Summary
PKR (protein kinase R) is a key component of the interferon antiviral defense pathway. Upon binding dsRNA, PKR undergoes autophosphorylation reactions that activate the kinase. PKR contains an N-terminal double-stranded RNA binding domain (dsRBD), which consists of two tandem dsRNA binding motifs, and a C-terminal kinase domain. We have used small angle X-ray and neutron scattering to define the conformation of latent PKR in solution. Guinier analysis indicates a radius of gyration of about 35 Å. The p(r) distance distribution function exhibits a peak near 30 Å with a broad shoulder extending to longer distances. Good fits to the scattering data require models that incorporate multiple compact and extended conformations of the two interdomain linker regions. Thus, PKR belongs to the growing family of proteins that contain intrinsically unstructured regions. We propose that the flexible linkers may allow PKR to productively dimerize upon interaction with RNA activators that have diverse structures.
Keywords: SAXS, SANS, protein kinase, innate immunity
Introduction
Protein kinase R (PKR) is an interferon-induced kinase that plays a key role in the innate immunity response to viral infection.1;2 The enzyme is synthesized in a latent form but it is activated by binding dsRNA to undergo autophosphorylation. The most well characterized cellular substrate of PKR is the alpha subunit of eukaryotic initiation factor eIF2. Phosphorylation of eIF2α inhibits protein synthesis in virally-infected cells. Thus, production of dsRNA during viral infection3 results in PKR activation and subsequent inhibition of viral and host protein synthesis. PKR also functions in the control of cell growth and proliferation and as a tumor suppressor protein 4;5
PKR is comprised of an N-terminal double-stranded RNA binding domain (dsRBD) and C-terminal kinase domain, with a central region of unknown function. The dsRBD consists of two tandem copies of the ~70 amino-acid dsRNA binding motif. In the NMR structure of the PKR dsRBD, each motif adopts the canonical αβββα fold.6 The folds of the two domains are quite similar, with a backbone RMSD of 2Å.6 In the X-ray structure of a complex of the PKR kinase domain with eIF2α, the catalytic domain has a typical protein kinase fold consisting of an N-terminal lobe that is mostly β sheet and a C-terminal lobe that is predominantly helical.7
The mechanism of PKR activation by dsRNA has intrigued researchers for decades and several models have been proposed.8 In the autoinhibition model, the latent enzyme exists in a closed conformation mediated by interaction of the dsRBD with the kinase domain that blocks substrate access. Binding of dsRNA activates PKR by inducing a conformational change that relieves the latent enzyme of inhibition. It has long been known that PKR is capable of dimerizing in the absence and the presence of dsRNA 9–13 and recent structural and biophysical data indicate a critical role for PKR dimerization in the activation process. The PKR kinase-eIF2α complex crystallizes as a back-to-back dimer with the interface formed by the kinase N-terminal lobes.7 Dimerization is sufficient to activate PKR in the absence of dsRNA.11 Fusion of a heterologous dimerization domain with the PKR kinase domain enhances autophosphorylation.14;15 Sedimentation velocity analysis of PKR binding to short dsRNAs supports an activation mechanism where the role of the dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization via the kinase domain.16 Hybrid autoinhibition/dimerization models have also been proposed where dsRNA binding induces a conformation change in PKR that leads to protein dimerization and activation.17;18
It is critical to develop structural models of the PKR holoenzyme in both inactive and active states to define the mechanism of activation. The existing structures of the dsRBD6;19 and kinase domain7 are insufficient to define the holoenzyme structure, how the domains interact, their relative orientations and how RNA binding is linked to productive dimerization and activation. The two dsRNA binding motifs are connected by an unstructured linker of ~ 20 amino acids.6 Furthermore, PKR contains a large (~90 residue) region between the dsRBD and kinase domain of unknown structure. This sequence has the signatures of an intrinsically disordered region (Figure 1).20;21 It has low sequence complexity with a high content of uncharged, polar residues and is particularly enriched in serines (22 residues). In NMR studies, this region appears unstructured or dynamic in TROSY-HSQC spectra.18 AFM images of PKR reveal up to three distinguishable structured regions linked by bridge-like stretches that adopt multiple structures.22 These results suggest that PKR may be a flexible protein that likely contains intrinsically disordered regions. Structural characterization of flexible multidomain proteins presents significant challenges for conventional X-ray diffraction and NMR approaches. Small angle scattering X-ray and neutron scattering (SAXS and SANS) provide low - resolution structural information on macromolecules in solution.23–25 When combined with high-resolution domain structures, small angle scattering data provide constraints that allow detailed structural modeling of multiprotein complexes or multidomain proteins. These approaches are being increasingly applied to flexible proteins that contain disordered segments.24;26–34 The experimental scattering curves from such systems comprise an ensemble – and time – average of the contributions from conformations present in solution. Recently, an approach was developed for structural analysis of flexible systems that uses a genetic algorithm to select an ensemble of structural models that best fit the experimental scattering data from a pool of randomly-generated structures.26
Figure 1.
Sequence alignment of PKR linker. The sequence of the second linker region in human PKR (residues 169–257) was aligned with the corresponding regions in R. norvegicus and M. auratus enzymes. The residues are colored as follows: acidic, blue; basic, red; polar uncharged, green; nonpolar, grey.
