematic. For all of the cases entered into the study, the histological diagnosis rendered by the original pathologist was confirmed by a review of the hematoxylin and eosin slides by the study dermatopathologist (R.W.S.). The misdiagnosed cases underwent a 3-tiered review. They were reviewed by the original pathologist. When the diagnosis was reversed, it underwent a consensus dermatopathology review at our institution (by at least 2 dermatopathologists). It was then subjected to review by the study dermatopathologist (distinct from the individuals indicated above), who confirmed the consensus final diagnosis. The composition of the 118 training set nevi studied was: 31 congenital intradermal nevi; 29 acquired intradermal nevi; 21 acquired dysplastic nevi; 17 congenital compound nevi; 15 acquired compound nevi; 4 junctional nevi; and 1 unclassified nevus. The histologic subtypes of the 416 training set melanomas were as follows: 198 superficial spreading melanomas; 135 nodular melanomas; 23 acral melanomas; 17 lentigo maligna melanomas; 16 desmoplastic melanomas; and 27 melanomas not otherwise classified. In the tissue set containing primary melanomas arising in association with a nevus, the breakdown of histologic subtypes for nevus and melanoma was as follows: 22 congenital nevi; 8 acquired nevi; 3 acquired dysplastic nevi; 1 acquired compound nevus; 1 acquired intradermal nevus; 1 congenital dysplastic nevus; 1 congenital compound nevus; 1 congenital intradermal nevus; 23 superficial spreading melanomas; 7 nodular melanomas; and 7 melanomas not otherwise classified. This analysis was approved by the Committee on Human Research, the UCSF Institutional Review Board.
Tissue Arrays.
Tissue microarrays were created as previously described using core diameters of 1.0 mm taken from the paraffin blocks (25, 26).
Immunohistochemistry.
Five markers (ARPC2, FN1, RGS1, SPP1, and WNT2) were selected for their potential utility as diagnostic markers using the following criteria: (i) they were differentially expressed in melanomas versus nevi as determined by statistical analysis of microarrays in a recent gene expression profiling study (12); and (ii) antibody reagents were available for testing. Expression of the selected markers was assessed using immunohistochemical analysis of TMAs and tissue sections. Slides were baked at 60°C for 30 min before staining, and deparaffinized and rehydrated by rinsing in xylene. The slides were then microwaved in 10 mM citrate buffer. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. In the case of WNT2, after washing with phosphate buffered saline (PBS), the slides were incubated at room temperature for 30 min with normal rabbit serum to reduce nonspecific background staining, and then washed with PBS. In the case of FN1, the slides were sequentially incubated with avidin and biotin blocking reagents. The primary antibody [goat polyclonal anti-WNT2 IgG (Biovision, 1:5 dilution); rabbit polyclonal anti-SPP1 IgG (Abcam, 1:200 dilution); rabbit polyclonal anti-FN1 IgG (Dako, 1:400 dilution); rabbit polyclonal anti-ARPC2 IgG (Upstate, 1:50 dilution); and chicken anti-RGS1 IgG (GeneTex, 1:100 dilution for TMAs and 1:50 dilution for tissue sections)] was then added and incubated overnight at 4°C. Biotinylated goat anti-rabbit, anti-chicken, or rabbit anti-goat IgG antibody (Vector Laboratories) was used as a secondary antibody for amplification, followed by incubation with ABC-HRP (Vector Laboratories) for 30 min, and DAB/hydrogen peroxide solution (Sigma). Slides were counterstained with hematoxylin and mounted with permount. In all of the validation sets, marker expression was examined using immunohistochemical analysis on 5-μM tissue sections. Positive controls for the various antibodies included: breast tumor (WNT2, SPP1); melanoma cell lines LOX and FEM (WNT2, ARPC2, SPP1); melanoma tissue sections (WNT2, FN1, ARPC2, RGS1, SPP1); normal kidney (FN1); thymus (RGS1); and non-Hodgkin lymphoma (RGS1). The technical negative control used for immunohistochemistry included the use of PBS instead of primary antibody, with all other conditions kept the same.
Evaluation of Immunohistochemical Staining.
The regions of most uniform staining were scored for each specimen. Expression of marker proteins was graded on cellular intensity using the following scale: no staining (0), weak staining (1), moderate staining (2), and intense staining (3). Intensity of marker expression was scored (where possible) both at the top and bottom of the neoplasm. For all lesions, the top was recorded in the junctional and/or papillary dermal region and the bottom at the deepest identified portion of the lesion. The arrays and sections were scored by the study dermatopathologist (R.W.S.) twice, and a third, consensus score was determined for any discrepant scoring for each marker.
