Abstract
Drosophila harbor substantial genetic variation for antibacterial defense, and investment in immunity is thought to involve a costly trade-off with life history traits, including development, life span, and reproduction. To understand the way in which insects invest in fighting bacterial infection, we selected for survival following systemic infection with the opportunistic pathogen Pseudomonas aeruginosa in wild-caught Drosophila melanogaster over 10 generations. We then examined genome-wide changes in expression in the selected flies relative to unselected controls, both of which had been infected with the pathogen. This powerful combination of techniques allowed us to specifically identify the genetic basis of the evolved immune response. In response to selection, population-level survivorship to infection increased from 15% to 70%. The evolved capacity for defense was costly, however, as evidenced by reduced longevity and larval viability and a rapid loss of the trait once selection pressure was removed. Counter to expectation, we observed more rapid developmental rates in the selected flies. Selection-associated changes in expression of genes with dual involvement in developmental and immune pathways suggest pleiotropy as a possible mechanism for the positive correlation. We also found that both the Toll and the Imd pathways work synergistically to limit infectivity and that cellular immunity plays a more critical role in overcoming P. aeruginosa infection than previously reported. This work reveals novel pathways by which Drosophila can survive infection with a virulent pathogen that may be rare in wild populations, however, due to their cost.
Author Summary
The fruit fly is commonly used as a model organism to understand the mechanistic nature of the immune response to bacterial pathogens. The fly is also commonly used to understand what immunity costs hosts in terms of other traits such as life span and reproductive success. Here, we examine these two questions together in flies selected for improved defense against the bacterium Pseudomonas aeruginosa. We show that selected flies develop from egg to adult more rapidly than unselected flies. It appears that the selected flies invest more heavily in a wing of the immune system that involves engulfment and walling off of invading bacteria. This investment can also explain the shift in developmental rate, as these two biological pathways are controlled by shared sets of genes. These latter two findings are counter to the conventional wisdom and reveal a costly, but effective, means for the fly to circumvent the virulence of Pseudomonas aeruginosa. This bacterium is normally deadly, as it has specific mechanisms to evade the host immune response. Our work is significant for demonstrating a pathway for flies to survive bacterial infection with Pseudomonas aeruginosa and for offering a reason why such a defense is not normally present in wild populations.
Introduction
It costs insects to invest in immunity. Highly immune Drosophila mate less and produce fewer offspring [1],[2], more immune bee colonies are less productive [3], and crickets with heightened immunity exhibit reduced sexual displays and longevity [4]. Recently, it has been shown that resource availability can also play a role in determining the strength and direction of these trade-offs between immunity and life history traits for insects [5]. While it is clear that individual insects vary with respect to their immune performance, only in the fly are we beginning to identify the genetic basis of this phenotypic variation [6]–[8]. With an understanding of which genetic changes confer enhanced immunity we can begin to elucidate how selection drives and balances investment into immunity in general and more specifically into different aspects of the immune response.
The innate immune response of insects is generally classified into cellular and humoral components [6], [9]–[11]. Cellular aspects of defence involve both phagocytosis by hemocytes and encapsulation of pathogens with biotoxic melanin. These aspects of the immune response are constitutively expressed and broad spectrum in target [12]. The key features of the humoral reaction, in contrast, are its inducibility upon exposure to infection and its specificity of response. Selective initiation of the Toll and/or the immune deficiency (Imd) pathways that depend on the specific pathogen, ultimately lead to the production and secretion of different sets of antimicrobial peptides (AMPs) [10], [12]–[14]. A recent study in the beetle, Tenebrio molitor, has suggested a challenge to the conventional wisdom, that the humoral response is the stronger partner of the two arms of the immune response. In the beetle, it appears that the cellular response clears the majority of infecting bacteria in the first hour after infection and that the humoral response acts secondarily to remove any persisting bacteria [15].