Here, we have used SAXS and SANS to define the conformation of latent PKR. It is useful to employ both methods. In SAXS, the high flux obtainable at synchrotron sources results in high precision scattering data but also may induce radiation damage. Although the flux of cold neutrons available at reactor sources is lower, an advantage of SANS is the absence of radiation damage. The scattering data and the p(r) distance distributions derived from the scattering data indicate that PKR adopts a range of compact and extended conformations. Using the coordinates of the dsRNA binding motifs and kinase domain, we refined structural models of the intact enzyme against the scattering data. The data do not fit well to structures obtained by rigid body modeling of the folded domains coupled with addition of the linkers. Good fits are obtained using models where flexibility of the linkers is accommodated by allowing for the contributions of multiple conformations to the experimental scattering profiles, indicating that PKR contains natively unstructured regions and adopts compact and extended conformations.
Results
Small angle scattering measurements require homogenous preparations and samples for SAXS and SANS analysis of PKR monomer were prepared to minimize contributions from dimer and higher aggregates. Our previous sedimentation equilibrium and velocity measurements have demonstrated that PKR exists as a homogeneous monomer at lower protein concentration, and at higher concentrations we detected a weak monomer - dimer equilibrium with Kd ~ 450 μM.11 Thus, scattering data were recorded at multiple PKR concentrations and detailed structural analysis was limited to low-concentration samples that contained the least amount of dimer. Higher aggregates were removed by gel filtration and subsequent ultrafiltration through 0.02 μm membranes immediately prior to analysis. Dynamic light scattering confirmed that the samples were free of higher molecular weight contaminants with a hydrodynamic radius consistent with a PKR monomer (data not shown).
Model-Independent Analysis
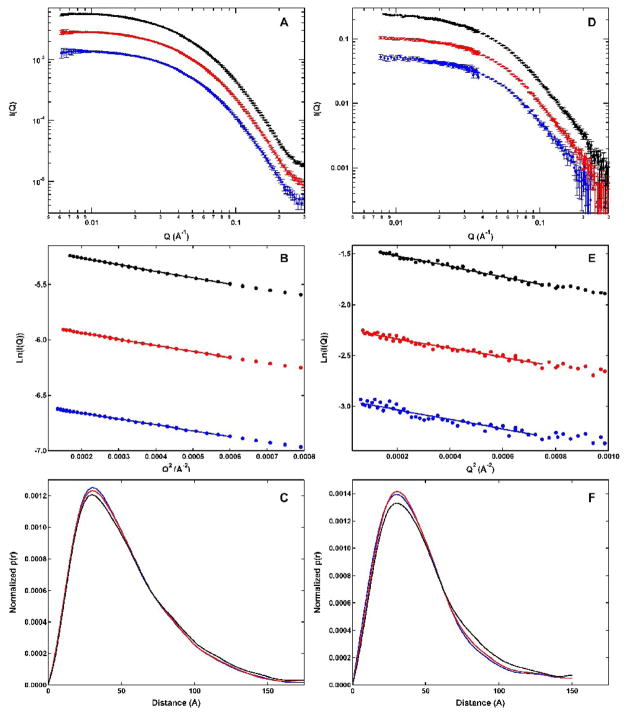
Figure 2A shows SAXS scattering profiles obtained over a concentration range of 1 to 4 mg/mL. The scattering curves are flat in the low Q range, confirming the absence of higher aggregates. The profiles obtained at different protein concentrations have the same shape, indicating that there is no dramatic increase in self-association with loading concentration over the range examined. Guinier analysis was used to quantitatively assess the effect of protein concentration on PKR association state and to define the radius of gyration (Rg). Figure 1B shows Guinier plots of the same SAXS datasets depicted in Figure 1A. The plots are linear over the Guinier region at low Q where Rg•Q < 1. Table 1 shows that values of Rg derived from linear fits to the Guinier plots are slightly concentration dependent, decreasing from 42.03 ± 0.16 Å at 4 mg/mL to 40.74 ± 0.29 Å at 2 mg/mL PKR, but then hardly decreasing to 39.92 Å ± 0.53 Å at 1 mg/mL. These results indicate that self-association of PKR is detectable above 2 mg/mL but, within error, the Rg values are concentration-independent at lower concentrations.
Figure 2.
Small angle scattering analysis of PKR. Protein concentrations: 1 mg/mL (blue), 2 mg/mL (red) and 4 mg/mL (black). A) SAXS scattering profiles. B) SAXS Guinier plots. The solid lines indicated regions used in the Guinier fits. C) SAXS p(r) distributions. D) SANS scattering profiles. E) SANS Guinier plots. The solid lines indicated the regions used in the Guinier fits. F) SANS p(r) distributions. Errors in A and D correspond to 1 stand deviation. Errors in B and C are not shown for clarity and are within the scatter of the data.
Table 1.
