Abstract
The integration of sensing elements with small groups of cells is a critical step toward miniaturization of cell cultivation and analysis. This paper describes the development of an optical, enzyme-based biosensor for local detection of hydrogen peroxide (H2O2) secreted by stimulated macrophages. Photolithographic patterning of horseradish peroxidase (HRP)-containing poly (ethylene glycol) (PEG) hydrogel microstructures was used to create sensing structures on the glass surface. Importantly, enzyme-entrapping hydrogel micropatterns did not support protein or cell deposition and allowed to guide attachment of macrophages next to the sensing elements. Amplex Red, a reagent that becomes fluorescent during HRP-catalyzed oxidation of H2O2, was either immobilized inside hydrogel elements alongside enzyme molecules or added into the cell culture media during cell activation. The production of H2O2 after mitogenic stimulation of macrophages resulted in appearance of fluorescence in the HRP-containing hydrogel microstructures, with fluorescence intensity being a strong function of analyte concentration. The novel cell culture system with integrated sensing elements described here may be enhanced in the future by incorporating additional biorecognition elements to enable multi-metabolite detection at the site of a cell.
1. INTRODUCTION
Traditional approaches for measuring cellular metabolism involve cultivation of cells in Petri dishes or micro-titer plates, followed by periodic collection of cells or culture media for off-chip analysis. These conventional methods require a large number of cells and/or reagent, reveal little about metabolism dynamics and are not amenable to multiplexing. Considerable emphasis has been placed on developing new bioanalytical methods for monitoring analytes at the site of the cells as opposed to the bulk solution. Wightman’s group pioneered the use of microelectrodes and developed fast scan cyclic voltammetry to monitor secretion of neurotransmitters from individual cells.(Troyer et al. 2002; Wightman 2006) Kennedy and co-workers have published a series of reports describing amperometric measurements of glucose consumption in individual islets of Langerhans.(Jung et al. 1999; Jung et al. 2000; Kauri et al. 2003) This group reported the existence of glucose concentration gradients at the distance of 10 to 20 μm from the cells. Others have described development of biosensors for in situ detection of nitric oxide released by endothelial cells.(Borgmann et al. 2006; Isik et al. 2006; Isik et al. 2007) Despite the considerable success in developing bioanalytical approaches to monitor cellular activity, integration of cells and biosensors remains a challenge. Micromanipulators or scanning probe approaches are frequently employed to position biosensors (e.g. microelectrodes) at the site of the cells, complicating the experiments and making them operator-dependent. In other cases cells seeded onto surfaces containing biosensors attach indiscriminately and eliminate the possibility of monitoring a well-defined, small population of cells.
Microfabrication techniques, originally developed for microelectronics industry, provide ideal means of miniaturization both biosensors and cell cultures.(Voldman et al. 1999; Folch et al. 2000; Park et al. 2003) Microfabrication has previously been employed to create integrated microsensor-cell culture platforms for on-chip monitoring of acidification (pH) and oxygen consumption.(Park et al. 2003; Sin et al. 2004) Reports describing the use of microfabricated sensors for detection of extracellular lactate and insulin have also appeared.(Cai et al. 2002; Cheng et al. 2006) Despite the use of microfabrication the methods for simple and effective integration of cells and miniature biosensors remain limited.
The present study explored the use of microfabricated biomaterial to define sites for cell attachment next to miniature biosensors. The biomaterial, PEG, is known to resist attachment of proteins or cells and is commonly employed to render surfaces non-fouling. Previously, we have demonstrated that micropatterning PEG hydrogels in a process similar to photolithography can be used to control placement of cells on the surface with micrometer-scale resolution.(Revzin et al. 2003; Revzin et al. 2005) Importantly, in addition to excellent non-fouling properties, PEG hydrogels has been shown to be an excellent matrix for encapsulation of functional biorecognition elements such as enzymes.(Pishko et al. 1991) While multiple reports described the use of microfabricated PEG hydrogel structures for biosensing in general, (Revzin et al. 2001; Seong et al. 2003; Koh et al. 2005; Allcock et al. 2006; Zguris et al. 2006; Lee et al. 2008), relatively few of these reports focused on the detection of cellular metabolites.(Zguris et al. 2006; Lee et al. 2008) Therefore, the goal in this study was to leverage dual functionality of PEG hydrogel microstructures as non-fouling surfaces and matrices for entrapment of enzymes in order to juxtapose cells with biosensors and subsequently detect extracellular metabolites. For our studies hydrogen peroxide (H2O2) was chosen. This metabolite is associated with oxidative stress and appears during inflammation as one of the reactive oxygen species (ROS). Given the central role of inflammation in a number of diseases including cancer, (Halliwell 2007) alcohol liver injury (Albano 2006) and diabetes,(Simmons 2006) it is important to detect ROS in general and H2O2 in particular.
In the present work, HRP-containing hydrogel microstructures were photolithographically patterned on glass substrates to create cell culture surfaces capable of detecting H2O2. When incubated with micropatterned surfaces, macrophages attached exclusively on the exposed glass regions and did not adhere to sensing microstructures. This allowed to juxtapose cells and biosensors, and enabled detection of macrophage-secreted H2O2 in the adjacent hydrogel structures. In the future, the cell culture/biosensor platform described here may be enhanced by incorporating multiple enzymes to enable multi-metabolite monitoring at the site of a small cell population. In addition, our approach may be easily extended to monitoring metabolic activity of cells organized into high-density array, and may therefore be used for high-throughput screening applications.