Here, in Drosophila melanogaster recently caught from the wild, we have artificially selected for defense against a virulent, opportunistic pathogen, Pseudomonas aeruginosa [16],[17]. In three highly resistant lines we have examined the relationship between correlated changes in life history and patterns of immune gene transcription. In contrast to traditional approaches that tend to compare gene expression of infected with uninfected flies, our microarray experiments have paired selected lines with unselected lines both post infection. The approach has lead to the identification of transcriptional changes that explain the evolved defense response instead of the genetic basis of the induced immune response. The evolved lines exhibited an effective genetic mechanism for defense against a highly virulent pathogen characterized by an increased transcriptional investment in cellular immunity. This genetic change was costly to females in particular in terms of longevity and fecundity. Antibacterial defense also correlated with an increase in developmental rate in both males and females, which was counter to expectation. Expression changes in a handful of genes that participate both in cellular immunity and host development provided a possible mechanism for this positive correlation through the action of pleiotropy.
Results
Antibacterial defense evolves rapidly in selected flies
Three independent lines stemming from a single base population were selected for improved defense against P. aeruginosa infection over 10 consecutive generations. Three additional populations, unexposed to infection, but reared with the same population size bottlenecks served as pair matched controls. In selected lines, the proportion of flies surviving P. aeruginosa infection rose from ∼15% at G1 to ∼30% by G3 (see Figure 1). Survival then increased again to ∼70% at G5 where it remained for the duration of the selection regime. There was a significant effect of selection at both G6 (treatment effect: F1,2 = 426.02, P<0.0023) and G10 (treatment effect: F1,2 = 117.44, P<0.0084), with selected lines showed significantly higher survivorship compared to corresponding controls. There was no sexual dimorphism in survivorship for these two generations G6 (sex effect: F1,4 = 1.68, P = 0.265) and G10 (treatment effect: F1,4 = 0.47, P = 0.531) nor was there any indication of sex-dependent evolution of survival, G6 (sex×treatment: F1,4 = 0.32, P = 0.601) and G10 (sex×treatment: F1,4 = 0.04, P = 0.843). The mean realized heritability of the evolved survival across the three lines was 16.7±1.3% (s.e.m). Unlike survivorship, the time it took for infected flies to die following infection did not change under the selection regime (data not shown). After the selection experiment, all fly lines were passaged without infection for a further 5 generations (G15). In the absence of selection, survival in the selected lines returned to pre-selection baseline levels and was no different from G15 controls (treatment effect: F1,2 = 0.5, P = 0.848) (Figure 1).
Figure 1. Average percentage survival of flies (male and female) at 48 hours post-infection of controls (open symbols) and lines selected for PA01 defense (solid symbols).
Survival was measured for every generation for selected lines and at G6 and G10 for control lines. Selection was halted at G10 before defense was assessed again at G15.
Selected flies have reduced lifespan and less viable offspring
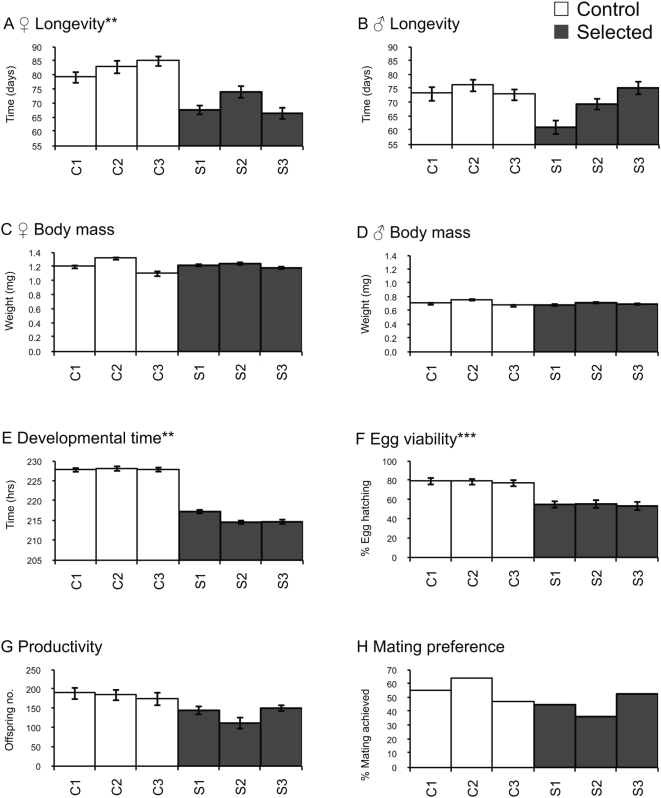
