Abstract
Rationale: Many of the interstitial lung diseases represent a diagnostic and therapeutic challenge because their clinical and even histologic features are often nonspecific. Likewise, the transcriptional signatures of most of them are unknown.
Objective: To compare the gene expression patterns from patients with idiopathic pulmonary fibrosis (IPF) hypersensitivity pneumonitis (HP), and nonspecific interstitial pneumonia (NSIP) using custom oligonucleotide microarrays.
Methods: We profiled lung biopsies from 15 patients with IPF, 12 with HP, and eight with NSIP. Labeled complementary ribonucleic acid was hybridized to a custom Affymetrix oligonucleotide DNA microarray using standard Affymetrix protocols. The custom array, Hu03, contained 59,619 probe sets representing an estimated 46,000 gene clusters.
Results: We identified statistically significant gene expression signatures that characterize HP and IPF. The HP gene expression signature was enriched for genes that are functionally associated with inflammation, T-cell activation, and immune responses, whereas the IPF signature was characterized by the expression of tissue remodeling, epithelial, and myofibroblast genes. We then compared these gene expression signatures to classify NSIP, a histologic pattern that is often difficult to differentiate consistently from HP and IPF. Two cases exhibited an IPF-like gene expression, another one could be more properly classified as HP, whereas others did not resemble HP or IPF, suggesting that they may represent idiopathic NSIP.
Conclusions: Our results underscore the value of gene expression signatures to classify the interstitial lung diseases and to understand pathogenic mechanisms, and suggest new ways to improve the diagnosis and treatment of patients with these diseases.
Keywords: global transcription analysis, interstitial lung diseases, lung fibrosis, microarrays, nonspecific interstitial pneumonia
Pulmonary fibrosis is the final common outcome of a diverse group of interstitial lung diseases (ILDs). A number of these disorders have known etiologies, arising from exposure to organic or inorganic particles or drugs, or they may have an association to collagen-vascular diseases (1). However, approximately 40 to 50% of these disorders are of unknown etiology and are classified as idiopathic interstitial pneumonias (IIPs) (2). The most common IIP is idiopathic pulmonary fibrosis (IPF), which represents a chronic and usually fatal lung disorder morphologically characterized by a usual interstitial pneumonia (UIP) pattern (2,3). Nonspecific interstitial pneumonia (NSIP) is a more recently recognized morphologic pattern characterized by varying degrees of inflammation and fibrosis. These changes are temporally uniform, suggesting that they have occurred over a single, relatively narrow time span (2,3). Importantly, NSIP can be difficult to differentiate consistently from hypersensitivity pneumonitis (HP) and UIP, and significant interobserver variability exists even among expert pathologists (4).
HP is a complex syndrome with variable clinical presentation and natural history, and represents a diagnostic challenge for clinicians and, in the chronic form, even for experienced pathologists. The diagnosis of HP requires a high index of suspicion and a combination of clinical, environmental, radiologic, physiologic, and pathologic findings (5, 6). Chronic HP often develops into fibrosis with destruction of the lung parenchyma, a process that mimics IPF (7).
It is widely believed that lung fibrosis is triggered by an injury that results in inflammation, which, if unresolved, is followed by fibroblast proliferation/activation and extracellular matrix accumulation. However, recent evidence suggests that this sequence is likely relevant for most ILDs, including HP, but probably not for IPF, which seems to represent primarily an epithelial/fibroblastic disorder (8,9).
To gain a better understanding of the molecular mechanisms that underlie the lung phenotypes in IPF, HP, and NSIP, we sought to identify and compare their gene expression “signatures” using genomewide, custom oligonucleotide microarrays. We hypothesized that these diseases exhibit different gene expression signatures that should be useful to distinguish these disorders and also provide insights regarding the divergent pathways leading to lung fibrosis. We also expected that specific signatures could lead to the identification of new biomarkers to aid in the diagnosis and classification of these diseases.
Our results indicate that IPF can be distinguished from HP by the expression of tissue remodeling and epithelial and myofibroblast genes, whereas HP is characterized by the expression of inflammatory genes representing T-cell activation and immune responses. Using these expression signatures, we were able to reclassify some of the NSIP cases as being closer to IPF or HP, whereas others were likely to represent idiopathic NSIP. Our results highlight the mechanistic differences between ILD subtypes and underscore the potential that transcriptional profiling has for the differential diagnosis of ILD.
METHODS
Fifteen patients with IPF, 12 with HP, and eight with NSIP-like pattern were included in this study. The study received approval from the local ethics committee, and informed consent was obtained from all subjects. Diagnosis of IPF and HP was performed as described elsewhere and confirmed by morphology (3, 10, 11). Diagnosis of NSIP was suspected in patients with an interstitial pneumonia that did not meet the clinical/radiologic criteria for other IIPs and was based primarily on histology (2, 3). Control samples included nonpathologic lung specimens obtained from trauma victims. None of the patients had been treated with corticosteroids/immunosuppressive drugs at the time of biopsy. Additional details are provided in the online supplement. One fragment of the lung samples was frozen in liquid nitrogen for RNA extraction and subsequent expression analysis by DNA microarrays.
RNA Extraction and DNA Microarray Hybridization
RNA extracted from lung tissue was converted into biotinylated cRNA and hybridized to a custom Affymetrix (Santa Clara, CA) oligonucleotide microarray (Hu03 containing 59,619 probe sets representing an estimated 46,000 gene clusters [see online supplement] using standard Affymetrix protocols). The generation of the gene expression data was performed as described (12).
Statistical Methods
Statistical analyses were performed using ScoreGenes software package (Jerusalem, Israel; available from http://www.cs.huji.ac.il/labs/compbio/scoregenes/) (13–15). The most informative genes were defined as those that pass the false-discovery rate of 5% (16) for t test or for threshold number of misclassifications (TNoM) method. To further assess the predictive power of the datasets, we used the Leave-One-Out Cross-Validation (LOOCV) statistical method. This method simulates removal of a single sample every trial and trains on the rest. The procedure is repeated until each sample is left out once and the number of correct and incorrect predictions is counted. For each p value, a classification score was calculated for every sample in a specific class (Reference 15 and the online supplement).
Functional Annotations
Enrichment of functional annotations was performed using National Institutes of Health DAVID and EASE online (http://apps1.niaid.nih.gov/david/) (17, 18). Statistical significance was determined using Fisher's exact score and EASE score (17). Additional details on statistical methods and functional annotations are provided in the online supplement.
Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) was performed as described (10). Cells were used for differential cell counts and fluorescence-activated cell sorter (FACS) analyses. Insulinlike growth factor binding protein 4 (IGFBP-4) was measured by ELISA following the instructions of the manufacturer (Diagnostic Systems Laboratories, Inc., Webster, TX). Results are expressed in micrograms of IGFBP-4/mg protein. Additional details are provided in the online supplement.
Immunohistochemistry
Tissue sections were treated as previously described (10). The following antibodies were used: Human anti–IGFBP-4 and N-cadherin polyclonal antibodies (Santa Cruz Biotech, Santa Cruz, CA), and human matrix metalloproteinase 1 (MMP-1) monoclonal antibody (Oncogene, San Diego, CA). The primary antibody was replaced by nonimmune serum for negative controls. Additional details are provided in the online supplement.
Flow Cytometric Analyses of Surface CD3, CD4, CD8, and CD69 on BAL Lymphocytes
BAL cells (1 × 106) were labeled with fluorescein isothiocyanate–conjugated anti-CD3, phycoerythrin-conjugated anti-CD4, PerCp anti-CD8, and fluorescein isothiocyanate anti-CD69 (Becton Dickinson, San Jose, CA). Flow cytometry was performed using a FACScan (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson). Additional details are provided in the online supplement.
RESULTS
Baseline Characteristics of Patients with IPF, HP, or NSIP
Demographic data, pulmonary function test results, and BAL differential cell counts are summarized in Table 1. All patients exhibited clinical, radiologic, and functional evidence of ILD, with variable degrees of dyspnea, decreased lung capacities, and hypoxemia at rest that worsened during exercise. In general, the patients with IPF were older, predominantly male, and more likely to have been cigarette smokers than the patients with HP or NSIP. By contrast, all patients with HP were female. This sex prevalence has been previously described in avian antigen-induced HP in Mexico (19). In the HP group, differential cell counts in BAL fluid were characterized by marked lymphocytosis, whereas in IPF, most BAL inflammatory cells were macrophages with a moderate increase in neutrophils and eosinophils.