2. MATERIALS AND METHODS
2.1 Materials
Poly (ethylene glycol) diacrylate (PEG-DA, MW 575), 2-hydroxy-2methyl-propiophenone (photoinitiator), 99.9% toluene, hydrogen peroxide (35 wt % solution in water), and horseradish peroxidase (HRP), Phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich (St Louis, MO). Amplex Red reagent was obtained from Invitrogen (Carlsbad, CA). 3-acryloxypropyl trichlorosilane was from Gelest, Inc. (Morrisville, PA). Dimethyla Sulfoxide (DMSO) was from Pierce (Rockford, IL). Phosphate buffered saline (PBS, 0.1M, pH 7.4) without calcium and magnesium was from Fisher Scientific and used to prepare aqueous solution. J774 Macrophages cell line was purchased from American Type Culture Collection (ATCC). All other chemicals were used without further purification.
2.2.2 Fabrication of PEG hydrogel microstructures
Amplex Red reagent was dissolved in analytically pure DMSO to reach the concentration of 5mM and was stored in a desiccants-loaded jaw at −20 °C. HRP stock solution was prepared by dissolving 10mg HRP in 1mL PBS to make the final concentration at 10mg/mL. Pre-polymer PEG hydrogel solution contained PEG-diacrylate (DA) (MW 575) and 2% (v/v) photoinitiator (2-hydroxy-2-methyl-propiophenone). In this study, enzymes or probe reagents were incorporated into PEG hydrogel precursor solution and immobilized onto glass surface using PEG hydrogel photolithography. Two pre-polymer solutions were prepared: HRP-PEG-DA precursor solution was prepared by mixing HRP stock solution (10%, v/v) with PEG-DA prepolymer solution (90%, v/v); Amplex Red/HRP/PEG-DA precursor solution was prepared by mixing Amplex Red stock solution (1%, v/v) with HRP-PEG-DA prepolymer solution (99%, v/v). The final concentrations of Amplex Red and HRP in the mixture PEG-DA precursor solution were 50 μM and 0.1 mg/mL, respectively.
The layout of the micropattern was drafted using AutoCAD (Autodesk Inc.) and was then converted into a transparency-based photomask (CAD Art Services, Portland, Oregon). The pattern was later transferred from a transparency onto a chrome-coated sodalime plate using standard photolithography and chrome etching protocols.
Before PEG gel immobilization, standard (75 mm × 25 mm) glass slides were modified with silane for PEG attachment following a protocol reported by us earlier.(Revzin et al. 2003) Sensing PEG hydrogel microstructures were fabricated according to previously reported protocols. Briefly, Amplex Red/HRP/PEG-DA polymer solution was spin-coated on the surface at 600 rpm for 5 seconds and exposed to UV with intensity 60 mW/cm2 for 1.2 second for cross-linking. After exposure, surfaces were immersed in DI water for 10 min to remove unpolymerized PEG. The height of hydrogel microstructures could be varied from 5 to 100 μm by controlling the spin-coating rate, spin-coating time, and solution viscosity. Hydrogel features with minimal resolution of 15μm are routinely patterned in our lab.
2.2.3 Cell seeding onto micropatterned surfaces
Mouse macrophage cells (J774A) were cultured at 37°C with 5% CO2 in phenol red-free Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The cells were grown in suspension culture in 50 ml bioreactor tubes (Techno Plastic Products) on a rolling apparatus (Stovall). The cells were passaged two times a week by centrifuging and re-suspending in fresh culture media.
Prior to cell seeding, glass slides with HRP-containing hydrogel micropatterns were diced into 0.5 × 0.5 in2 pieces using a diamond scribing pen and these smaller glass pieces were placed into P60 Petri dishes. 1 ml of cell suspension at 106 cells/m was introduced into a Petri dish, allowing cells to sediment and interact with the micropatterned surface. Silicone gaskets (diameter: 6 mm; depth: 1mm) were used to decrease the required volume of cell suspension to 50 μl. After 30 minutes of incubation cell suspension was aspirated and replaced with fresh DMEM. In the process of cell seeding, macrophages were able to attach only on silane-modified glass regions, becoming localized next to but not on top of enzyme-carrying hydrogel microstructures (see Figure 1).
Figure 1.