To assess the fitness cost of evolved defense in the selected flies, six life-history traits representing major aspects of host fitness were measured at G9. Longevity was quantified by rearing virgin males and females separately and then recording their time to death in days. A general linear model demonstrated there was no sex or sex×treatment effect on longevity (data not shown). While there was no effect of selection on longevity (Figure 2B) in males (t2 = 1.70, P = 0.14) in the absence of infection, a significant reduction (t2 = 4.07, P<0.01) in average lifespan of female flies was observed in selected flies relative to control flies (Figure 2A). A general linear model demonstrated there was no sex or sex×treatment effect on body mass (data not shown). The mean body mass for selected female (1.21±0.010 g, Figure 2C) and male (0.71±0.008 g, Figure 2D) flies were not different (data not shown) from their respective controls, 1.20±0.013 g and 0.69±0.007 g. Selected flies developed from egg to eclosion (Figure 2D) on average ∼12 hours faster (t2 = 13.0, P<0.01) than controls. Mean egg viability (Figure 2F) of the selection lines (54% egg hatch) was lower (t 2 = 73.1, P<0.001) than that of controls (78%). Number of offspring produced from a single mating between a pair of virgin flies was recorded as female productivity. The mean number of offspring produced (Figure 2G) in selected lines, in contrast, did not differ when compared to controls (t2 = 3.3, P = 0.08). To assess the effect of selection on male attractiveness, a selected male and a control male were allowed to compete for a female from the base population. The mating success of male flies from selected lines did not differ compared with controls (F1,1 = 0.68, P = 0.56).
Figure 2. Life-history traits of control (open bars) and selected (black bars) lines measured at G9.
Line means are plotted±sem. * P-value<0.05, ** P-value<0.01, *** P-value<0.001.
Selected flies show changes in gene expression relative to infected controls
Both selected and control lines were infected at G10 and their RNA was extracted for transcriptional profiling experiments. This comparison specifically revealed the changes in expression due to selection for defense. This is in contrast to the traditional approach of comparing infected lines to uninfected, where the question is instead about which genes are induced after infection. A total of 414 (337 up, 77 down) transcripts showed shared patterns of altered expression in all three lines after selection (Figure 3). Expression profiles of S1 and S2 were most similar to one another. Approximately, 69 immune related genes were significantly up-regulated in at least 2 of the 3 selected lines and 46 of these genes showed similar increases across all three lines (Table S1). Eighteen genes with known roles in either the cellular or humoral immune response showed parallel changes in expression in at least 2 of the 3 selected lines (Table 1).
Figure 3. Venn diagram of number of transcripts that show significant expression changes (up-regulated/down-regulated) across the three lines.
Table 1. Fold change of up-regulated immune genes in selected lines compared to respective controls.
| Flybase Gene ID | NAME | Microarrays | qPCR validation | |||||
| S1 | S2 | S3 | S1 | S2 | S3 | |||
| Humoral response | ||||||||
| PGRP | ||||||||
| FBgn0043578 | PGRP-SB1 | 1.98 | 2.01 | 2.19 | ||||
| FBgn0035806 | PGRP-SD | 1.71 | 1.96 | 1.67 | 1.68*** | 2.05*** | 1.56* | |
| FBgn0043575 | PGRP-SC2 | 1.81 | 1.76 | |||||
| Antimicrobial peptides | ||||||||
| FBgn0052282 | drosomycin-4 | 1.71 | 1.49 | 1.40 | 1.24 | 1.64* | 1.25 | |
| FBgn0035434 | drosomycin-5 | 1.53 | 1.48 | 1.39 | ||||