Concentration dependence of Rg and Io.a
| Protein Concentration (mg/mL) | SAXS Rg (Å) | SANS Rg (Å) | SAXS I0b | SANS I0 (cm−1)b |
|---|---|---|---|---|
| 1 | 39.92 ± 0.53 | 37.39 ± 1.39 | 0.00142 | 0.0529 |
| 2 | 40.74 ± 0.29 | 36.62 ± 0.75 | 0.00294 | 0.106 |
| 4 | 42.03 ± 0.16 | 39.33 ± 0.33 | 0.00583 | 0.242 |
Parameters obtained from Guinier analysis as depicted in figure 1.
Errors for SAXS and SANS I0 values are approximately 5%.
Table 1 also shows the scattering intensity extrapolated to zero angle (I0) obtained from the Guinier analysis. The value of I0 is predicted to be proportional to protein concentration and molecular weight. As expected, I0 increases with protein concentration such that for the SAXS data the ratios I0 (2 mg/mL)/I0 (1 mg/mL) = 2.07 and I0 (4 mg/mL)/I0 (2 mg/mL) = 1.99. Mass action PKR self-association would tend to increase these ratios above two. The near-linear relationship of I0 and protein concentration indicates that PKR does not undergo substantial association in these experiments. Although it is possible to calculate molecular weights from I0 using appropriate standards, we have found that values based on this method are somewhat unreliable because they depend on absolute protein concentrations, which are difficult to measure. In contrast, molecular weights obtained by analytical ultracentrifugation are not dependent on absolute concentration and our previous studies have demonstrated that PKR is homogenous and monomeric at low protein concentrations.11
The pair distance distribution function, p(r) was obtained by indirect Fourier transformation of the scattering data using the program GNOM.35 Figure 2C shows SAXS p(r) curves, normalized by I0 to allow comparison of data obtained at different protein concentrations. The distributions are smooth and highly asymmetric, with maxima near 30 Å and tails extending to longer distances. Typically, multidomain proteins with long linkers exhibit extended tails in p(r) functions.24 Thus, the shape of these distributions indicating that PKR populates highly extended conformations and is consistent with the presence of flexible linker regions that adopt multiple conformations. There are slight changes in the shapes of the curves with protein concentration. As the concentration is decreased from four to two mg/mL, the amplitude of the peak near 30 Å increases and the shoulder at longer distances decreases. There is a very slight increase in the peak upon further reducing the protein concentration from two to one mg/mL, but the distributions at longer distances are superimposable. These slight changes in the shapes of the p(r) distributions with protein concentration are consistent with the Guinier analysis and indicate that some PKR self-association is detectable by SAXS above two mg/mL. Consistent with these observations, the PKR samples at one and two mg/ml are expected to contain only about 6 and 11% dimer, respectively, based on the Kd ~ 450 μM.11 Given the low amount of dimer present, these lower concentration samples are appropriate for detailed structural analysis.
As indicated in figure 2D–F, analogous small angle scattering measurements were performed with neutrons. Although the SANS profiles are slightly noisier than the corresponding SAXS data, their shapes are similar and do not vary appreciably with protein concentration. The Rg values derived from the SANS Guinier analysis are slightly lower than those obtained by SAXS. Although SANS data reported here were collected in 100% D2O buffer to enhance the contrast, this difference is not likely due to solvent effects because we obtain similar Rg values for PKR from SANS experiments performed in D2O and H2O buffers (J. Wong and James Cole, unpublished observations). The difference in Rg may be due to the effects of hydration: in SAXS the hydration layer has higher electron density than the bulk solvent and contributes to the scattering whereas in SANS the hydration layer is invisible.36;37
The p(r) distributions derived from the SANS data closely resemble the SAXS distributions with a peak at shorter internuclear distances and a broad tail extending to longer distances. The peak near 30 Å increases in amplitude upon decreasing the PKR concentration from 4 to 2 mg/mL; however, the distributions obtained at two and one mg/mL are nearly superimposable. Thus, as observed in the SAXS studies, there is no concentration dependence in the range of 1 to 2 mg/mL and these SANS profiles are essentially free from contributions of PKR oligomers. The SANS studies thus confirm the SAXS data in demonstrating that PKR populates highly extended conformations.
Structural Modeling of PKR
Based on the p(r) distributions, we have pursued structural models where PKR consists of three folded domains connected by linkers that adopt multiple conformations. Detailed structural analysis of PKR based on SAXS and SANS is particularly challenging given this flexibility. Here we use a recently-developed EOM approach26 that treats the folded domains as rigid units and uses a genetic algorithm to select an ensemble of conformers that best agree with the data from a large pool of models where the conformations of the flexible linkers are randomly varied. We focused on the SAXS data, which are of higher precision, using the highest concentration samples (2 mg/mL) where self-association is not detectable. Similar results were obtained using the 1 mg/mL SAXS data as well as the 1 and 2 mg/mL SANS data (data not shown).