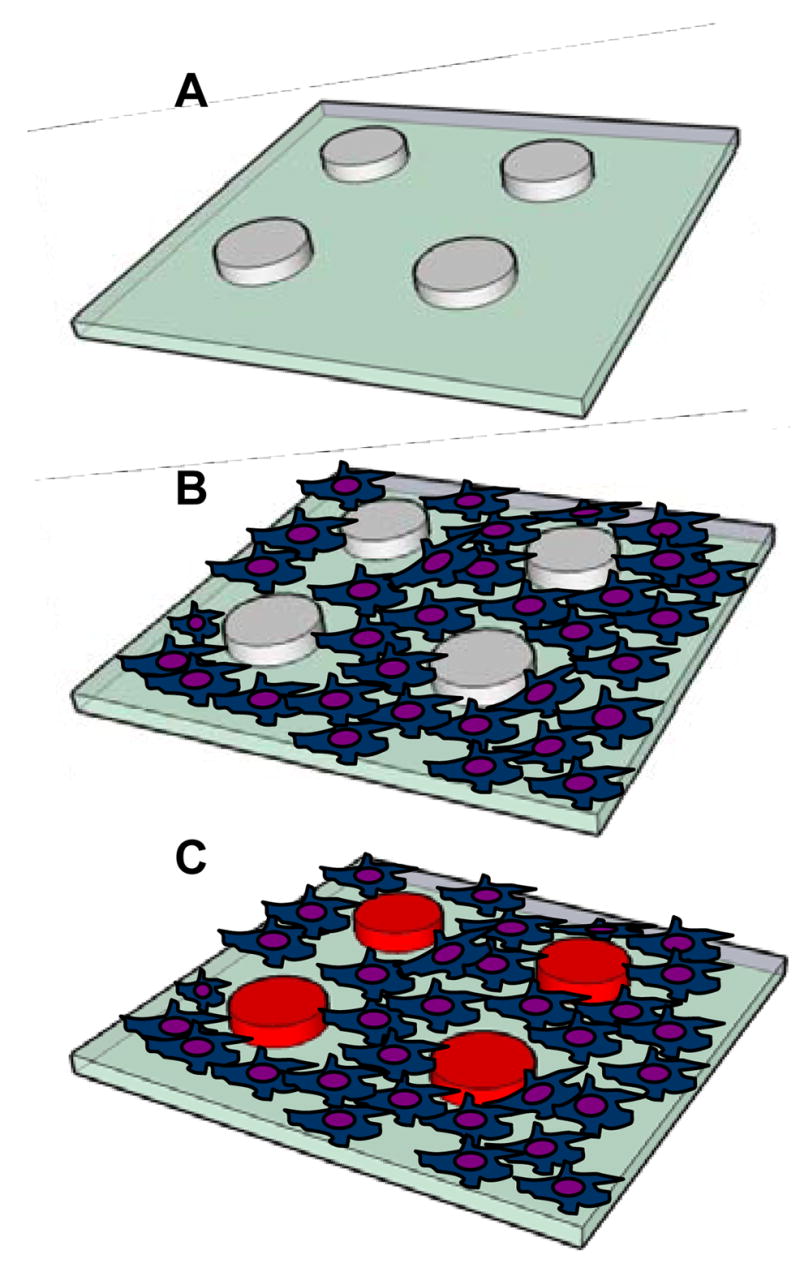
Culturing cells with sensing hydrogel micropatterns. (A) HRP-containing PEG hydrogel micropatterns are fabricated on glass substrate. (B) Encapsulation of enzyme molecules does not diminish cell-resistive properties of PEG hydrogel. When incubated with micropatterned substrates macrophages attach next to sensing hydrogel structures. (C) Macrophages release H2O2 upon mitogenic stimulation. Release of this metabolite in the presence of Amplex Red results in an appearance of fluorescence signal in the adjacent PEG hydrogel structures.
2.2.4 Detection of macrophage-secreted H2O2 using hydrogel microstructures
In order to induce production of H2O2 macrophages residing near HRP-containing PEG hydrogel structures were exposed for to PMA, a mitogenic stimulant dissolved in cell culture, for 1 or 3h. In order to detect presence of the secreted metabolite 50 μM Amplex Red was added into cell culture medium after cell activation. As result, HRP-carrying hydrogel structures were expected to have a fluorescence signal corresponding in intensity to concentration of secreted H2O2. Appearance of the fluorescence signal in the hydrogel microstructures was monitored using a microscope as described below and was determined to occur rapidly (on the scale of minutes). In most of the experiments reported here, fluorescence images of sensing hydrogel structures were acquired 5 min after introducing Amplex Red into the Petri dish containing cells and hydrogel micropatterns. The calibration curves were constructed to convert fluorescence intensity signal into analyte concentration. In order to construct a calibration curve, HRP-containing hydrogel micropatterns without cells were exposed to H2O2 ranging in concentration from 0 to 20 μM and corresponding fluorescence signals were recorded as described below. All experiments were performed in triplicate (n=3) for statistical significance. 1 μM H2O2 was chosen to serve as a reference point; hence, the calibration curve for H2O2 was easily normalized. In order to account for experimental variability, fluorescence signal in response to 1 μM H2O2 was used to normalize other data points of the calibration curve. When detecting H2O2 from macrophages we performed a parallel control experiment where HRP-containing hydrogel elements were challenged with 1 μM H2O2 without cells.