Imaging Analysis of Immunohistochemical Staining.
The tissue arrays stained for WNT-2 were scanned and images were captured using the Carl Zeiss Mirax Scan and Axiovision 4.5 image processing system (Carl Zeiss MicroImaging). Regions with an identifiable melanocytic lesion were selected for evaluation, including the top and bottom areas of the lesion, when available. Characterization of the immunohistochemical staining was calculated by applying a segmentation feature with 4 different phase measurement masks recognizing background (no tissue), extracellular matrix and stroma, nuclei (hematoxylin stained), and brown-immunostained cells. The intensity was calculated by masking out all areas not selected by the brown phase threshold and calculating the integrated optical densitometry of brown within the remaining regions. The value was divided by the area in pixels squared of the brown phase to calculate the mean densitometric intensity of the regions selected. In the digital imaging analysis, WNT2 intensity scores assigned by the study dermatopathologist were compared with their corresponding mean densitometric intensity scores within the training set via Spearman rank-order correlation analysis.
Statistical Analysis.
The diagnostic accuracy of each marker's intensity was assessed with univariate logistic regression. The diagnostic accuracy of the combination of the intensity of all five markers was assessed with multivariate logistic regression, and analyzed using ROC curves and by calculating the AUC, as well as specificities and sensitivities. Both in the case of top-to-bottom comparisons and in the validation set containing melanomas arising in nevi, the diagnostic accuracy was analyzed via the Fisher exact test. The difference between intensity of marker immunostaining in the nevus base versus the melanoma base was tested for each marker using the Mann–Whitney test (corrected for tied observations). The difference between intensity of marker immunostaining in the lesion junctional zone and base was compared between nevus and melanoma and tested for each marker using the Mann–Whitney test. All P-values reported are 2-sided.
Two different classification schemes were used to distinguish melanomas from nevi among the 534 lesions in the training set: a standard classification tree generated by a freely available software program (rpart) (27), and a diagnostic algorithm obtained from MDMS, tailored specifically to our data. The rpart program generated a classification tree using four discriminatory parameters. The tailored diagnostic algorithm was trained to exploit the heterogeneity of nevus subtypes. This algorithm first confirmed a lesion with dysplastic or Spitzoid features as actually being either a dysplastic or Spitz nevus, and then diagnosed all non-confirmed lesions as either a different type of benign nevus or as a melanoma. The tailored algorithm also encompassed top-to-bottom difference scores and marker intensity scores. In the top-to-bottom analysis, a lesion was classified as benign by each marker if its top intensity score exceeded its corresponding bottom intensity score; otherwise the lesion was classified as malignant. This tailored diagnostic algorithm was applied uniformly to the lesions in all four independent validation sets. When implementing both multi-marker classification schemes, no preference was given to achieving sensitivity versus specificity (i.e., they were weighted as equally important). When derived from the application of this algorithm, the AUC values calculated were based on binary diagnoses, possessing a single cut-point, and refer to the area enclosed within the polygon whose northwest corner point is defined by that single cut-point.
Acknowledgments.
We thank Dr. Frederick Baehner and Loretta Chan of the Cancer Center Tissue Core for assistance with marker immunostaining, Ken Pratt of Carl Zeiss for the use of the Mirax scanner and Axiovision software, and Rosie Casella for manuscript preparation. This work was supported by the Herschel and Diana Zackheim Endowment Fund, American Cancer Society Grant RSG-03–247-01-MGO, and National Institutes of Health Grants CA114337 and CA122947.