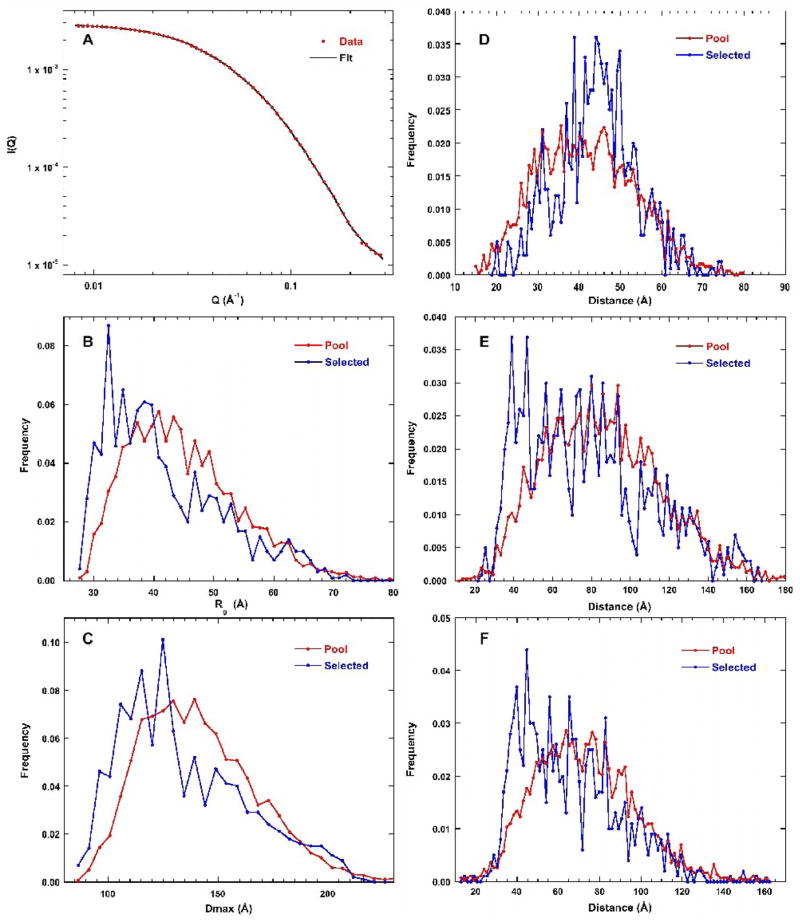
Figure 3A shows that a very good fit to the SAXS profile is obtained using EOM, with χ2 = 0.414 for the optimized ensemble containing 20 structures. The quality of fit deteriorates upon reducing the size of the ensemble. The value of χ2 rises to 0.491 for an ensemble size of 6 and χ2 = 2.25 when the ensemble is reduced to a single structure. Thus, the best fit to the SAXS data requires a superposition of multiple conformations of the linker regions, suggesting that PKR is flexible.
Figure 3.
EOM analysis of PKR SAXS data. A) Scattering profile. The red points are the experimental data (2mg/mL) and the solid black line is the fit. Error bars are omitted for clarity and correspond to those shown in figure 2A. For details see Materials and Methods. B) Rg distribution. The red line is the pool and the blue line corresponds to the ensemble of structures selected by the genetic algorithm. The same color convention is used for B–F. C) Dmax distribution. D) Distribution of dsRBM1 - dsRBM2 distances. E) Distribution of dsRBM1 - kinase distances. F) Distribution of dsRBM2 - kinase distances.
The distribution of structures selected by the genetic algorithm was analyzed using several descriptors. In figure 3B, the distribution of Rg of the selected conformations is compared to the starting pool. The distribution of the starting pool is a broad and asymmetric distribution with a peak near 40 Å and extending to a large Rg of about 70 Å. The selected distribution is biased towards more compact structures, with a peak near 30–35 Å and a broad tail to longer distances. The distribution of maximum distances (Dmax) shows similar trends (Figure 3C). The pool is a broad asymmetric distribution with a peak near 125 Å and the selected structures are biased towards less extended conformations. The shapes of these distributions are insensitive to truncation of the scattering data at the high-Q range to 0.19 Å−1 and the low Q range to 0.014Å −1, indicating that the EOM analysis is stable (data not shown).
These results suggest weak interdomain interactions may be inducing PKR to adopt more compact conformations. We have tested for specific interdomain interactions by generating distributions of the distances between the centers of mass for each of the three domains for the pool and for the selected. For each of the interdomain distance plots (Figure 3D–F), the pool distribution has a shape that appears similar to a Gaussian distribution. In the case of the dsRBM1-dsRBM2 distribution, the selected structures mostly overlay the pool with perhaps a slight bias to longer distances. Thus, the linker connecting these domains is essentially unstructured.
As expected for domains connected by a much longer linker, the distribution of dsRBM2-kinase domain distances extends out further (Figure 3E). Interestingly, the selected structures are biased to shorter distances relative to the pool, with a peak near 40 Å and a tail extending out to longer distances. Similarly, the distribution of dsRBM1-kinase distances is also biased to shorter interdomain distances relative to the pool (Figure 3F). These results suggest that one or both of the dsRBMs interact with the kinase. For multidomain proteins containing identical domains, previous simulations have shown that the EOM method cannot readily discriminate which of the domains interact.26 Given that dsRBM1 and dsRBM2 have nearly identical structures, it is likely that the method cannot determine which of the two domains is interacting with the kinase.