| FBgn0034407 | Diptericin B | 2.02 | 1.77 | 1.90 | ||||
| Toll pathway | ||||||||
| FBgn0030926 | Persephone | 1.48 | 1.66 | 1.36 | ||||
| FBgn0000533 | Easter | 1.24 | 1.24 | 1.23 | ||||
| Cellular response | ||||||||
| Recognition and phagocytosis | ||||||||
| FBgn0014033 | Scavenger receptor class C, type I | 2.46 | 1.74 | 1.94 | 1.86*** | 1.30* | 1.52 | |
| FBgn0041182 | Tep II | 2.13 | 1.39 | 2.06 | 1.38* | 1.15 | 1.36* | |
| FBgn0028545 | nimC1 | 1.84 | 1.96 | 1.60 | ||||
| FBgn0039484 | Eater | 1.92 | 2.02 | 1.83 | 1.80** | 1.88*** | 2.09** | |
| FBgn0027562 | CG10345 | 1.57 | 1.16 | |||||
| FBgn0035090 | CG2736 | 1.90 | 1.24 | |||||
| FBgn0041183 | Tep I | 4.96 | 1.54 | |||||
| FBgn0043792 | CG30427 | 1.19 | 1.57 | 1.48 | ||||
| FBgn0039687 | CG7593 | 1.33 | 1.14 | |||||
| FBgn0035993 | CG3891 | 2.64 | 1.60 | 1.87 | ||||
| Melanization and coagulation | ||||||||
| FBgn0033367 | CG8193 | 1.51 | 1.32 | 1.32 | ||||
| FBgn0000165 | Black cells | 2.70 | 1.96 | 1.99 | 1.99* | 1.46** | 1.51 | |
*: P-value<0.05.
**: P-value<0.01.
***: P-value<0.001.
For cases where multiple transcripts are present for the same gene, an average was taken. qPCR validation of 6 genes is shown next to the microarrays data.
Humoral immunity contributes to the evolved defense
Three peptidoglycan-recognition protein (PGRP) genes showed up-regulation in at least two of the three selected lines (Table 1). Both PGRP-SB1 and PGRP-SD are produced in the fat body and are only induced upon infection. PGRP-SB1 codes for a bactericidal amidase [18], while PGRP-SD, which functions as a receptor for gram-positive bacteria is involved in Toll activation [19]. PGRP-SC2 is a predicted amidase and was up-regulated in S2 and S3 [20]. Three AMP genes belonging to two families are also up-regulated in selected flies (Table 1). Drosomycin-4 and -5, which are primarily antifungal and target gram-positive bacterium, showed increased expression in all three selected lines [21]. Diptericin B, which has previously been shown to be stimulated upon P. aeruginosa infection, showed the strongest expression changes among AMP genes [22]. Both persephone and easter which encode serine endopeptidases and that regulate the Toll signalling pathway [13] were significantly up-regulated in all selected lines (Table 1).
Cellular immunity contributes heavily to the evolved defense
In previous studies examining the expression profiles of infected flies in response to a range of pathogens, including Pseudomonas, the humoral response dominates in terms of numbers of responsive genes (Table 2). Here, as best seen by the ratio of the number of humoral/cellular responding genes, the nature of evolved defence has shifted toward the cellular. The cellular genes responding to selection in this study are associated with both recognition/phagocytosis and melanization/coagulation. Many of these genes (N = 8) were up-regulated in all three selected lines (Table 1).
Table 2. A comparison of the number of genes involved in humoral and cellular immunity upon infection from various microarrays studies.
| Reference | Bacterial strain | Humoral (#genes) | Cellular (#gene) | Humoral/Cellular (ratio of #) | |||||
| AMP | PGRP | Toll/Imd | Sum | Recognition/Phagocytosis | melanization/coagulation | Sum | |||
| [7] | E. coli and M. luteus | 15 | 7 | 9 | 31 | 2 | 9 | 11 | 2.8 |
| [8] | E. coli and M. luteus | 15 | 5 | 5 | 25 | 1 | 0 | 1 | 25.0 |
| [53] | P. aeruginosa PA14 | 13 | 1 | 2 | 16 | 2 | 0 | 2 | 8.0 |
| This study* | P. aeruginosa PA01 | 3 | 3 | 2 | 8 | 10 | 2 | 12 | 0.67 |
*: based on genes with shared expression in 2/3 lines.