TABLE 1.
BASELINE DEMOGRAPHIC, CLINICAL, PHYSIOLOGIC, AND BRONCHOALVEOLAR LAVAGE CHARACTERISTICS OF THE THREE STUDY GROUPS
| Variable | IPF (n = 15) | HP (n = 12) | NSIP (n = 8) |
|---|---|---|---|
| Age | 59.3 ± 7.9 | 45.6 ± 10.5 | 45.9 ± 11.1 |
| Sex, male/female | 11/4 | 0/12 | 1/7 |
| Duration of symptoms before diagnosis, mo | 23.4 ± 20.6 | 18.3 ± 14.3 | 19.3 ± 12.9 |
| Smoking status | |||
| Never | 9 | 12 | 6 |
| Former | 3 | 0 | 2 |
| Current | 3 | 0 | 0 |
| Clubbing | 12/15 | 3/12 | 4/8 |
| FVC, % predicted | 56.7 ± 13.5 | 60.8 ± 19.4 | 53.4 ± 17.5 |
| PaO2, mm Hg* | 51.5 ± 10.7 | 53.7 ± 6.6 | 51.4 ± 7.3 |
| BAL macrophages, % | 80.6 ± 6.8 | 31.9 ± 18.4 | 54.2 ± 10.9 |
| BAL lymphocytes, % | 13.5 ± 6.9 | 66.6 ± 18.6 | 44.5 ± 12.2 |
| BAL neutrophils, % | 3.5 ± 1.3 | 0.7 ± 1.2 | 0.5 + 0.8 |
| BAL eosinophils, % | 2.1 ± 2.5 | 0.8 ± 1.1 | 0.9 + 1.6 |
Definition of abbreviations: BAL = bronchoalveolar lavage; HP = hypersensitivity pneumonitis; IPF = idiopathic pulmonary fibrosis; NSIP = nonspecific interstitial pneumonia.
Normal values at Mexico City altitude: 67 ± 3 mm Hg.
Gene Expression Patterns Distinguish IPF and HP
IPF is a disease of unknown etiology, and by far the most severe ILD with very poor prognosis. HP is an ILD of known etiology, although its pathogenic mechanisms are not completely understood. Although considered a more benign ILD, some patients with chronic HP also evolve to end-stage lung fibrosis and die of the progressive destruction of the lung parenchyma. We therefore compared the gene expression patterns of these diseases, and asked whether we could find genes specifically expressed in each disease. A well-acknowledged challenge with microarray data is the asymmetry between the number of samples and the number of parameters measured. This asymmetry may lead to spurious p values regardless of which scoring system is used.
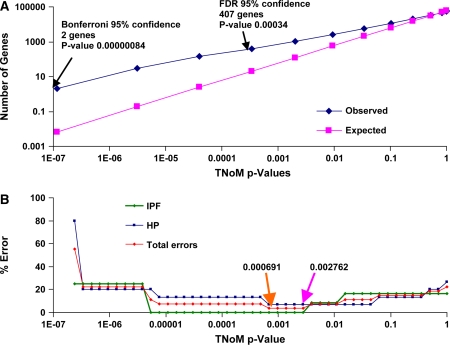
To address this challenge, we performed overabundance analysis, which revealed that the actual number of genes for every p value was substantially higher than would be expected under the null hypothesis that the separation of the samples is random (Figure 1A). Using the nonparametric TNoM scoring method (see Methods here and in the online supplement), we identified two genes that had TNoM score p values that passed Bonferroni correction and 407 genes that passed a significance threshold controlling the false-discovery rate at 5% as previously described (16). To identify the genes that best distinguished IPF from HP, we performed LOOCV for every TNoM score p value (Figure 1B). Minimal error rates were observed at TNoM score p values of 0.002762 and 0.000691 (Figure 1B) where classification errors were 1 and 0 for misclassifying HP as IPF and IPF as HP, respectively, strongly suggesting that the signature distinguishing these two diseases is significant. Unsupervised clustering also distinguished the diseases, as did Student's t test (data not shown).
Figure 1.
Hypersensitiviy pneumonitis (HP) and idiopathic pulmonary fibrosis (IPF) can be distinguished using gene expression profiling. (A) Overabundance analysis of the actual and expected number of genes that significantly distinguish whole lung gene expression between patients with IPF or HP and healthy subjects. The x axis denotes the number of genes (probes) in log scale, and the y axis shows the p value (threshold number of misclassifications [TNoM]) in log scale. Actual number of genes is significantly more abundant than would be expected (pink line). (B) Leave-One-Out Cross-Validation (LOOCV) classification graph. The x axis denotes the TNoM p value in log scale, and the y axis denotes the error rate in percentages. The pink arrow is the highest p value that gets the lowest error rate. The orange arrow is the lowest p value that gets the lowest error rate. FDR = false-discovery rate.
The Gene Expression Signatures That Characterize HP or IPF Are Composed of Distinctly Different Functional Groups
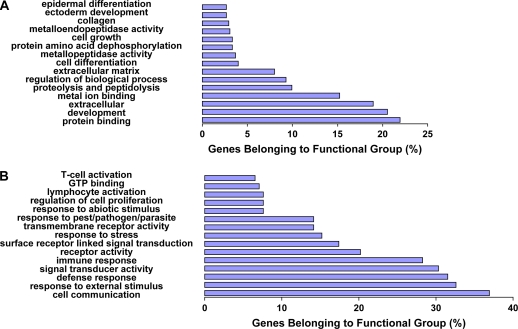
To better understand the mechanistic nature of the genes that distinguish IPF from HP, we looked for Gene Ontology functional annotations that were enriched in genes that were higher in IPF lungs or in HP lungs using the National Institutes of Health DAVID and EASE online (http://apps1.niaid.nih.gov/david/) (17, 18). This analysis was only performed on transcripts that could be found in LocusLink (http://www.ncbi.nlm.nih.gov/projects/LocusLink/). Most of the genes that were significantly increased in IPF were enriched for genes involved in development, extracellular matrix structure and turnover, and cellular growth and differentiation. The 354 upregulated genes that include the complete list of statistically significant annotations are listed in Table E1 on the online supplement. Some relevant upregulated genes are shown in Table 2. Furthermore, in addition to the statistically significant p values, many of these genes were associated with the relevant annotations (Figure 2A), suggesting that this represents a biologically meaningful phenomenon. The functional profile was completely different in HP lungs. The 595 genes that were overexpressed in HP compared with IPF and had locus-link information were enriched with a variety of host defense and inflammatory annotations (Table E2 and Figure 2B). Some relevant upregulated genes are shown in Table 3.
TABLE 2.