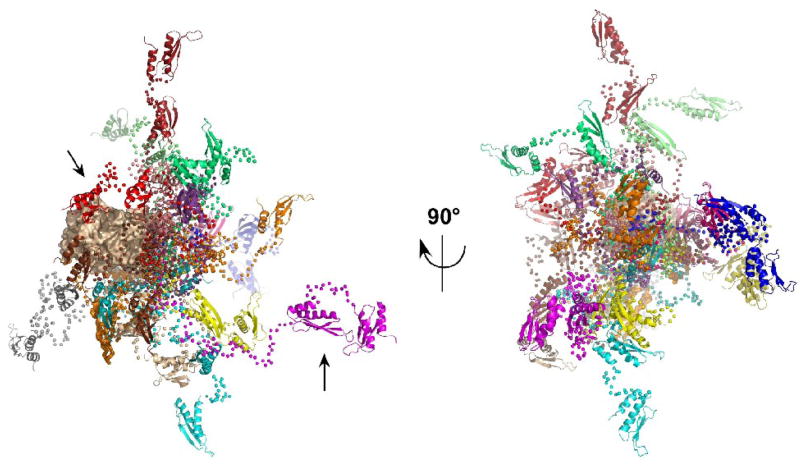
Although it is not possible to infer specific conformations from ensemble analysis, it is useful to examine the range of structures that are selected by EOM from the random pool. Figure 4 shows an overlay of 20 representative conformers selected by EOM aligned on the kinase domain. Because the linker connects to the kinase via the N-lobe, in most of the conformers the dsRNA binding motifs are closer to the N-lobe than to the C-lobe. When viewed end-on (Figure 4, right), there appears to be no strongly preferred orientations for the dsRNA binding motifs. As expected from the distance distributions, some conformers contain highly extended linkers (e.g., magenta, Rg= 75.8 Å) such that the dsRNA binding motifs are far from kinase whereas others are more compact where dsRBM1 or dsRBM2 is close to the kinase domain (e.g., red, Rg= 31.6 Å).
Figure 4.
Overlay of PKR structures selected by EOM. The 20 PKR conformations from the ensemble selected by EOM analysis of the 2 mg/mL SAXS data are superimposed by alignment on the kinase domain. The kinase domain is shown in a tan surface representation, the dsRBM1 and dsRBM2 are shown in ribbon representation and the Cα atoms in the flexible linkers are shown as spheres. Each of the 20 conformations is depicted in a different color. In the view on the left, the kinase domain is oriented with the C-lobe on the left and the N-lobe to the right. The arrows indicate representative compact (red) and extended (magenta) conformers. The ensemble on the right is rotated by −90° about the y-axis such that the kinase N-lobe points out of the page.
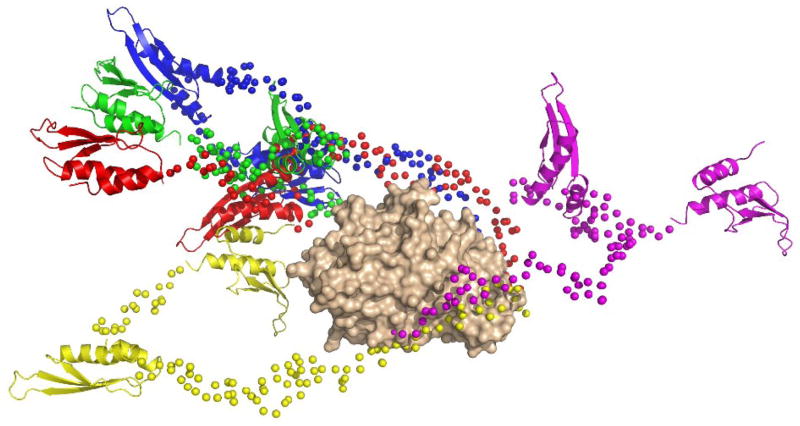
We have confirmed that structural modeling of PKR against SAXS data requires a superposition of multiple structures by attempting to fit the data to a single structure using the program BUNCH. In this approach, an initial model is built containing the folded domains connected by flexible linker residues and simulated annealing is performed to define the optimal positions and orientations of the folded domains and configurations of the linkers that best fit the experimental data. Five independent simulated annealing runs were performed using the same SAXS data set and domain structures that were employed in EOM. The values of χ2 obtained with BUNCH range from 1.50 to 2.10, which are significantly above the value of χ2 = 0.414 obtained with EOM using an ensemble size of 20, but in agreement with the EOM result obtained with an ensemble size of one. The PKR conformations generated by BUNCH are not similar to each other (Figure 5). In four cases, the dsRNA binding domains are extended out beyond the kinase C-lobe and in one case they are located near the N-lobe. However, the Rg values are very close, ranging from 39.2 to 41.5 Å for the group. Presumably, these partially extended structures are required to fit the average Rg of the SAXS data and represent averages lying between the more fully extended and compact structures observed in figure 4.
Figure 5.
Overlay of structures generated by simulated annealing runs using BUNCH. The five PKR conformations from BUNCH analysis of the 2 mg/mL SAXS data are superimposed by alignment on the kinase domain. The kinase domain is shown in a tan surface representation, the dsRBM1 and dsRBM2 are shown in ribbon representation and the Cα atoms in the flexible linkers are shown as spheres. The orientation corresponds to the right-hand representation in figure 4.