Footnotes
Conflict of interest statement: M.K.-S. has stock in Melanoma Diagnostics, Inc. J.R.M. has ownership interest in MDMS.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Crowson AN, Magro CM, Mihm MC., Jr . The Melanocytic Proliferations. New York: Wiley-Liss; 2001. Chapter 10. [Google Scholar]
- 3.Piepkorn MW, et al. A multiobserver, population-based analysis of histologic dysplasia in melanocytic nevi. J Am Acad Dermatol. 1994;30:707–714. doi: 10.1016/s0190-9622(08)81499-5. [DOI] [PubMed] [Google Scholar]
- 4.Farmer ER, Gonin R, Hanna MP. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol. 1996;27:528–531. doi: 10.1016/s0046-8177(96)90157-4. [DOI] [PubMed] [Google Scholar]
- 5.Corona R, et al. Interobserver variability on the histopathologic diagnosis of cutaneous melanoma and other pigmented skin lesions. J Clin Oncol. 1996;14:1218–1223. doi: 10.1200/JCO.1996.14.4.1218. [DOI] [PubMed] [Google Scholar]
- 6.Barnhill RL, et al. Atypical Spitz nevi-tumors: Lack of consensus for diagnosis, discrimination from melanoma, and prediction of outcome. Hum Pathol. 1990;30:513–520. doi: 10.1016/s0046-8177(99)90193-4. [DOI] [PubMed] [Google Scholar]
- 7.Brochez L, et al. Inter-observer variation in the histopathological diagnosis of clinically suspicious pigmented skin lesions. J Pathol. 2002;196:459–466. doi: 10.1002/path.1061. [DOI] [PubMed] [Google Scholar]
- 8.Busam KJ, et al. Expression of melan-A (MART1) in benign melanocytic nevi and primary cutaneous malignant melanoma. Am J Surg Pathol. 1998;22:976–982. doi: 10.1097/00000478-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Fullen DR, Reed JA, Finnerty B, McNutt NS. S100A6 preferentially labels type C nevus cells and nevic corpuscles: Additional support for Schwannian differentiation of intradermal nevi. J Cutan Pathol. 2001;28:393–399. doi: 10.1034/j.1600-0560.2001.028008393.x. [DOI] [PubMed] [Google Scholar]
- 10.King R, Googe PB, Weilbaecher KN, Mihm MC, Jr, Fisher DE. Microphthalmia transcription factor expression in cutaneous benign, malignant melanocytic, and nonmelanocytic tumors. Am J Surg Pathol. 2001;25:51–57. doi: 10.1097/00000478-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Kucher C, et al. Expression of Melan-A and Ki-67 in desmoplastic melanoma and desmoplastic nevi. Am J Dermatopathol. 2004;26:452–457. doi: 10.1097/00000372-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Haqq C, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alizadeh AA, et al. Distinct types of diffuse B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 14.van 't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 15.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 16.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 17.Lossos IS, et al. Prediction of survival in diffuse large B-cell lymphoma based on expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 18.Chen HY, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 19.Troxel DB. Pitfalls in the diagnosis of malignant melanoma: Findings of a risk management panel study. Am J Surg Pathol. 2003;27:1278–1283. doi: 10.1097/00000478-200309000-00012. [DOI] [PubMed] [Google Scholar]
- 20.Terranova VP, Williams JE, Liotta LA, Martin GR. Modulation of the metastatic activity of melanoma cells by laminin and fibronectin. Science. 1984;226:982–985. doi: 10.1126/science.6505678. [DOI] [PubMed] [Google Scholar]
- 21.Lacovara J, Cramer EB, Quigley JP. Fibronectin enhancement of directed migration of B16 melanoma cells. Cancer Res. 1984;44:1657–1663. [PubMed] [Google Scholar]
- 22.Philip S, Bulbule A, Kundu GC. Osteopontin stimulates tumor growth and activation of promatrix metalloproteinase-2 through nuclear factor-kappa B-mediated induction of membrane type 1 matrix metalloproteinase in murine melanoma cells. J Biol Chem. 2001;276:44926–44935. doi: 10.1074/jbc.M103334200. [DOI] [PubMed] [Google Scholar]
- 23.Philip S, Kundu GC. Osteopontin induces nuclear factor kappa B-mediated promatrix metalloproteinase-2 activation through I kappa B/IKK signaling pathways, and curcumin (diferulolylmethane) downregulates these pathways. J Biol Chem. 2003;278:14487–14497. doi: 10.1074/jbc.M207309200. [DOI] [PubMed] [Google Scholar]
- 24.You L, et al. An anti-Wnt-2 monoclonal antibody induces apoptosis in malignant melanoma cells and inhibits tumor growth. Cancer Res. 2004;64:5385–5389. doi: 10.1158/0008-5472.CAN-04-1227. [DOI] [PubMed] [Google Scholar]
- 25.Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1996;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 26.Kashani-Sabet M, et al. NF-kappa B in the vascular progression of melanoma. J Clin Oncol. 2004;22:617–623. doi: 10.1200/JCO.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 27.Therneau TM, Atkinson EJ. An introduction to recursive partitioning using rpart routines. Mayo Clinic Report #61. 1997 Sep 3; [Google Scholar]