UPREGULATED GENES IN IDIOPATHIC PULMONARY FIBROSIS
| Gene Name | Locus-Link No. | TNoM p Value | t Test p Value |
|---|---|---|---|
| NM_017680: asporin (LRR class 1; ASPN), mRNA | 54829 | 4.0382E-05 | 0.00028842 |
| NM_033285: tumor protein p53 inducible nuclear protein 1 (TP53INP1), mRNA | 94241 | 0.00033652 | 2.5438E-06 |
| NM_000029: angiotensinogen (serine [or cysteine] proteinase; AGT), mRNA | 183 | 0.00033652 | 3.6249E-06 |
| NM_000861: histamine receptor H1 (HRH1), mRNA | 3269 | 0.00033652 | 1.6201E-05 |
| Hs.31386: secreted frizzled-related protein 2 | 6423 | 0.00033652 | 0.00020623 |
| NM_001792: cadherin 2, type 1, N-cadherin (neuronal) (CDH2), mRNA | 1000 | 0.00033652 | 0.00026976 |
| NM_005214: cytotoxic T-lymphocyte–associated protein 4 (CTLA4), mRNA | 1493 | 0.00033652 | 0.00039581 |
| NM_000577: interleukin-1 receptor antagonist (IL1RN), transcript variant 3, mRNA | 3557 | 0.00033652 | 0.00194308 |
| NM_033326: SRY (sex-determining region Y)-box 6 (SOX6), mRNA | 55553 | 0.00033652 | 0.0019824 |
| NM_004425: extracellular matrix protein 1 (ECM1), transcript variant 1, mRNA | 1893 | 0.00033652 | 0.00223442 |
| NM_001847: collagen, type IV, α6 (COL4A6), transcript variant A, mRNA | 1288 | 0.00033652 | 0.00342796 |
| NM_006475: osteoblast-specific factor 2 (fasciclin I-like; OSF-2), mRNA | 10631 | 0.00033652 | 0.00359433 |
| NM_130445: collagen, type XVIII, α1 (COL18A1), transcript variant 2, mRNA | 80781 | 0.00033652 | 0.00457581 |
| NM_002423: matrix metalloproteinase 7 (matrilysin, uterine; MMP7), mRNA | 4316 | 0.00033652 | 0.00486756 |
| NM_002991: chemokine (C-C motif) ligand 24 (CCL24), mRNA | 6369 | 0.00033652 | 0.0099309 |
| NM_000582: secreted phosphoprotein 1 (osteopontin, SPP1), mRNA | 6696 | 0.00201911 | 2.6178E-05 |
| Hs.156369: tenascin N | 63923 | 0.00201911 | 7.8339E-05 |
| NM_005268: gap junction protein, β5 (connexin 31.1; GJB5), mRNA | 2709 | 0.00201911 | 8.237E-05 |
| NM_000494: collagen, type XVII, α1 (COL17A1), transcript variant long, mRNA | 1308 | 0.00201911 | 8.792E-05 |
| NM_032888: collagen, type XXVII, α1 (COL27A1), mRNA | 85301 | 0.00201911 | 0.00012678 |
| Hs.9914: follistatin | 10468 | 0.00201911 | 0.0001307 |
| NM_022097: hepatocellular carcinoma antigen gene 520 (LOC63928), mRNA | 63928 | 0.00201911 | 0.00014831 |
| NM_001723: bullous pemphigoid antigen 1 (BPAG1), transcript variant 1e, mRNA | 667 | 0.00201911 | 0.00020654 |
| NM_006142: stratifin (SFN), mRNA | 2810 | 0.00201911 | 0.00030087 |
| NM_152999: six transmembrane epithelial antigen of prostate 2 (STEAP2), mRNA | 261729 | 0.00201911 | 0.00039041 |
| Hs.306692: epithelial membrane protein 1 | 2012 | 0.00201911 | 0.00042533 |
| NM_014799: hephaestin (HEPH), transcript variant 2, mRNA | 9843 | 0.00201911 | 0.00051385 |
| Hs.512555: collagen, type XIV, α1 (undulin) | 7373 | 0.00201911 | 0.00058874 |
| Hs.398100: mucin 6, gastric | 4588 | 0.00201911 | 0.00087085 |
| NM_003289: tropomyosin 2 (β; TPM2), mRNA | 7169 | 0.00201911 | 0.00090799 |
| NM_002546: tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin; TNFRSF11B), mRNA | 4982 | 0.00201911 | 0.00106604 |
| NM_002785: pregnancy-specific β1-glycoprotein 11 (PSG11), mRNA | 5680 | 0.00201911 | 0.00148639 |
| NM_005786: serologically defined colon cancer antigen 33 (SDCCAG33), mRNA | 10194 | 0.00201911 | 0.00134524 |
| NM_002421: matrix metalloproteinase 1 (interstitial collagenase; MMP1), mRNA | 4312 | 0.00201911 | 0.00180712 |
| NM_014476: α-actinin-2–associated LIM protein (ALP), mRNA | 27295 | 0.00201911 | 0.00185632 |
| NM_153756: fibronectin type III domain containing 5 (FNDC5), mRNA | 252995 | 0.00201911 | 0.00225902 |
| NM_004385: chondroitin sulfate proteoglycan 2 (versican; CSPG2), mRNA | 1462 | 0.00201911 | 0.00284683 |
| NM_001552: insulinlike growth factor binding protein 4 (IGFBP4), mRNA | 3487 | 0.00201911 | 0.003664 |
| Hs.443625: collagen, type III, α1 | 1281 | 0.00201911 | 0.00486324 |
| NM_000599: insulinlike growth factor binding protein 5 (IGFBP5), mRNA | 3488 | 0.00201911 | 0.00619535 |
| NM_001855: collagen, type XV, α1 (COL15A1), mRNA | 1306 | 0.00201911 | 0.00778896 |
| NM_004363: carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5), mRNA | 1048 | 0.00201911 | 0.00834501 |
| Hs.169849: myosin binding protein C, slow type | 4604 | 0.00928793 | 0.00042598 |
| NM_003882: WNT1 inducible signaling pathway protein 1 (WISP1), transcript variant 1, mRNA | 8840 | 0.00928793 | 0.0004304 |
| Hs.172028: a disintegrin and metalloproteinase domain 10 | 102 | 0.00928793 | 0.00046155 |
| Hs.58324: a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) | 11096 | 0.00928793 | 0.00105437 |
| Hs.56255: keratin 6 irs4 | 121391 | 0.00928793 | 0.00155258 |
| Hs.443567: transitional epithelia response protein | 29914 | 0.00928793 | 0.00190983 |
| NM_001793: cadherin 3, type 1, P-cadherin (placental; CDH3), mRNA | 1001 | 0.00928793 | 0.00217983 |
| Hs.352156: a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 14 | 140766 | 0.00928793 | 0.00277865 |
| NM_015444: Ras-induced senescence 1 (RIS1), mRNA | 25907 | 0.00928793 | 0.00397939 |
| Hs.28005: transforming growth factor, β receptor I (activin A receptor type II–like kinase, 53 kD) | 7046 | 0.00928793 | 0.00403024 |
| NM_002273: keratin 8 (KRT8), mRNA | 3856 | 0.00928793 | 0.00463512 |
| NM_007159: sarcolemma-associated protein (SLMAP), mRNA | 7871 | 0.00928793 | 0.00467059 |
| NM_053025: myosin, light polypeptide kinase (MYLK), transcript variant 1, mRNA | 4638 | 0.00928793 | 0.00584138 |
| NM_000640: interleukin-13 receptor, α2 (IL13RA2), mRNA | 3598 | 0.00928793 | 0.00666183 |
| NM_012134: leiomodin 1 (smooth muscle; LMOD1), mRNA | 25802 | 0.00928793 | 0.00847851 |
| NM_000094: collagen, type VII, α1 (COL7A1), mRNA | 1294 | 0.00928793 | 0.00937536 |
| NM_001202: bone morphogenetic protein 4 (BMP4), transcript variant 1, mRNA | 652 | 0.00928793 | 0.00953431 |
Definition of abbreviation: TNoM = threshold number of misclassifications.
Figure 2.
The distribution of differentially expressed genes among functional categories. (A) Genes that had locus-link information and were significantly increased in IPF compared with HP. (B) Genes that were increased in HP compared with IPF. Note the large percentage of genes that belong to the distinct functional annotations. All enrichments are statistically significant (Fisher's exact score p value < 0.05).
TABLE 3.