Discussion
A comprehensive picture of how dsRNA binding activates PKR to undergo autophosphorylation and phosphorylate eIF2α requires structures of the full-length enzyme in both latent and activated forms. As a first step in this endeavor, we have used small angle X-ray and neutron scattering to develop a structural model for latent PKR. Typically, it is not possible to construct a unique three-dimensional structure using one-dimensional small angle scattering data from unoriented samples in the absence of additional constraints. This problem is compounded for flexible systems where the observed scattering may arise from an ensemble of interconverting structures. In modeling PKR, we have constrained the conformations of the dsRBD and kinase to agree with the previously determined domain structures6;7;19 and considered alterative models for the interdomain linkers. Our analysis demonstrates that PKR is flexible and contains two intrinsically unstructured regions. Good fits to the scattering data require models that incorporate multiple conformations of the linker regions to account for the flexibility of the protein. The structures contained within the pool selected by the genetic algorithm are quite diverse and include a range of extended and compact conformations. Our results are consistent with previous NMR studies indicate that the PKR interdomain regions are unstructured or in rapid exchange among multiple conformations6;18 and AFM images that reveal up to three structured regions linked by bridge-like stretches that adopt multiple structures.22 These results are also supported by equilibrium chemical denaturation experiments that reveal that the folded domains of PKR are only minimally stabilized in the context of the full length enzyme relative to the isolated domains.38
Although the PKR interdomain linkers are unstructured, there is a clear bias in the distribution of dsRBD-kinase domain distances towards more compact conformations in the selected structures relative to the random pool, suggesting weak attractive interactions between these regions of the protein. This observation is consistent with sedimentation velocity experiments where we showed that dsRBD and kinase domain constructs bind weakly, with K ≈ 4 × 103 M−1.38 Using a worm-like chain model for the linker region,39 this intermolecular binding constant corresponds to an intramolecular equilibrium constant of Ki ≈ 1–10. These calculations suggest that PKR exists in an equilibrium between open and closed states, in agreement with the small angle scattering experiments. Additional evidence for a weak interaction between the dsRBD and kinase comes from NMR chemical shift perturbation measurements.19;40 Our structural model for PKR is not consistent with an autoinhibition model8 which postulates that latent PKR is locked into a stable closed conformation that blocks access to the kinase active site.
In previous SANS analysis of PKR, Rg values of 49 Å and 44 Å were reported for a latent PKR K296R mutant9 and a phosphorylated PKR construct,41 respectively. These values are somewhat larger than the parameters reported here. Note that the earlier studies were performed at only a single protein concentration and it was reported that both preparations were predominantly dimeric. Structural models were developed for several oligomeric, phosphorylated PKR preparations and complexes of PKR with an antibody, but flexibility was not considered.41
Our results demonstrate that PKR belongs to the family of proteins that contain intrinsically disordered regions. Intrinsically disordered proteins and multidomain proteins that contain large intrinsically disordered segments are common and the abundance of such proteins is enhanced in higher organisms. 20;21 As is typical of intrinsically disordered regions, the long linker between the PKR dsRBD and kinase domain has low sequence complexity with a high content of polar residues. The N-terminal region is enriched in acidic residues and C-terminus is basic (Figure 1). Interestingly, the length of the linker varies considerably among different species, from about 90 residues in the human enzyme down to 28 residues in M. auratus PKR.42 Thus, the overall length of the linker is unlikely to play a critical role in the PKR activation mechanism. The C-terminal basic region is fairly well conserved among species containing linkers of different lengths (Figure 1) and may play a role in modulating PKR dimerization (see below). In addition to its function in translational regulation, PKR participates in numerous signal transduction pathways.43 Possibly, the longer linkers may contain additional interaction motifs that function in signaling.
We propose that the two flexible linker regions in PKR may allow the enzyme to productively interact with RNA activators that have diverse structures. In the dimerization model for PKR activation, binding to RNA serves to bring the kinase domains into proximity to allow dimerization, leading to enzymatic activation. The interferon-γ mRNA activates PKR through a pseudoknot in its 5′ untranslated region.44 Other highly-structured RNAs that activate PKR include the 3′-untranslated regions of several cytoskeletal muscle mRNAs,45 the mRNA of P23/TCTP46 and the expanded CUG repeats found in the 3′-region of the DMPK gene.47 The secondary structure elements present in these complex RNA activators may reduce the accessible dsRNA regions required for PKR binding, thereby preventing PKR monomers from binding to adjacently regions of dsRNA. Flexible tethers would permit dimerization of kinase domains in cases where the dsRBDs of two bound PKR monomers are bound at more distal regions of the RNA activators.