A Zeiss 200M epi-fluorescence microscope (Carl Zeiss MicroImaging, Inc. Thornwood, NY) equipped with an AxioCam MRm (CCD monochrome, 1300 × 1030 pixels) was used in order to detect fluorescence signal from hydrogel microstructures. During image acquisition objectives, camera and fluorescence filters were computer controlled through a PCI interface. A time-lapse function was set up to capture images at the rate of one frame per minute. Image acquisition and fluorescence analysis were carried out using AxioVision software (Carl Zeiss MicroImaging, Inc. Thornwood, NY). In this work, fluorescence associated with Amplex Red reagent was detected using excitation 550/25nm/emission 605/70nm filter. The excitation light source was a 120-W mercury lamp (X-cite 120, EXFO, Mississauga, Ontario, Canada). To acquire quality fluorescence images with right-scaled intensity and to avoid photobleaching, neutral density filters (ND) were inserted in the excitation light path based on the sample brightness. Usually, the same ND filter was usually utilized to a group of experiments requiring the intensity comparison. In cases when ND filters had to be modified the intensity was converted to a consistent scale based on their attenuation. All images were recorded under a 10 x objective.
Production of H2O2 by activated macrophages was also verified using a standard microplate reader. In these experiments, 200 μL of macrophage suspension at concentration of one million cells/mL were added into wells of a 96-well plate. The cells were then activated with PMA dissolved in DMEM for 1 or 3h. Cells without stimulant served a negative control. After the desired incubation time at 37°C with 5% CO2, 2 μL of 5mM Amplex Red were added into each well. After 5 min interval plates were analyzed using a fluorescence microplate reader (with excitation/emission wavelength of 563 ± 5 nm/587 ± 5 nm) (Safire2TM, Tecan Inc).
3. RESULTS AND DISSCUSSION
We are reporting a strategy for intimate integration of cells with miniature biosensors. In this approach, enzyme-containing PEG hydrogel microstructures were photolithographically patterned on glass substrates to define regions conducive to cell adhesion. When seeded on the micropatterned surfaces, macrophages attached on glass domains in the immediate vicinity of sensing hydrogel structures (see Figure 1). Entrapment of HRP in hydrogel microstructures allowed to detect oxidative burst (H2O2 production) of activated macrophages.
3.1 Non-fouling properties of enzyme-carrying hydrogel microstructures
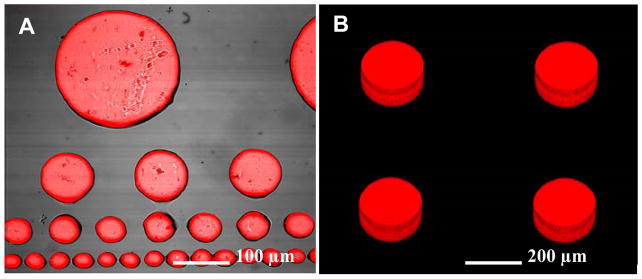
Surface micropatterning with PEG hydrogels has been used extensively to create well-defined substrates for engineering cellular microenvironment.(Revzin et al. 2003; Suh et al. 2004) In parallel, hydrogel microstructures carrying enzymes or cells have been proposed for use as biosensors.(Koh et al. 2002; Koh et al. 2003; Koh et al. 2005) The goal of the present study was to demonstrate that hydrogel microstructures can serve a dual purpose of defining and detecting cellular microenvironment. Figure 2 shows representative HRP-containing PEG hydrogel micropatterns after incubation with 10 μM of H2O2. As seen from Figure 2A, dimensions of the hydrogel biosensors could be controlled from 20 to 500 μm diameter by photolithographic patterning. A compilation of focal plane slices presented in Figure 2B underscores three-dimensionality of the hydrogel structures (thickness ~ 40 μm) and the uniform distribution of the enzyme molecules within these sensing structures.
Figure 2.
(A) HRP-containing PEG hydrogel micropattern challenged with 10μM H2O2 in the presence of Amplex Red. (B) Z-stack image of hydrogel micropatterns underscoring three-dimensionality of the structures and uniform distribution of HRP-molecules inside the gel.
It should be noted that in experiments described in Figure 2 and 3, Amplex Red was entrapped inside hydrogel microstructures alongside HRP molecules. However, we found that encapsulated Amplex Red molecules became auto-fluorescent over time in cell culture media, likely due to degradation of this reagent into fluorescent byproducts (e.g. resorufin). Therefore, when detecting cell-secreted H2O2, Amplex Red was added into cell culture media prior to imaging of the HRP-carrying hydrogel structures.
Figure 3.
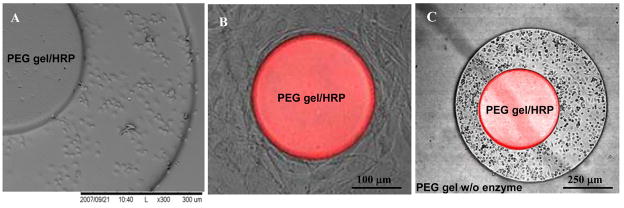
(A) SEM image of HRP-containing hydrogel micropattern after incubation with macrophages. The cells attached exclusively on exposed glass regions and did not bind to hydrogel microstructures. (B) Attachment of 3T3 fibroblasts around HRP-containing hydrogel microstructure 250 μm in diameter. Addition of 10 μM H2O2 and 50 μM Amplex Red resulted in appearance of fluorescence in the gel. This image highlights dual role of hydrogel structures as sensing elements and non-fouling biomaterials. (C) Two sets of hydrogel microstructures fabricated in alignment on the same surface. HRP-containing cylindrical elements 250 μm in diameter were fabricated inside an array of enzyme-free hydrogel wells 1 mm in diameter. After addition of 10 μM H2O2 signal appeared only from HRP-containing gel structures. Both sets of microstructures were non-fouling and macrophage attachment was seen only on glass regions.