UPREGULATED GENES IN HYPERSENSITIVITY PNEUMONITIS
| Gene Name | Locus-Link No. | TNoM p Value | t Test p Value |
|---|---|---|---|
| NM_005516: major histocompatibility complex, class I, E (HLA-E), mRNA | 3133 | 3.1063E-06 | 8.581E-08 |
| NM_002985: chemokine (C-C motif) ligand 5 (CCL5), mRNA | 6352 | 3.1063E-06 | 1.1854E-07 |
| Hs.351874: major histocompatibility complex, class II, DOα | 3111 | 3.1063E-06 | 2.0498E-07 |
| Hs.368409: major histocompatibility complex, class II, DP β1 | 3115 | 3.1063E-06 | 2.6687E-06 |
| NM_014387: linker for activation of T cells (LAT), mRNA | 27040 | 3.1063E-06 | 2.9103E-06 |
| NM_000732: CD3D antigen, δ polypeptide (TiT3 complex; CD3D), mRNA | 915 | 4.0382E-05 | 1.1558E-06 |
| NM_002209: integrin, αL (antigen CD11A [p180], lymphocyte function–associated antigen 1; α polypeptide; ITGAL), mRNA | 3683 | 4.0382E-05 | 1.3539E-06 |
| NM_002198: interferon regulatory factor 1 (IRF1), mRNA | 3659 | 4.0382E-05 | 2.2803E-06 |
| NM_005601: natural killer cell group 7 sequence (NKG7), mRNA | 4818 | 4.0382E-05 | 2.5394E-06 |
| NM_001767: CD2 antigen (p50), sheep red blood cell receptor (CD2), mRNA | 914 | 4.0382E-05 | 3.4159E-06 |
| NM_001558: interleukin-10 receptor, α (IL10RA), mRNA | 3587 | 4.0382E-05 | 1.0847E-05 |
| NM_006120: major histocompatibility complex, class II, DMα (HLA-DMA), mRNA | 3108 | 4.0382E-05 | 1.141E-05 |
| NM_002118: major histocompatibility complex, class II, DMβ (HLA-DMB), mRNA | 3109 | 4.0382E-05 | 1.3627E-05 |
| NM_000246: MHC class II transactivator (MHC2TA), mRNA | 4261 | 4.0382E-05 | 2.1963E-05 |
| NM_001109: a disintegrin and metalloproteinase domain 8 (ADAM8), mRNA | 101 | 4.0382E-05 | 3.0077E-05 |
| NM_000565: interleukin-6 receptor (IL6R), transcript variant 1, mRNA | 3570 | 4.0382E-05 | 3.5004E-05 |
| NM_004355: CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated; CD74) | 972 | 4.0382E-05 | 4.5692E-05 |
| NM_000206: interleukin-2 receptor, γ (severe combined immunodeficiency; IL2RG), mRNA | 3561 | 4.0382E-05 | 5.3729E-05 |
| NM_001778: CD48 antigen (B-cell membrane protein; CD48), mRNA | 962 | 4.0382E-05 | 0.00019398 |
| NM_002339: lymphocyte-specific protein 1 (LSP1), mRNA | 4046 | 4.0382E-05 | 0.0004061 |
| NM_006144: granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3; GZMA), mRNA | 3001 | 0.00033652 | 3.5408E-06 |
| NM_003874: CD84 antigen (leukocyte antigen; CD84), mRNA | 8832 | 0.00033652 | 2.1163E-05 |
| Hs.181244: major histocompatibility complex, class I, A | 3105 | 0.00033652 | 2.3716E-05 |
| NM_000265: neutrophil cytosolic factor 1 (47 kD, chronic granulomatous disease, autosomal 1; NCF1), mRNA | 4687 | 0.00033652 | 2.7125E-05 |
| NM_001768: CD8 antigen, α polypeptide (p32) (CD8A), transcript variant 1, mRNA | 925 | 0.00033652 | 3.6982E-05 |
| NM_004355: CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated; CD74) | 972 | 0.00033652 | 3.7304E-05 |
| Hs.387679: major histocompatibility complex, class II, DQ α1 | 3117 | 0.00033652 | 7.4311E-05 |
| Hs.387787: killer cell lectinlike receptor subfamily K, member 1 | 22914 | 0.00033652 | 0.00010199 |
| NM_003019: surfactant, pulmonary-associated protein D (SFTPD), mRNA | 6441 | 0.00033652 | 0.00015899 |
| NM_016326: chemokine-like factor (CKLF), transcript variant 3, mRNA | 51192 | 0.00033652 | 0.00027442 |
| NM_001242: tumor necrosis factor receptor superfamily, member 7 (TNFRSF7), mRNA | 939 | 0.00033652 | 0.00038993 |
| NM_012292: minor histocompatibility antigen HA-1 (HA-1), mRNA | 23526 | 0.00033652 | 0.00046328 |
| NM_000878: interleukin-2 receptor, β (IL2RB), mRNA | 3560 | 0.00033652 | 0.00531621 |
| NM_003879: CASP8 and FADD-like apoptosis regulator (CFLAR), mRNA | 8837 | 0.00033652 | 0.00648208 |
| NM_002984: chemokine (C-C motif) ligand 4 (CCL4), mRNA | 6351 | 0.00201911 | 6.3196E-06 |
| NM_002416: chemokine (C-X-C motif) ligand 9 (CXCL9), mRNA | 4283 | 0.00201911 | 5.4339E-05 |
| NM_004221: natural killer cell transcript 4 (NK4), mRNA | 9235 | 0.00201911 | 9.8112E-05 |
| NM_021181: SLAM family member 7 (SLAMF7), mRNA | 57823 | 0.00201911 | 0.00016428 |
| NM_000063: complement component 2 (C2), mRNA | 717 | 0.00201911 | 0.00024703 |
| NM_172369: complement component 1, q subcomponent, gamma polypeptide (C1QG), RNA | 714 | 0.00201911 | 0.00040263 |
| NM_003467: chemokine (C-X-C motif) receptor 4 (CXCR4), mRNA | 7852 | 0.00201911 | 0.00043211 |
| NM_001079: ζ-chain (TCR)–associated protein kinase 70 kD (ZAP70), mRNA | 7535 | 0.00201911 | 0.00061774 |
| NM_020125: SLAM family member 8 (SLAMF8), mRNA | 56833 | 0.00201911 | 0.00062281 |
| NM_001565: chemokine (C-X-C motif) ligand 10 (CXCL10), mRNA | 3627 | 0.00201911 | 0.00109636 |
| NM_004513: interleukin 16 (lymphocyte chemoattractant factor; IL16), transcript variant 1, RNA | 3603 | 0.00201911 | 0.00188205 |
| NM_005781: activated Cdc42-associated kinase 1 (ACK1), mRNA | 10188 | 0.00201911 | 0.00213368 |
| NM_007261: leukocyte membrane antigen (CMRF-35H), mRNA | 11314 | 0.00201911 | 0.00214496 |
| NM_153461: interleukin-17 receptor C (IL17RC), transcript variant 1, mRNA | 84818 | 0.00201911 | 0.0026955 |
| NM_000733: CD3E antigen, ɛ polypeptide (TiT3 complex; CD3E), mRNA | 916 | 0.00201911 | 0.00294939 |
| NM_021798: interleukin-21 receptor (IL21R), transcript variant 1, mRNA | 50615 | 0.00201911 | 0.003582 |
| NM_001953: endothelial cell growth factor 1 (platelet-derived; ECGF1), mRNA | 1890 | 0.00201911 | 0.00438273 |
| NM_000579: chemokine (C-C motif) receptor 5 (CCR5), mRNA | 1234 | 0.00928793 | 9.6163E-05 |
| NM_019111: major histocompatibility complex, class II, DR α (HLA-DRA), mRNA | 3122 | 0.00928793 | 0.00036044 |
| NM_003820: tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator; TNFRSF14), mRNA | 8764 | 0.00928793 | 0.00041457 |
| Hs.409934: major histocompatibility complex, class II, DQ β1 | 3119 | 0.00928793 | 0.00055793 |
| NM_006926: surfactant, pulmonary-associated protein A2 (SFTPA2), mRNA | 6436 | 0.00928793 | 0.0007712 |
| NM_015259: inducible T-cell costimulator ligand (ICOSL), mRNA | 23308 | 0.00928793 | 0.00281981 |
| NM_001774: CD37 antigen (CD37), mRNA | 951 | 0.00928793 | 0.00288764 |
| NM_001784: CD97 antigen (CD97), transcript variant 2, mRNA | 976 | 0.00928793 | 0.00541578 |
| NM_014312: cortical thymocyte receptor (X. laevis CTX) like (CTXL), mRNA | 23584 | 0.00928793 | 0.00621325 |
| NM_003810: tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10), mRNA | 8743 | 0.00928793 | 0.00811714 |
For definition of abbreviation, see Table 2.
IPF Can Be Distinguished from HP by the Expression of Proliferation, Tissue Remodeling, and Myofibroblast Genes
IPF lungs strongly express genes of a variety of extracellular matrix components, as well as muscle-specific genes and genes involved in cell motility and muscle contraction, suggesting significant activity in tissue remodeling and reorganization (Table 2). Likewise, many of these genes are implicated in epithelial cell adhesion/migration. Genes encoding extracellular matrix molecules that were highly expressed in IPF samples included many collagens (types III, IV, VII, XIV, XV, XVII, XVIII, and XXVII), tenascin N, versican, and asporin. Other extracellular matrix components increased in IPF lungs included a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif (ADAMTS)-14 and ADAMTS-5 (aggrecanase-2), and two MMPs, MMP-1 and MMP-7.
Several muscle-specific and actin-binding genes were upregulated, including myosin-binding protein C, myosin heavy polypeptide 11, tropomyosin 2, calponin 1, leiomodin 1, light polypeptide kinase of myosin, and α-actinin-2–associated LIM protein. Likewise, several other genes related to cell motility and muscle contraction were also increased.
Two genes involved in the Wnt pathway, the WNT1-inducible signaling pathway protein 1 (WISP-1) and the secreted frizzled-related protein 2, were increased in IPF lungs.