There is evidence that the unstructured linker between the dsRBD and kinase may affect PKR activation and dimerization. It has long been know that PKR can dimerize9–13 and dimerization in the absence of dsRNA is sufficient to induce autophosphorylation at higher protein concentrations.11;18 PKR constructs containing the flexible linker and kinase domain are also capable of dsRNA-independent activation18 whereas the isolated kinase domain exists as a monomer38 and does not undergo activation in the absence of dsRNA.18 The mechanism by which the linker modulates PKR dimerization is not clear but most likely involves the most conserved, C-terminal basic region. In the crystal structure of the PKR kinase domain-eIF2α complex, the dimer interface is formed by the kinase N-terminal lobes.7 Intrinsically disordered proteins often fold into an ordered structure upon binding a target.20;21 Thus, the linker region may contribute to the dimer interface in the context of full length PKR. Similarly, this region may become ordered upon RNA binding. Further structural studies of dimeric PKR and PKR-RNA complexes will be required to better define the role of the flexible regions of PKR in the activation mechanism.
Materials and Methods
All reagents used were reagent grade purchased from Fisher Scientific except as noted. Unphosphorylated PKR was expressed and purified as previously described.11 Samples for SAXS were prepared in a buffer containing 20 mM HEPES, 200 mM NaCl, 1 mM TCEP, 0.1 mM EDTA, 5% glycerol, pH 7.5 and SANS samples were in 20 mM HEPES, 75 mM NaCl, 0.1 mM TCEP, 0.1 mM EDTA, pH 7.5 in D20. In both cases, samples were filtered through 0.02 μm filters (Whatman Anatop 10) and centrifuged at 14K RPM for 10 minutes immediately prior to measurement. SAXS data were collected at beamline 18ID at the Advanced Photon Source.48 configured with a sample-detector distance of 2.3 meters with a range of Q= 0.006 to 0.37 Å−1 at an X-ray wavelength of 1.03 Å. The sample was flowed though a 1 mm ID quartz capillary maintained at 4°C so that protein was exposed to x-rays for less than 100 ms.49 Data obtained using a 2-fold attenuated beam overlaid scans recorded using an unattenuated beam, confirmed the absence of radiation damage. Data were reduced to I vs. Q using the BioCAT Igor Pro macros. SANS data were collected at the NG3 and NG7 instruments at the NCNR using sample-detector distances of 11m and 1.75 m to give a range of Q = 0.006 to 0.34 Å−1 at a wavelength of 6 Å. Samples were loaded into 2mm (D2O) and 1 mm (H2O) pathlength quartz cells maintained at 6°C. Data reduction was performed using the NCNR Igor Pro macro package.50 I vs. Q data were subsequently corrected for buffer scattering and incoherent scattering from hydrogen in the sample.
Guinier analysis was performed using the low Q portion of the data where Rg•Q < 1. The program GNOM35 was used to calculate the p(r) pair distance distribution function by an indirect Fourier transform of the scattering data over the range Q ~ 0.014 to 0.22 Å−1. The value of p(r) was constrained to be 0 at r=0 but was unconstrained at the maximum distance (Dmax). The shapes of the p(r) curves were essentially independent of the value of Dmax used in the transform. Values of Dmax = 175 and 150 Å were used for the SAXS and SANS data, respectively.
Structural modeling of PKR was performed using two methods with the 2 mg/mL SAXS data over a range Q ~ 0.008–0.30 Å−1. In the EOM package,26 the folded domains are treated as treated as rigid units and a genetic algorithm is used to select an ensemble of conformers that best agree with the data from a large pool of models where the conformations of the flexible linkers are randomly varied. The coordinates of the kinase domain were obtained from the crystal structure of the PKR kinase domain-EIF2α complex (PDB 2A1A)7 and coordinates for dsRBM1 and dsRBM2 were obtained from the NMR structure of the dsRBD (PDB 1QU6). A missing loop within the kinase domain (residues 338 to 354) was modeled into the structure using ArchPRED.51 Two flexible regions were identified between dsRBM1 and dsRBM2 (residues 80–100)6 and between dsRBM2 and the kinase domain (residues 169 to 257)18 based on NMR studies. A pool of trial structures was generating assuming a random conformation for the two flexible linkers. Control experiments demonstrated that a pool size of 3000 was sufficient based on the value of χ2. The optimized ensemble was selected with a genetic algorithm using an ensemble size of 20 with 50 ensembles for 1,000 generations with 50 repetitions. Distributions of Rg and Dmax were obtained with the EOM software and a perl script was written to generate distributions of interdomain distances. Alternatively, the program BUNCH52 was used to define a single optimal arrangements of the three structured domains and linker regions using a simulated annealing protocol. Protein structures were visualized using Pymol (Delano Scientific, Palo Alto, CA, www.pymol.org).