Effective juxtaposing of small groups of cells with sensing PEG hydrogel micropatterns requires that enzyme-entrapping hydrogel structures remain non-fouling. To demonstrate that incorporation of enzymes did not adversely affect non-fouling properties of PEG hydrogels, HRP-containing PEG precursor solution was photopatterned on glass substrates and incubated with marophages or fibroblasts. These cell types are known to robustly produce matrix proteins and then attach on variety of surfaces via endogenous matrix proteins. As shown in Figure 3(A,B), upon interaction with glass substrates containing HRP-PEG micropatterns, macrophages and fibroblasts selectively attached on glass regions with limited or no adhesion observed on PEG domains. Importantly, introduction of exogenous H2O2 at 10 μM concentration into culture media resulted in optical (fluorescence) signal appearing from sensing hydrogel microstructures and pointed to retention of enzymatic activity of entrapped HRP. In addition, a multi-step PEG fabrication process could be employed to micropattern enzyme-carrying as well as enzyme-free hydrogel structures on the same surface. Figure 3C demonstrates one example where two different prepolymer solutions and two photomasks were used to create sensing HRP-containing PEG structures integrated into non-sensing (enzyme-free) PEG layer. As seen from Figure 3C, only circular (200 μm diameter) HRP-carrying hydrogel element shows fluorescence response when challenged with H2O2. The enzyme-free PEG hydrogel layer shows no fluorescence and may be used as a negative control in metabolite detection experiments. Overall, Figure 3 highlights the non-fouling nature of enzyme-carrying hydrogel microstructures and points to the possibility of integrating cells with sensing hydrogel micropatterns.
3.3 Characterization of sensing hydrogel microstructures
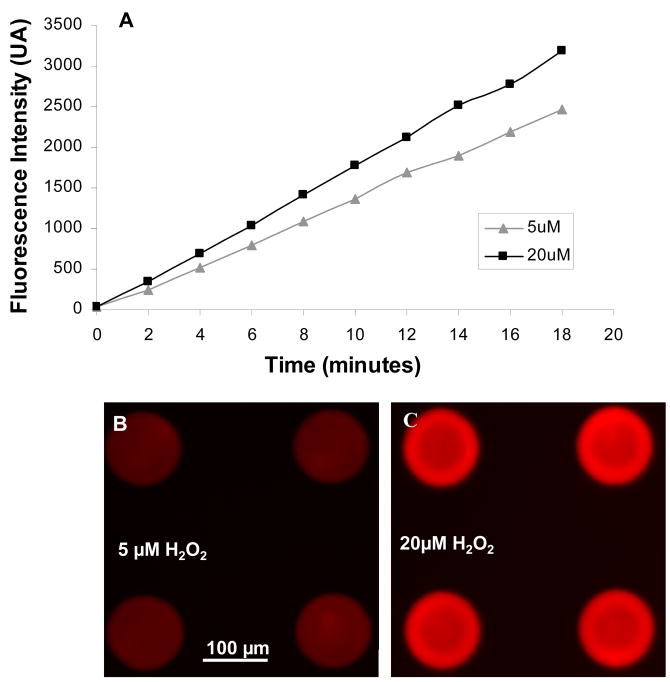
In our initial experiments Amplex Red was entrapped inside hydrogel microstructures along with HRP. However, Amplex Red is irreversibly oxidized during the HRP catalyzed breakdown of H2O2(Towne et al. 2004), therefore, immobilized probe was expected to loose its ability to sense an analyte over extended periods of time. An alternative approach was to introduce soluble Amplex Red into cell culture media during detection. Figure 4 shows a set of experiments were HRP-containing hydrogel micropatterns were exposed to 5 and 20 μM H2O2 in the presence of 50 μM Amplex Red. As seen from these data, fluorescence intensity of sensing elements challenged with a constant concentration of analyte increased as a function of time. Similar behavior of Amplex Red was reported previously (Kim et al. 2005). Based on the data presented in Figure 4, we chose to record fluorescence intensity after 5 min of interaction between sensing hydrogel structures and Amplex Red. This time was short enough to potentially capture dynamics of H2O2 secretion by live cells, yet was long enough to result in a strong signal (~40% of maximum). Others have reported suing similar time frame for detecting fluorescence signals based on Amplex Red oxidation (Rupcich et al. 2003; Li et al. 2006) (Zhou et al. 1997). This point is underscored by Figure 4(B,C) that shows a difference in fluorescence signal after incubation of hydrogel sensors with 5 vs. 20 μM of H2O2 for 5 min.
Figure 4.
(A) Time-dependent increase in fluorescence intensity of hydrogel microstructures in the presence of 5 or 20 μM H2O2 and 50 μM Amplex Red. (B–C) Fluorescence images of hydrogel microstructures after 5 min incubation with 5 μM H2O2 (B) and 20 μM H2O2 (C) in the presence of Amplex Red. When sensing for cell-secreted H2O2 images were collected 5 min after introduction of Amplex Red into cell culture medium.