A number of genes implicated in tissue fibrogenesis were upregulated in IPF lungs when compared with HP. Among them, angiotensinogen was one of the most overexpressed. Likewise, the profibrotic cytokine osteopontin, IGFBP-4 and IGFBP-5, and the interleukin (IL)-13 receptor were increased.
Several epithelial-related genes were upregulated in IPF lungs. These include keratins 6 and 8, mucin 6, stratifin, which regulates cell growth and is also a tumor suppressor gene, and extracellular matrix protein 1, a gene located in chromosome 1q21 (close to the epidermal differentiation complex), which is believed to play an important role in epithelial cell differentiation (20). N-cadherin, a member of a large family of calcium-dependent cell adhesion molecules, was also increased. Taken together, these data indicate that IPF involves profound changes within epithelial cells of the lung and in the connective tissue, which together mediate morphologic restructuring of the lung.
Localization of IGFBP-4, N-Cadherin, and MMP-1 Immunoreactive Proteins in IPF Lungs
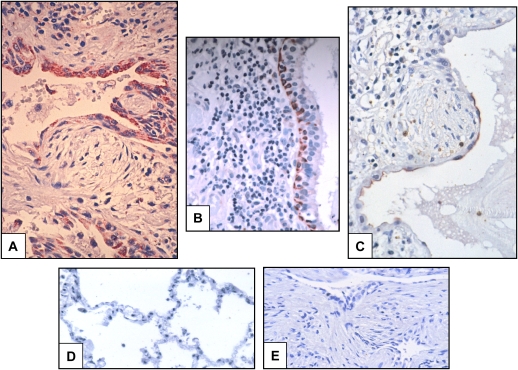
To establish whether the information obtained using gene expression profiling reflects protein expression, we selected three representative genes found elevated in the IPF lungs (Table 2). The cellular sources of N-cadherin, IGFBP-4, and MMP-1 were examined by immunohistochemistry on tissue sections of both IPF and control lungs. As shown in Figure 3, MMP-1 was detected in hyperplastic type 2 pneumocytes, whereas IGFBP-4 was found expressed by alveolar and bronchiolar (basal) epithelial cells. N-cadherin was detected in flattened alveolar epithelial cells covering fibroblast foci. MMP-1 was also revealed in macrophages in HP lungs (not shown). These findings strongly suggest that the gene expression levels correlate with protein expression in the tissues studied and support the notion that epithelial cells are highly active in IPF.
Figure 3.
Immunolocalization of insulinlike growth factor binding protein 4 (IGFBP-4), N-cadherin, and matrix metalloproteinase 1 (MMP-1) in IPF and normal lungs. (A) MMP-1 immunoreactive protein was found in alveolar epithelial cells. It can be noticed that the enzyme is virtually absent in fibroblastic foci (original magnification, 40×). (B) IGFBP-4 immunoreactive bronchiolar basal epithelial cells in IPF lungs (40×). (C) N-cadherin immunoreactive protein was observed in alveolar epithelial cells usually covering fibroblastic foci (40×). (D) Normal lungs were usually negative for the three molecules as exemplified for N-cadherin (40×). (E) A negative control omitting the primary (in this case, IGFBP-4) antibody (40×). Results illustrate the analysis of six IPF and five control lungs.
To explore this further, we quantified IGFBP-4 in BAL fluids of patients with IPF or HP as well as suitable control subjects. As shown in Figure 4, IGFBP-4 protein was undetectable in BAL from control individuals, and was significantly elevated in BAL from patients with IPF compared with patients with HP (2.84 ± 1.35 vs. 1.17 ± 0.33 μg/mg protein; p < 0.01).
Figure 4.
ELISA for IGFBP-4 protein in bronchoalveolar lavage (BAL) fluid samples. The protein was not detected in BAL from healthy individuals (controls). An increased concentration of IGFBP-4 was detected in BAL obtained from patients with IPF compared with patients with HP. The data represent the mean ± SD. For the IPF samples, p < 0.01.
HP Is Characterized by the Upregulation of Genes Related to T-Cell Activation and Immune Responses
In HP lungs, microarray analyses identified a variety of genes typically associated with inflammation, T-cell activation, and immune responses (Table 3). Genes related to T-lymphocyte activation included Src-like adaptor 2, CD2, components of the T-cell receptor complex (CD3-D, and CDR-E), and the α chain of CD8. Likewise, major histocompatibility complex (MHC) class II transactivator, the master regulator of MHC class II expression, and several genes encoding MHC class I and II molecules were upregulated.
Several chemokines were increased in HP, including CXCL9 and CXCL10. These recruit activated T and natural killer cells through the chemokine receptor CXCR3 and have been associated with T-helper type 1 (Th1) immune responses (21). CXCR4 and CCR5 and their ligands CCL5 and CCL4 were also upregulated, suggesting that the recruiting/homing program for lung lymphocytes involves multiple chemokines. Another increased gene was CD84, which is associated with follicular T-helper cells, a third major subset of nonpolarized effector T cells that provides help to B cells (22).
Also, several genes involved in host defense were overexpressed, including granzyme A, which reflects the presence of activated CD8 T lymphocytes (see below). Other immune genes increased in HP included several members of the tumor necrosis factor superfamily, IL-16 and several cytokine receptors, including IL-2R, IL-6R, IL-10R, IL-17R, and IL-21R.
FACS Analysis of BAL Cells Confirmed the Presence of Activated T Cells in HP but Not in IPF
Patients with HP showed a significant increase in total T lymphocytes as well as in the number of activated CD4+ and CD8+ T cells in the BAL when compared with BAL cells from patients with IPF (Table 4).
TABLE 4.
FLUORESCENCE-ACTIVATED CELL SORTER ANALYSIS OF LYMPHOCYTES OBTAINED BY BRONCHOALVEOLAR LAVAGE FROM PATIENTS WITH HYPERSENSITIVITY PNEUMONITIS OR IDIOPATHIC PULMONARY FIBROSIS
| HP (n = 7) | IPF (n = 9) | p Value | |
|---|---|---|---|
| Total cells, ×106 | 30.1 ± 14.6 | 10.8 ± 4.2 | < 0.01 |
| Lymphocytes, % | 59.9 ± 13.2 | 14.7 ± 7.9 | < 0.01 |
| Total CD3 T cells, ×106 | 21.7 ± 6.6 | 1.6 ± 0.7 | < 0.01 |
| Total CD4/CD69, ×106 | 10.1 ± 3.6 | 0.8 ± 0.4 | < 0.01 |
| Total CD8/CD69, ×106 | 7.3 ± 4.2 | 0.7 ± 0.4 | < 0.01 |
Definition of abbreviations: HP = hypersensitivity pneumonitis; IPF = idiopathic pulmonary fibrosis.
HP and IPF Gene Expression Signatures Identify NSIP Diagnostic Subgroups
We then analyzed the patients with NSIP in the context of the molecular signatures that distinguish IPF from HP. To avoid overfitting, we opted to use the larger set of genes (1,058 genes; TNoM p value, 0.002762; Figure 1B) with the minimal error rate for classification purposes. We used the 12 HP cases and the 15 UIP samples as the training set. The NSIP-like samples were classified according to them. Figure 5A shows the Infogram of these 1,058 genes. The test samples are in the middle, whereas IPF lungs are on the left and HP on the right. Samples were ranked by their classification score. Samples in the test set closest to IPF classify as IPF, whereas samples closest to HP classify as HP. The classification graph reveals that, in the training set, all IPF samples were correctly classified as IPF, whereas one HP sample was classified as IPF. In the test set, one NSIP sample was classified as HP (Figure 5B). This patient had a history of previous exposure to home birds, a hobby common in Mexico (where the samples were collected), but without cause–effect relationship, positive precipitins, or apparent exposure to other organic particles. Two other NSIP samples were classified as IPF, supporting the notion that fibrotic NSIP may be undistinguishable from UIP. The other five NSIP samples did not classify as either IPF or HP, suggesting that they indeed represent true idiopathic NSIP. Four of these cases were evaluated by a panel of experts (American Thoracic Society workshop on NSIP) and classified as definite (three cases) or probable (one case) NSIP.
Figure 5.
Infogram and classification score. (A) Infogram of 1,058 genes that made the least classification errors by LOOCV. The expression levels for each gene were normalized to the geometric mean of all the samples for each gene. Increased genes are shown in progressively brighter shades of yellow, and decreased genes are shown in progressively darker shades of blue. Genes shown in gray are not different between the groups. The genes were ranked according to their significance level. (B) Classification score of every sample in the dataset. Samples below the zero line were considered as usual interstitial pneumonia. Samples above the separation line were classified as HP. Note that only one HP sample was misclassified. NSIP = nonspecific interstitial pneumonia.