The complement related, Thioester-containing proteins (Tep)1 and Tep2 function as opsonins that bind to pathogen surface to promote the detection and phagocytosis of the invading microbes [23]. Tep2 has previously been shown to be required for effective phagocytosis of Gram-negative bacterium E. coli [24]. Two phagocyte specific receptor molecules Scavenger receptor class C type 1 (SR-C1) and eater, which are found on hemocyte surface that bind to a broad range of pathogens [25],[26], are up-regulated in all selected lines. Nimc1 is another phagocytosis gene, which is structurally related to phagocytosis receptors such as eater and Draper, plays an important role in both phagocytosis and development as they are efficient in removing microbes as well as apoptotic cells [27]. Annotation of CG10345 and CG2736 suggest they have cell adhesion and scavenger receptor activities [28]. CG30427, CG7593 and CG3891 are genes required for phagocytosis [24],[28] and CG7593, CG8193 [24] and Black cells [29] have monophenol monooxygenase activity and are essential for the production of melanin from tyrosine (Table 1).
Discussion
The rapid response to selection by G5, indicates that the initial population of D. melanogaster harbored substantial additive genetic variation for defense against P. aeruginosa infection. The proportion of surviving individuals in the selected population, however, did not increase above 80% despite continued selection pressure. This in combination with the rapid decrease in population survivorship after selection was removed also suggests the presence of antagonistic pleiotropy and/or physiological constraints at work. Corresponding reductions in fitness attributes in selected flies, namely female longevity and fecundity also provide evidence of a trade-off. Such negative correlations between immunity and other aspects of host fitness are predicted [30] and well-documented in the literature [1],[2],[4],[31].
The consistent correlated increase in antibacterial defense and developmental rate in the selected lines was, however, surprising. An elevated investment in immune defense predicts a lengthening of the development processes caused by the depletion of essential nutrients [32]. Indeed the direction of this predicted trade-off has been confirmed in a selection experiment for sexual competitiveness in Drosophila [33] and virus resistance in moths [32]. Here the increase in developmental rate occurred without a reduction in body mass that may be attributed to a lack of competition for food under laboratory conditions. An examination of the transcriptional profiles of our selected lines revealed expression changes in a number of genes that have dual roles in both development and immunity. We, therefore, propose that pleiotropy between developmental and cellular immune processes and the multi-tasking functional role of hemocytes may underlie the shift toward faster development.
The Toll signaling pathway, which is an essential component of humoral immunity, also plays a key role in dorsal-ventral pattern formation in Drosophila embryos [34],[35]. The signal for dorsal-ventral axis formation is conveyed by serine proteases and Easter, which is the last serine protease in a cascade that modifies the transmembrane Toll receptor and leads to activation of the pathway [36],[37]. The process of melanization requires the activation of prophenoloxidase (PPO) to PO. The activation of PPO and Easter are negatively regulated by a single serine protease inhibitor (serpin27) [36],[38]. Transcriptional profiling of our selected lines showed that four POs genes and Easter were up-regulated in all lines. The decrease in developmental time can thus be explained in part by the selection for PPO activation, which would consequently activate Easter and alter the timing of the dorsal-ventral axis formation in the embryo [38].
In addition to patrolling the hemolymph for invading microorganisms, the hemocytes are known to play important roles during embryonic development. Hemocytes are the prominent producer of embryonic basement membrane proteins including proteoglycan papilin and the major connective tissue collagen IV [39],[40], both of which are up-regulated in all selected lines. Hemocytes migrate along conserved pathways in the embryo and shape various tissues by removing apoptotic cells and depositing extracellular matrix. Hemocyte migration and number are both tightly controlled [40]. In Drosophila, the number of hemocytes is shown to influence the outcome of the infection specifically, greater numbers of circulating hemocytes confer greater immunity [41],[42]. We found that the selected flies evolved a greater investment in cellular immunity that could translate into increases in hemocyte number and/or activity. This in turn could also alter the rate of development in selected flies.