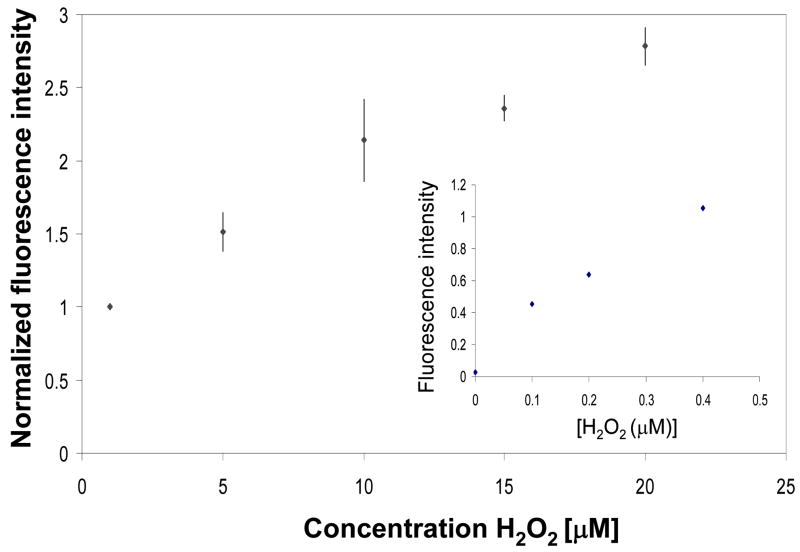
The calibration curve of [H2O2] vs. fluorescence intensity was constructed in order to quantify the amount of metabolite produced by macrophages. The fluorescence signal increased linearly over the range of 0 to 20μM as shown in Figure 5, indicating that the concentration-dependant response could be derived from the micropatterned PEG hydrogel structures. Similar detection range was reported by Zhou et al. for a system where HRP and Amplex Red were present in solution (Zhou et al. 1997). It should be noted that when constructing calibration curve shown in Figure 5 absolute fluorescence intensity signals where normalized by the sensor signal due to 1μM H2O2. We found this normalization procedure necessary in order to account for experiment-to-experiment differences in absolute fluorescence signals. When detecting endogenous H2O2 from macrophages, there was always a control experiment where the same hydrogel micropatterns without cells where exposed to 1μM H2O2 providing a reference point.
Figure 5.
Calibration curve correlating fluorescence intensity of the hydrogel microstructures to concentration of H2O2. Because of experiment-to-experiment variability in optical signal, all fluorescence intensity values were divided (normalized) by the signal for 1μM H2O2. Inset shows signal vs. analyte relationship for low concentrations of H2O2.
3.4 Detecting H2O2 production by macrophages using sensing hydrogel microstructures
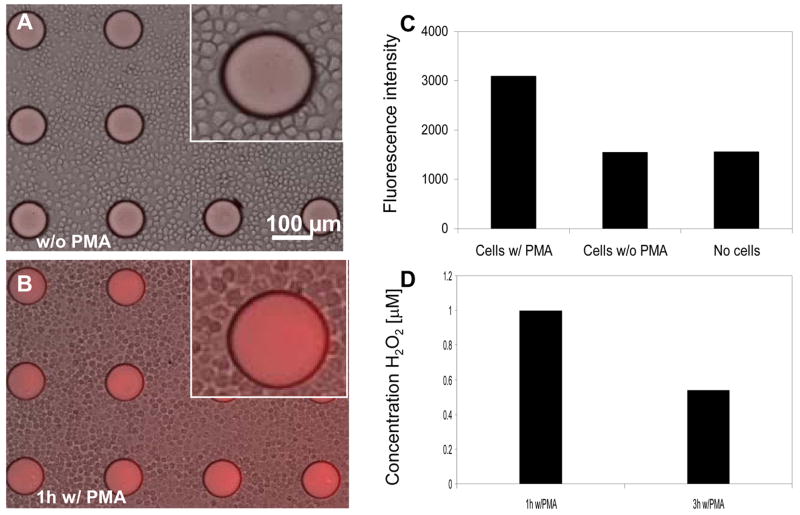
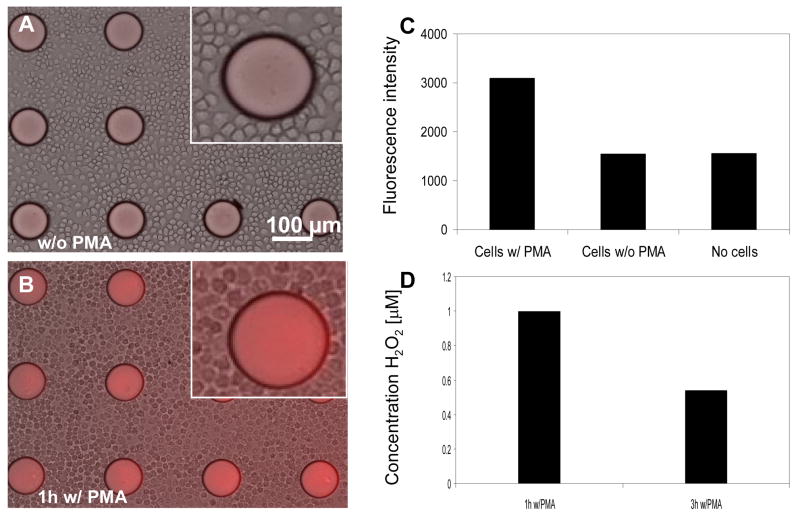
To investigate the possibility of detecting endogenous H2O2, macrophages were introduced into the HRP-containing PEG hydrogel microstructures and stimulated with a mitogen, PMA, for either 1h or 3h. This protocol was expected to result in oxidative burst and release of H2O2 from macrophages. Upon activation of cells, Amplex Red was introduced into cell culture medium and the fluorescence signal appearing in sensing hydrogel structures was recorded. Figure 6A shows a representative brightfield/fluorescence image of unactivated macrophages cultured with hydrogel micropatterns in the presence of Amplex Red. While some fluorescence is observed, this should be considered as non-specific background signal that is most likely due to auto-oxidation of Amplex Red into fluorescent resorufin in the ambient environment. This point is underscored by the image of macrophages after 3h incubation (Figure 6B) that clearly shows higher fluorescence intensity in the hydrogel microstructures. Analysis of fluorescence intensity from these images, presented in Figure 6C, shows that PMA activation of macrophages led to more than 3-fold increase in signal observed in hydrogel sensing elements, compared to macrophages not exposed to this stimulant. In addition, incubation of HRP-containing hydrogel micropatterns with Amplex Red but without cells resulted in fluorescence signals that were comparable to signals from unactivated macrophages (Figure 6C). These results highlight the connection between activation of macrophages and the appearance of H2O2 signal in the adjacent hydrogel biosensors. In addition, standard spectrafluoremetric analysis employing soluble HRP/Amplex Red (Zhou et al. 1997) was used to validate production of H2O2 by macrophages exposed to stimulant and lack of H2O2 secretion when the stimulant was absent.