DISCUSSION
This study used microarrays to identify disease-specific gene expression signatures and to directly compare gene expression profiles from various ILDs of known and unknown etiology on a genomewide scale. Our results demonstrate that gene expression patterns clearly distinguish IPF from HP, and indicate that these disease-specific gene expression signatures are biologically meaningful and provide significant information on the mechanisms underlying these diseases.
IPF is a specific and progressive form of chronic interstitial pneumonia limited to the lung and characterized by epithelial cell injury and activation followed by a fibroproliferative response and subsequent scarring with only mild to moderate signs of inflammation. However, this disease has generally been considered a chronic, insidious, inflammatory disease, and consequently, research in this disorder has primarily focused on its inflammatory component and, more recently, on its fibroproliferative component.
Our findings indicate that IPF lungs do not exhibit a “typical” inflammatory pattern (characterized by leukocyte/lymphocyte genes, and mediators such as cytokines). In this study, only CCL24 was found to be significantly upregulated when compared with HP lungs. In a previous study, CCL7 was found to be upregulated in IPF in comparison with other forms of IIPs (23). These differences may reflect different sensitivities of the gene array platforms used.
By contrast, it is precisely the absence of many of these proinflammatory lymphoid/leukocyte-associated genes that distinguishes IPF from HP. Instead, IPF is characterized by the expression of genes that indicate an active tissue remodeling program, including extracellular matrix and basement membrane components as well as by a large number of myofibroblast/smooth muscle cell–associated and epithelial cell–related genes. Thus, several fibrillar and nonfibrillar collagens were increased in IPF. One of these is type VII collagen, the major structural component of the anchoring fibrils at the dermal–epidermal junction in the skin (24). Interestingly, these anchoring fibrils have been described in fibrotic lungs where their presence seems to correlate with the severity of the fibrosis (25). Other upregulated collagens included type XV and type XVIII, which are expressed in specialized basement membranes, and their C-terminal parts, endostatin and restin, have been reported to have a strong antiangiogenic effect (26). Another overexpressed gene is transmembrane collagen XVII, an epithelial adhesion molecule that belongs to the group of collagenous transmembrane proteins, which has been recently implicated in the regulation of keratinocyte migration (27). Interestingly, collagen XXVII, a new member of the vertebrate fibrillar collagen family, was also upregulated. This peculiar collagen is expressed by a variety of epithelial cell layers in developing tissues, including lung (28).
MMP-1 and MMP-7 were also upregulated. As previously shown, MMP-7 seems to play an important role in the pathogenesis of IPF (29).
Angiotensinogen, which is produced by fibroblasts from fibrotic but not from normal tissues (30, 31), was strongly upregulated, supporting the notion that this mediator may be important in the pathogenesis of the disease. Angiotensin has been implicated not only in profibrotic activity as a result of induction of extracellular matrix accumulation but also in alveolar epithelial cell apoptosis, an event considered one of the earliest alterations in IPF (9).
To confirm the RNA profiling analyses, we selected three genes overexpressed in IPF for additional studies at the protein level, and all of them were mainly localized in epithelial cells. IGFBP-4 belongs to a family of at least six specific high-affinity IGF binding proteins that modulate the local bioavailability of IGF-1, a fibroblast mitogen and stimulator of collagen synthesis by fibroblasts, which may also contribute to the fibrogenic process (32, 33). IGFBP-5 gene expression was also upregulated, and it has been recently reported that this protein together with IGFBP-3 are increased in IPF lungs and localized to fibroblasts surrounding airspaces, and to epithelial cells in bronchioles and alveoli (34). The functional roles of IGFBP are poorly understood. They usually inhibit IGF-1 effects by blocking binding to its receptor, but they can also enhance IGF-stimulated cell growth (35, 36). Moreover, it has been suggested that IGFBP-3 may increase the bioactivity of IGF-1, a process that may contribute to lung fibrosis (37). More recently, it was found that IGFBP-3 and IGFBP-5 induce extracellular matrix production by normal lung fibroblasts (34).
Another important finding in IPF lungs was the overexpression of N-cadherin, a mesenchymal cadherin preferentially expressed in migratory cells and in connective tissue. In our study, this molecule was localized in flattened and attenuated alveolar epithelial cells usually overlaying fibroblastic foci and occasionally in hyperplastic type 2 pneumocytes. N-cadherin belongs to a family of calcium-dependent cell adhesion molecules that mediate cell–cell adhesion and also modulate cell migration and tumor invasiveness, thus showing the opposite effect of E-cadherin (38). Actually, it has been hypothesized that a cadherin switch (i.e., a change from E- to N-cadherin expression), is involved not only in the detachment and migration of epithelial cells during embryonic development but also in the transition from a benign to an invasive, malignant tumor phenotype (38, 39). Interestingly, WISP-1 and the secreted frizzled-related protein 2 were also increased in IPF (40). WISP-1 belongs to a recently described connective tissue growth factor/cysteine-rich 61/nephroblastoma (CCN) family of multifunctional signaling regulators that comprises six cysteine-rich proteins, including connective tissue growth factor (41), and is strongly increased in aggressive fibromatosis (42). WISP-1 can activate the antiapoptotic Akt/protein kinase B signaling pathway protecting cells from p53-dependent cell death (43). Collagen type XVIII, another gene overexpressed in IPF lungs, has been recently shown to colocalize with Wnt molecules, and its frizzled-containing N-terminal fragment may be implicated in Wnt signaling (44). In addition, several target genes of the canonic WNT/β-catenin pathway, such as MMP-7 and osteopontin, were also increased in IPF lungs, further supporting the concept that this pathway may be aberrantly induced in IPF but not in HP. Moreover, we have recently demonstrated that, in IPF, osteopontin colocalizes with MMP-7 in alveolar epithelial cells, and application of weakest-link statistical models to microarray data suggests a significant interaction between both molecules (45).
The functional enrichment of IPF lungs with genes associated generally with development and cellular differentiation strongly suggests that a global process involving a change in cellular phenotypes occurs in IPF but not in HP. Such a process may contribute to a transition of epithelial cells to a mesenchymal phenotype. Recently, while this article was under review, findings suggesting an epithelial–mesenchymal transition in IPF have been reported (46).
Our findings on IPF gene expression contrast strongly with those of HP. In the latter disease, the main finding is a strong T-cell response that reflects a strong proinflammatory mechanism. Importantly, similar findings have been made in some chronic cases in which septal fibrosis was present in the biopsy. Taken together, these results support the existence of at least two different routes for developing diffuse pulmonary fibrosis. One of them is through an inflammatory pathway represented by HP, where there is an early, clearly defined phase of alveolitis, which may be followed by a late fibrotic phase. The second is represented by IPF (9) and may be mainly an epithelial/fibroblastic pathway, which may not include a “typical” inflammatory phase.
HP lungs showed a strong inflammatory component, with many genes of infiltrating activated T cells, including cytotoxic T lymphocytes and natural killer cells. The presence of activated CD8 T cells strongly suggests a viral component in this disease, a possibility that has been suggested previously (47, 48). Although the factors that trigger the onset of HP have not been defined yet, genetic susceptibility associated with the MHC (49) and/or viral infections may participate. Common respiratory viruses are often present in the lower airways of patients with HP, and viral infection strongly augments both the early and late inflammatory responses in experimental HP (47, 48). Interestingly, CCL5, which is commonly induced by acute respiratory viral infection, was also upregulated in the HP lungs (19, 50). In addition, CCL5 has been found expressed in the cell membranes of granuloma cells from noncaseating granulomas, which also characterize HP (51). On the other hand, the upregulation of the chemokines CXCL9 and CXCL10, known to be IFN-γ− inducible and ligands of CXCR3, supports the hypothesis that HP is mediated by a Th1-like cell-mediated host response.
The term “nonspecific interstitial pneumonia” is currently applied for lung biopsy specimens from some patients with IIPs that do not fit into any well-defined histopathologic pattern. It is important to emphasize that NSIP was originally described by the absence of morphologic characteristics of other IIPs rather than by exhibiting distinguishing features of its own. Although NSIP is often idiopathic, the same morphologic pattern has been associated with a number of other diseases, including collagen vascular diseases, drug-induced pneumonitis, and importantly, HP (2, 52, 53). Therefore, finding an NSIP pattern should prompt the clinician to carefully consider potentially causative exposure or the presence of a systemic disease. In general, idiopathic NSIP represents a problem for the clinician because its clinical features have been poorly described and thus there is as yet no distinctive clinical description for these patients. Morphologically, two subtypes of NSIP are recognized: cellular and fibrotic. Differential diagnosis of cellular NSIP includes HP, and differential diagnosis of fibrotic NSIP includes primarily UIP but also fibrotic forms of other types of ILDs, including advanced forms of HP (54). Our findings indicating that the transcriptional pattern of three of the eight NSIP-like cases behaves as UIP or HP support this notion.