Acknowledgments
We thank Liang Guo assistance in the SAXS data collection and Pascal Lapierre for computation assistance. This work was supported by grant number AI-53615 from the National Institutes of Health to J.L.C. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center RR-08630. The neutron scattering studies utilized facilities supported in part by the National Science Foundation under Agreement No. DMR-0454672. The content of this publication is solely the responsibility of the authors and does not necessarily reflect the official views of the National Center for Research Resources or the National Institutes of Health, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–528. [Google Scholar]
- 2.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 3.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 5.Meurs E, Galabru J, Barber GN, Katze MG, Hovanessian AG. Tumor supressor function of interferon-induced double-stranded RNA activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpick BW, Graziano V, Schneider D, Maitra RK, Lee X, Williams BRG. Characterization of the solution complex between the interferon-induced double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 10.Langland JO, Jacobs BL. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr=66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 11.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 12.Patel RC, Stanton P, McMillan NM, Williams BR, Sen GC. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 14.Ung TL, Cao C, Lu J, Ozato K, Dever TE. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20:3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vattem KM, Staschke KA, Wek RC. Mechanism of activation of the double-stranded-RNA-dependent protein kinase, PKR: role of dimerization and cellular localization in the stimulation of PKR phosphorylation of eukaryotic initiation factor-2 (eIF2) Eur J Biochem. 2001;268:3674–3684. doi: 10.1046/j.1432-1327.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 16.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–60. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 18.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 19.Nanduri S, Rahman F, Williams BRG, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 21.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 24.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 25.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struct Biol. 2007;17:562–71. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–64. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 27.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. Conformation of the HIV-1 Gag protein in solution. J Mol Biol. 2007;365:812–24. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moncoq K, Broutin I, Craescu CT, Vachette P, Ducruix A, Durand D. SAXS study of the PIR domain from the Grb14 molecular adaptor: a natively unfolded protein with a transient structure primer? Biophys J. 2004;87:4056–64. doi: 10.1529/biophysj.104.048645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taraban M, Zhan H, Whitten AE, Langley DB, Matthews KS, Swint-Kruse L, Trewhella J. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J Mol Biol. 2008;376:466–81. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsutakawa SE, Hura GL, Frankel KA, Cooper PK, Tainer JA. Structural analysis of flexible proteins in solution by small angle X-ray scattering combined with crystallography. J Struct Biol. 2007;158:214–23. doi: 10.1016/j.jsb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47:10345–53. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama S, Fujisawa T, Ishimori K, Morishima I, Aono S. Activation mechanisms of transcriptional regulator CooA revealed by small-angle X-ray scattering. J Mol Biol. 2004;341:651–68. doi: 10.1016/j.jmb.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 33.Buey RM, Monterroso B, Menendez M, Diakun G, Chacon P, Hermoso JA, Diaz JF. Insights into molecular plasticity of choline binding proteins (pneumococcal surface proteins) by SAXS. J Mol Biol. 2007;365:411–24. doi: 10.1016/j.jmb.2006.09.091. [DOI] [PubMed] [Google Scholar]
- 34.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–78. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenyuk AV, Svergun DI. GNOM–a program package for small-angle scattering data processing. J Appl Cryst. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svergun DI, Richard S, Koch MH, Sayers Z, Kuprin S, Zaccai G. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc Natl Acad Sci USA. 1998;95:2267–72. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins SJ. X-ray and neutron scattering analyses of hydration shells: a molecular interpretation based on sequence predictions and modelling fits. Biophys Chem. 2001;93:129–139. doi: 10.1016/s0301-4622(01)00216-2. [DOI] [PubMed] [Google Scholar]
- 38.Anderson E, Cole JL. Domain Stabilities in Protein Kinase R (PKR): Evidence for Weak Interdomain Interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 40.Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, Halperin J, Wagner G. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J Mol Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabel F, Wang D, Madern D, Sadler A, Dayie K, Daryoush MZ, Schwahn D, Zaccai G, Lee X, Williams BR. Dynamic flexibility of double-stranded RNA activated PKR in solution. J Mol Biol. 2006;359:610–623. doi: 10.1016/j.jmb.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 42.Zhu R, Zhang YB, Zhang QY, Gui JF. Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol. 2008;82:6889–901. doi: 10.1128/JVI.02385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 45.Nussbaum JM, Gunnery S, Mathews MB. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 2002;30:1205–12. doi: 10.1093/nar/30.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bommer UA, Borovjagin AV, Greagg MA, Jeffrey IW, Russell P, Laing KG, Lee M, Clemens MJ. The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA. 2002;8:478–96. doi: 10.1017/s1355838202022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischetti R, Stepanov S, Rosenbaum G, Barrea R, Black E, Gore D, Heurich R, Kondrashkina E, Kropf AJ, Wang S, Zhang K, Irving TC, Bunker GB. The BioCAT undulator beamline 18ID: a facility for biological non-crystalline diffraction and X-ray absorption spectroscopy at the Advanced Photon Source. J Synchrotron Rad. 2004;11:399–405. doi: 10.1107/S0909049504016760. [DOI] [PubMed] [Google Scholar]
- 49.Fischetti RF, Rodi DJ, Mirza A, Irving TC, Kondrashkina E, Makowski L. High-resolution wide-angle X-ray scattering of protein solutions: effect of beam dose on protein integrity. J Synchrotron Rad. 2003;10:398–404. doi: 10.1107/s0909049503016583. [DOI] [PubMed] [Google Scholar]
- 50.Kline SR. Reduction and Analysis of SANS and USANS Data using Igor Pro. J Appl Cryst. 2006;39:895–900. [Google Scholar]
- 51.Fernandez-Fuentes N, Zhai J, Fiser A. ArchPRED: a template based loop structure prediction server. Nucleic Acids Res. 2006;34:W173–6. doi: 10.1093/nar/gkl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89 doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]