Figure 6.
(A–B) Brightfield/fluorescence images of microphages adherent around HRP-containing PEG microstructures without (A) and with (B) PMA incubation. Exposure of cells to PMA was expected to trigger H2O2. (C) Fluorescence signal from sensing hydrogel elements incubated with macrophages (+) PMA, macrophages (−) PMA and no cells. These results show that minimal or no H2O2 release was observed in unactivated macrophages. (D) Using calibration curves presented in the previous Figure, concentration of macrophage-secreted H2O2 was determined after 1h and 3h of incubation with stimulant PMA.
Interestingly, we noted a difference in levels of H2O2 production by macrophages exposed to PMA (stimulant) for 1 vs. 3h. The fluorescence signal detected in HRP-containing hydrogel micropatterns was converted to analyte concentration using a calibration curve presented in Figure 5. The results of this experiment presented in Figure 6D point to ~2 fold higher H2O2 production (1 μM vs. 0.55 μM) in the case of shorter stimulation. While this observation is counterintuitive at first sight, it may be explained by a transient nature of oxidative burst whereby a high initial H2O2 production is followed by attenuation of levels of this metabolite. We are currently carrying out experiments to further test dynamics of oxidative burst of macrophages upon activation.
Overall, hydrogel micropatterns allowed to detect changes in levels of macrophage-secreted H2O2 in situ, in the Petri dish, without the need for media collection followed by off-line analysis. Placement of miniature biosensors next to cells, demonstrated in this manuscript, is envisioned as a step toward real-time monitoring of metabolites produced by small groups of cells or individual cells.
4. Conclusion
In this paper PEG hydrogel micropatterns were employed for the dual purpose of controlling cell attachment and detecting secreted cellular products. The enzyme-carrying hydrogel structures were found to retain their non-fouling properties and did not support attachment of macrophages or fibroblasts. This enabled precise and reproducible placement of cells adjacent to the biosensing structures and contributed to intimate integration of cells with biosensors. In a proof-of-concept experiment, HRP-containing hydrogel micropatterns were used to detect oxidative burst (H2O2 release) from activated macrophages. The non-fouling and sensing hydrogel micropatterns described here may be used in the future to detect metabolic activity of single cells arranged into high-density array format. In addition, the multi-step hydrogel photopatterning process may be used in the future to immobilize multiple sensing elements at the site of the cells, enabling detection of multiple metabolites in parallel.