The most important obstacle in the differential diagnosis of IPF/UIP is NSIP, which differs clinically by a more favorable response to corticosteroid therapy and a better prognosis (51, 55, 56). When fibrosis is prominent in NSIP, the changes can usually be difficult to distinguish from UIP. In addition, areas resembling NSIP are present in many UIP cases in both biopsy and explant specimens, and they are extensive in some of them (57). These findings suggest that an NSIP-like pattern may be found even randomly in the biopsy specimens of many patients with IPF/UIP, making the identification of the latter problematic. Supporting this point of view, it has been demonstrated that patients with IPF may exhibit histologic variability in their lung specimens when biopsy samples are taken from multiple lobes (58, 59). Interestingly, patients with discordant UIP—that is, UIP morphology in one part of the lung and NSIP in the other—show survival, clinical, and physiologic features similar to those with concordant UIP. Moreover, the prognosis in both patients with concordant and discordant UIP is significantly worse than that of the patients with concordant NSIP (NSIP in multiple biopsies).
Although our study includes the largest number of human samples reported in microarray analysis of IPF (or any other nonmalignant lung disease), the sample size still prevents generalization of our results. However, given the statistical magnitude of the differences between IPF and HP, the magnitude and statistical significance of the difference in functional groups enriched within each disease gene expression signature, and the biological plausibility of the results, it is unlikely that larger numbers of samples will change our conclusions significantly. Although, the results of this study should be tested on other independent datasets, the identification of statistically significant, biologically meaningful, disease-specific gene expression signatures suggests that we could use these signatures to predict whether patients would respond to immunosuppressive therapy or to future specific antifibrotic therapies. Currently, it is recommended (with little scientific support) that a therapeutic trial with corticosteroids and some immunosuppressive drugs would be administered to any patient with IPF/UIP and definitely to patients with the other more traditional inflammatory ILDs. Our data support the view that gene expression signatures can be a better predictor of patient responsiveness to antiinflammatory therapy, thus preventing the side effects of an unindicated therapeutic trial from other patients. In addition, when specific antifibrotic therapies may be ready to be tested in clinical trials in the near future, the best way to select patients may be to use their tissue functional gene expression signatures.
In summary, genomewide gene expression profiling of individual IPF, HP, and NSIP lungs has allowed the identification of genetic signatures for these diseases. This may speed up the understanding of the molecular mechanisms underlying these disorders and may also potentially aid in their differential diagnosis. Advances in such diagnoses could potentially be invaluable to, first, distinguish HP from IPF, and second, to provide a more accurate diagnosis in cases currently diagnosed as NSIP.
Supplementary Material
Acknowledgments
The authors thank Jennifer Kane for technical support.
Supported in part by National Institutes of Health grant HL 073745-01, and by a generous donation from the Simmons family (N.K.), and by DGAPA, UNAM IN215003 (A.P.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200504-644OC on September 15, 2005
Conflict of Interest Statement: M.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.E. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.R.W. was an employee of Eos Biotechnology from November 1998–February 2003. She has filed patents on the lung fibrosis microarray data, and owns shares in Protein Design Labs. K.W. is currently employed by and has stock and stock options in Protein Design Labs, which is the parent company of Eos Biotechnology, an original sponsor of the research. N.A. was an employee of Eos Biotechnology from January 2000–March 2003. She has filed patents on the lung fibrosis microarray data, and owns shares in Protein Design Labs. N.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.Z. was an employee of Eos Biotechnology from September 2000–March 2003. He owns shares in Protein Design Labs, a company that acquired Eos Biotechnology. A provisional patent application was filed by Eos Biotechnology on some data contained in this article. He is currently employed by Neurocrine Biosciences, which has no financial interests in the contents of this article.
References
- 1.Schwarz ML, King TE, Raghu G. Approach to the evaluation and diagnosis of interstitial lung disease. In: King TE, Schwarz ML, editors. Interstitial lung disease. Ontario, BC, Canada: B.C. Decker; 2003. pp. 1–30.
- 2.American Thoracic Society/European Respiratory Society. American Thoracic Society/European Respiratory Society international multidisciplinary classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med 2002;165:277–304. [DOI] [PubMed] [Google Scholar]
- 3.Katzenstein ALA, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med 1998;157:1301–1315. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson AG, Colby TV, du Bois RM, Hansell DM, Wells AU. The prognostic significance of the histologic pattern of interstitial pneumonia in patients presenting with the clinical entity of cryptogenic fibrosing alveolitis. Am J Respir Crit Care Med 2000;162:2213–2217. [DOI] [PubMed] [Google Scholar]
- 5.Patel AM, Ryu JH, Reed CE. Hypersensitivity pneumonitis: current concepts and future questions. J Allergy Clin Immunol 2001;108:661–670. [DOI] [PubMed] [Google Scholar]
- 6.Ramírez-Venegas A, Sansores RH, Pérez Padilla R, Carrillo G, Selman M. Utility of a provocation test for diagnosis of chronic pigeon breedeŕs disease. Am J Respir Crit Care Med 1998;158:862–869. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Padilla R, Salas J, Chapela R, Sánchez M, Carrillo G, Pérez R, Sansores R, Gaxiola M, Selman M. Mortality in Mexican patients with chronic pigeon breeders lung compared to those with usual interstitial pneumonia. Am Rev Respir Dis 1993;148:49–53. [DOI] [PubMed] [Google Scholar]
- 8.Selman M, King TE, Pardo A. Idiopathic pulmonary fibrosis: prevailing and evolving hypothesis about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:134–136. [DOI] [PubMed] [Google Scholar]
- 9.Pardo A, Selman M. Molecular mechanisms of pulmonary fibrosis. Front Biosci 2002;7:d1743–d1761. [DOI] [PubMed] [Google Scholar]
- 10.Pardo A, Barrios R, Gaxiola M, Segura-Valdez L, Carrillo G, Estrada A, Mejia M, Selman M. Increase of lung neutrophils in hypersensitivity pneumonitis is associated with lung fibrosis. Am J Respir Crit Care Med 2000;161:1698–1704. [DOI] [PubMed] [Google Scholar]
- 11.American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS) and the European Respiratory Society (ERS). Am J Respir Crit Care Med 2000;161:646–664. [DOI] [PubMed] [Google Scholar]
- 12.Henshall SM, Afar DE, Hiller J, Horvath LG, Quinn DI, Rasiah KK, Gish K, Willhite D, Kench JG, Gardiner-Garden M, et al. Survival analysis of genome-wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Res 2003;63:4196–4203. [PubMed] [Google Scholar]
- 13.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol 2002;27:125–132. [DOI] [PubMed] [Google Scholar]
- 14.Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M. Blood transcriptional signatures of multiple sclerosis: unique gene expression of disease activity. Ann Neurol 2004;55:410–417. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol 2000;7:559–583. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
- 17.Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [PubMed] [Google Scholar]
- 19.Selman M. Hypersensitivity pneumonitis: a multifaceted deceiving disorder. Clin Chest Med 2004;25:531–547. [DOI] [PubMed] [Google Scholar]