Acknowledgments
The authors thank Dr. Louie’s lab from the department of biomedical engineering of University of California, Davis for providing assistance with confocal microscope and spectrophotometer. We also thank Dr. Henrich’s laboratory for help in cultivating macrophages. The work was supported by an NIH grant (R21CA126716) awarded to AR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albano E. Proceedings of the nutrition society. 2006;65(3):278–290. doi: 10.1079/pns2006496. [DOI] [PubMed] [Google Scholar]
- Allcock HR, Phelps MVB, Barrett EW, Pishko MV, Koh WG. Chemistry Of Materials. 2006;18(3):609–613. [Google Scholar]
- Borgmann S, Radtke I, Erichsen T, Blochl A, Heumann R, Schuhmann W. Chembiochem. 2006;7(4):662–668. doi: 10.1002/cbic.200500399. [DOI] [PubMed] [Google Scholar]
- Cai X, Klauke N, Glidle A, Cobbold P, Smith GL, Cooper JM. Anal Chem. 2002;74:908–914. doi: 10.1021/ac010941+. [DOI] [PubMed] [Google Scholar]
- Cheng W, Klauke N, Sedgwick H, Smith GL, Cooper JM. Lab On A Chip. 2006;6(11):1424–1431. doi: 10.1039/b608202e. [DOI] [PubMed] [Google Scholar]
- Folch A, Toner M. Annu Rev Biomed Eng. 2000;2:227. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Biochem J. 2007;401(1):1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- Isik S, Castillo J, Blochl A, Csoregi E, Schuhmann W. Bioelectrochemistry. 2007;70(1):173–179. doi: 10.1016/j.bioelechem.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Isik S, Schuhmann W. Angewandte Chemie-International Edition. 2006;45(44):7451–7454. doi: 10.1002/anie.200601708. [DOI] [PubMed] [Google Scholar]
- Jung SK, Gorski W, Aspinwall CA, Kauri LM, Kennedy RT. Anal Chem. 1999;71:3642–3649. doi: 10.1021/ac990271w. [DOI] [PubMed] [Google Scholar]
- Jung SK, Kauri LM, Qian WJ, Kennedy RT. J Bio Chem. 2000;9:6642–6650. doi: 10.1074/jbc.275.9.6642. [DOI] [PubMed] [Google Scholar]
- Kauri LM, Jung SK, Kennedy RT. BBRC. 2003;304:371–377. doi: 10.1016/s0006-291x(03)00595-3. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim B, Yadavalli VK, Pishko MV. Anal Chem. 2005;77(21):6828–33. doi: 10.1021/ac0507340. [DOI] [PubMed] [Google Scholar]
- Koh WG, Revzin A, Pishko MV. Langmuir. 2002;18:2459–2462. doi: 10.1021/la0115740. [DOI] [PubMed] [Google Scholar]
- Koh WG, Itle LJ, Pishko MV. Analytical Chemistry. 2003;75(21):5783–5789. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- Koh WG, Pishko M. Sensors And Actuators B-Chemical. 2005;106(1):335–342. [Google Scholar]
- Lee S, They BL, Cote GL, Pishko MV. Sensors And Actuators B-Chemical. 2008;128(2):388–398. [Google Scholar]
- Li W, Jin W. TALANTA. 2006;70(2):251–256. doi: 10.1016/j.talanta.2006.02.033. [DOI] [PubMed] [Google Scholar]
- Park TH, Shuler ML. Biotechnol Prog. 2003;19:243–253. doi: 10.1021/bp020143k. [DOI] [PubMed] [Google Scholar]
- Pishko MV, Michael AC, Heller A. Anal Chem. 1991;63:2268–2272. doi: 10.1021/ac00020a014. [DOI] [PubMed] [Google Scholar]
- Revzin A, Russell RJ, Yadavalli VK, Koh WG, Deister C, Hile DD, Mellott MB, Pishko MV. Langmuir. 2001;17:5440–5447. doi: 10.1021/la010075w. [DOI] [PubMed] [Google Scholar]
- Revzin A, Sekine K, Sin A, Tompkins RG, Toner M. Lab Chip. 2005;5:30–37. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]
- Revzin A, Tompkins RG, Toner M. Langmuir. 2003;19:9855–9862. [Google Scholar]
- Rupcich N, Brennan J. ANALYTICA CHIMICA ACTA. 2003;500(1–2):3–12. [Google Scholar]
- Seong GH, Heo J, Crooks RM. Anal Chem. 2003;75:3161–3167. doi: 10.1021/ac034155b. [DOI] [PubMed] [Google Scholar]
- Simmons RA. Free Radic Biol Med. 2006;40(6):917–22. doi: 10.1016/j.freeradbiomed.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Sin A, Chin K, Jamil MF, Kostov Y, Rao G, Shuler ML. Biotechnol Prog. 2004;20:338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- Suh KY, Seong J, Khademhosseini A, Laibinis PE, Langer R. Biomaterials. 2004;25(3):557–563. doi: 10.1016/s0142-9612(03)00543-x. [DOI] [PubMed] [Google Scholar]
- Towne V, Will M, Oswald B, Zhao Q. ANALYTICAL BIOCHEMISTRY. 2004;334(2):290–296. doi: 10.1016/j.ab.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Troyer KP, Heien M, Venton BJ, Wightman RM. Cur Opin Biotech. 2002;6:696–703. doi: 10.1016/s1367-5931(02)00374-5. [DOI] [PubMed] [Google Scholar]
- Voldman J, Gray ML, Schmidt MA. Annu Rev Biomed Eng. 1999;1:401–425. doi: 10.1146/annurev.bioeng.1.1.401. [DOI] [PubMed] [Google Scholar]
- Wightman RM. Science. 2006;311:1570–1574. doi: 10.1126/science.1120027. [DOI] [PubMed] [Google Scholar]
- Zguris J, Pishko MV. Sensors And Actuators B-Chemical. 2006;115(1):503–509. [Google Scholar]
- Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. Anal Biochem. 1997;253(2):162–8. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]