- 20.Smits P, Poumay Y, Karperien M, Tylzanowski P, Wauters J, Huylebroeck D, Ponec M, Merregaert J. Differentiation-dependent alternative splicing and expression of the extracellular matrix protein 1 gene in human keratinocytes. J Invest Dermatol 2000;114:718–724. [DOI] [PubMed] [Google Scholar]
- 21.Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol 2001;2:102–107. [DOI] [PubMed] [Google Scholar]
- 22.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol 2004;173:68–78. [DOI] [PubMed] [Google Scholar]
- 23.Choi ES, Jakubzick C, Carpenter KJ, Kunkel SL, Evanoff H, Martinez FJ, Flaherty KR, Toews GB, Colby TV, Kazerooni EA, et al. Enhanced monocyte chemoattractant protein-3/CC chemokine ligand-7 in usual interstitial pneumonia. Am J Respir Crit Care Med 2004;170:508–515. [DOI] [PubMed] [Google Scholar]
- 24.Rattenholl A, Pappano WN, Koch M, Keene DR, Kadler KE, Sasaki T, Timpl R, Burgeson RE, Greenspan DS, Bruckner-Tuderman L. Proteinases of the bone morphogenetic protein-1 family convert procollagen VII to mature anchoring fibril collagen. J Biol Chem 2002;277:26372–26378. [DOI] [PubMed] [Google Scholar]
- 25.Kawanami O, Ferrans VJ, Roberts WC, Crystal RG, Fulmer JD. Anchoring fibrils: a new connective tissue structure in fibrotic lung disease. Am J Pathol 1987;92:389–410. [PMC free article] [PubMed] [Google Scholar]
- 26.Tomono Y, Natio I, Ando K, Yonezawa T, Sado Y, Hirakawa S, Arata J, Okigaki T, Ninomiya Y. Epitope-defined,onoclonal antibodies against multiplexin collagens demonstrate that type XV and XVIII collagens are expressed in specialized basement membranes. Cell Struct Funct 2002;27:9–20. [DOI] [PubMed] [Google Scholar]
- 27.Tasanen K, Tunggal L, Chometon G, Bruckner-Tuderman L, Aumailley M. Keratinocytes from patients lacking collagen XVII display a migratory phenotype. Am J Pathol 2004;164:2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boot-Handford RP, Tuckwell DS, Plumb DA, Rock CF, Poulsom R. A novel and highly conserved collagen (pro(alpha)1(XXVII)) with a unique expression pattern and unusual molecular characteristics establishes a new clade within the vertebrate fibrillar collagen family. J Biol Chem 2003;278:31067–31077. [DOI] [PubMed] [Google Scholar]
- 29.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA 2002;99:6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Ramos C, Joshi I, Zagariya A, Pardo A, Selman M, Uhal BD. Human lung myofibroblast-derived inducers of alveolar epithelial apoptosis identified as angiotensin peptides. Am J Physiol 1999;277:L1158–L1164. [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi Y, Takagi K, Hara M, Fukasawa C, Sugiura T, Nishimagi E, Harigai M, Kamatani N. Angiotensin II in the lesional skin of systemic sclerosis patients contributes to tissue fibrosis via angiotensin II type 1 receptors. Arthritis Rheum 2004;50:216–226. [DOI] [PubMed] [Google Scholar]
- 32.Bonner JC, Brody AR. Cytokine-binding proteins in the lung. Am J Physiol 1995;268:L869–L878. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein RH, Poliks CF. Stimulation of collagen formation by insulin and insulin-like growth factor I in cultures by human lung fibroblasts. Endocrinology 1989;124:964–970. [DOI] [PubMed] [Google Scholar]
- 34.Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol 2005;166:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 1995;16:3–34. [DOI] [PubMed] [Google Scholar]
- 36.Hamon GA, Hunt TK, Spencer EM. In vivo effects of systemic insuline-like growth factor-I alone and complexed with insulin-like growth factor binding protein-3 on corticosteroid suppressed wounds. Growth Regul 1993;3:53–56. [PubMed] [Google Scholar]
- 37.Aston C, Jagirdar J, Lee TC, Hur T, Hintz RL, Rom WN. Enhanced insulin-like growth factor molecules in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 1995;151:1597–1603. [DOI] [PubMed] [Google Scholar]
- 38.Christofori G. Changing neighbours, changing behaviour: cell adhesion molecule-mediated signalling during tumour progression. EMBO J 2003;22:2318–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol 2000;148:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh JC. Specificity of WNT-receptor interactions. Front Biosci 2004;9:1333–1338. [DOI] [PubMed] [Google Scholar]
- 41.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet 2004;3:363:62–64. [DOI] [PubMed] [Google Scholar]
- 42.Skubitz KM, Skubitz AP. Gene expression in aggressive fibromatosis. J Lab Clin Med 2004;143:89–98. [DOI] [PubMed] [Google Scholar]
- 43.Su F, Overholtzer M, Besser D, Levine AJ. WISP-1 attenuates p53-mediated apoptosis in response to DNA damage through activation of the Akt kinase. Genes Dev 2002;16:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elamaa H, Snellman A, Rehn M, Autio-Harmainen H, Pihlajaniemi T. Characterization of the human type XVIII collagen gene and proteolytic processing and tissue location of the variant containing a frizzled motif. Matrix Biol 2003;22:427–442. [DOI] [PubMed] [Google Scholar]
- 45.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dakhama A, Hegele RG, Laflamme G, Israel-Assayag E, Cormier Y. Common respiratory viruses in lower airways of patients with acute hypersensitivity pneumonitis. Am J Respir Crit Care Med 1999;159:1316–1322. [DOI] [PubMed] [Google Scholar]
- 48.Gudmundsson G, Monick MM, Hunninghake GW. Viral infection modulates expression of hypersensitivity pneumonitis. J Immunol 1999;162:7397–7401. [PubMed] [Google Scholar]
- 49.Camarena A, Juarez A, Mejia M, Estrada A, Carrillo G, Falfan R, Zuñiga J, Navarro C, Granados J, Selman M. Major histocompatibility complex and TNF-α gene polymorphisms in pigeon breeder's disease. Am J Respir Crit Care Med 2001;163:1528–1533. [DOI] [PubMed] [Google Scholar]
- 50.Glass WG, Rosemberg HF, Murphy PM. Chemokine regulation of inflammation during acute viral infection. Curr Opin Allergy Clin Immunol 2003;3:467–473. [DOI] [PubMed] [Google Scholar]
- 51.Oki M, Ohtani H, Kinouchi Y, Sato E, Nakamura S, Matsumoto T, Nagura H, Yoshie O, Shimosegawa T. Accumulation of CCR5+ T cells around RANTES+ granulomas in Crohn's disease: a pivotal site of Th1-shifted immune response? Lab Invest 2005;85:137–145. [DOI] [PubMed] [Google Scholar]
- 52.Travis WD, Matsui K, Moss J, Ferrans VJ. Idiopathic nonspecific interstitial pneumonia: prognostic significance of cellular and fibrosing patterns: survival comparison with usual interstitial pneumonia and desquamative interstitial pneumonia. Am J Surg Pathol 2000;24:19–33. [DOI] [PubMed] [Google Scholar]
- 53.Arakawa H, Yamada H, Kurihara Y, Nakajima Y, Takeda A, Fukushima Y, Fujioka M. Nonspecific interstitial pneumonia associated with polymyositis and dermatomyositis: serial high-resolution CT findings and functional correlation. Chest 2003;123:1096–1103. [DOI] [PubMed] [Google Scholar]
- 54.Katzenstein ALA, Fiorelli RF. Nonspecific interstitial pneumonia/ fibrosis: histologic features and clinical significance. Am J Surg Pathol 1994;18:136–147. [PubMed] [Google Scholar]
- 55.Bjoraker JA, Ryu JH, Edwin MK, Myers JL, Tazelaar HD, Schroeder DR, Offord KP. Prognostic significance of histopathologic subsets in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 1998;157:199–203. [DOI] [PubMed] [Google Scholar]
- 56.Daniil ZD, Gilchrist FC, Nicholson AG, Hansell DM, Harris J, Colby TV, du Bois RM. A histologic pattern of nonspecific interstitial pneumonia is associated with a better prognosis than usual interstitial pneumonia in patients with cryptogenic fibrosing alveolitis. Am J Respir Crit Care Med 1999;160:899–905. [DOI] [PubMed] [Google Scholar]
- 57.Katzenstein AL, Zisman DA, Litzky LA, Nguyen BT, Kotloff RM. Usual interstitial pneumonia: histologic study of biopsy and explant specimens. Am J Surg Pathol 2002;26:1567–1577. [DOI] [PubMed] [Google Scholar]
- 58.Flaherty KR, Travis WD, Colby TV, Toews GB, Kazerooni EA, Gross BH, Jain A, Strawderman RL, Flint A, Lynch JP, et al. Histopathologic variability in usual and nonspecific interstitial pneumonias. Am J Respir Crit Care Med 2001;164:1722–1727. [DOI] [PubMed] [Google Scholar]
- 59.Monaghan H, Wells AU, Colby TV, du Bois RM, Hansell DM, Nicholson AG. Prognostic implications of histologic patterns in multiple surgical lung biopsies from patients with idiopathic interstitial pneumonias. Chest 2004;125:522–526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.