The hallmark of the humoral immune response is the production of AMPs as regulated by the Toll and Imd pathways. The signaling cascades that lead to AMP activation are well studied and it is now generally accepted that whether one or both pathways respond to infection depends on the specific pathogen [43]. Shared components that exist in both pathways also provide for some level of cross-regulation [21],[44],[45]. Gene knockout studies have found that flies deficient for either Toll or Imd pathways are more susceptible to P. aeruginosa infection than the wild type [46]. We compared the transcriptome of selected flies to that of controls during early infection in an attempt to identify mechanisms for limiting the initiation and the early progression of P. aeruginosa infection. Components of the Toll pathway including persephone and PGRP-SD were up-regulated in all selected lines. AMP genes from both pathways including drosomycin (Toll) and diptericin (Imd), showed similar patterns of expression increase across all lines. Our data indicate that the Toll and Imd pathways work synergistically as part of the evolved defense against Pseudomonas aeruginosa.
P. aeruginosa synthesize an extensive collection of virulence associated factors that suppress the host immune defense. Drosophila hemocytes, which are the target of several P. aeruginosa toxins, are impaired by the bacterium leading to suppression of phagocytosis [22],[47]. We found a strong involvement of cellular immunity in selected lines that appears to have overcome this immune suppressive effect, possibly acting very early in the infection process [12],[15] before toxins could be produced. All major aspects of cellular immunity including recognition, phagocytosis and melanization are involved in fighting the bacterium. The comprehensive list of cellular immune genes begins with opsonins and surface receptors that recognize and phagocytose bacteria. An array of lysosomal enzymes, proteases, lipases and DNases was up-regulated in selected flies that are involved in the break down of the bacterium in the phagosome (Table S1). Melanization and coagulation genes, including PO genes, which produce melanin that physically impede the growth of intruding microorganisms [14], are up-regulated in selected flies. The conserved pattern of cellular immunity gene expression among the selected lines emphasizes the crucial role of hemocytes in suppressing P. aeruginosa. This also suggests that the synergistic activation of phagocytosis, AMP production and melanization together in selected flies is the best strategy in limiting bacterial infection [41],[42].
The selected flies have evolved mechanisms to overcome the immune suppressive effects of P. aeruginosa that involve a substantial mobilization of cellular immunity as well as investment in the humoral response. We think we see greater evidence of a cellular component in our study as compared to previous work with Pseudomonas as well as other pathogens due to a combination of both methodology and the role of selection. First, it is important to remember that our control lines were also infected and so we are focusing only on the evolved aspects of the response. Evolution of greater investment into the cellular response may be the most effective means of pathogen control. This is in keeping with recent experimental work showing the efficacy of the cellular response over the humoral in early clearing of systemic infections [12],[15]. It may also be that given the inducible nature of the humoral response that it is already operating at the upper limits of its functionality determined by cellular constraints instead of lack of genetic variation. In either case, the investment in both aspects of immunity has come at a cost particularly for females in terms of longevity and fecundity. Both selected males and females also exhibit accelerated development that may be due to changes in expression of shared gene sets in both processes and the multifunctional role of hemocytes. These experiments have revealed highly effective mechanisms of defense available to genetically diverse flies that are nonetheless unsustainable in the absence of continuous pathogen pressure due to their cost.
Materials and Methods
Fly and bacterial culture
Brisbane (BNE) base stock was founded from 26 females D. melanogaster caught around the University of Queensland St Lucia campus in August 2006. The flies were treated with 0.5% penicillin and streptomycin in the diet for one generation [48] and then passaged without antibiotic for more than 10 generations before the start of the selection experiment. A large inbred population was maintained as the base stock and reared on standard yellow corn meal medium.
P. aeruginosa PA01 was cultured as in LB medium supplemented with 100 mg ml−1 ampicillin at 37 °C [49]. For infection, the concentration of an overnight bacterial culture was adjusted to an OD of 0.5±0.05 measured spectrometrically at 600 nm. The culture was then diluted 100 fold using sterile LB. This OD was determined at the start of the selection experiments to achieve a population kill rate of 80–90%.
Selection regime
The base stock was split into 3 control and 3 selected lines. These replicate populations were used to test the reproducibility of the selection given the genetic variation present in the base population. Selected lines were infected each generation with PAO1 and the survivors allowed to produce the subsequent generation. Selection was applied for 10 generations. For each round of selection, 8 sub-replicate populations consisting of 20 flies each per gender (160 flies per gender per line per generation) were infected with P. aeruginosa PA01. Mated flies aged to 4–7 days old were anaesthetized with CO2 and infected as previously described by dipping a sterile needle in the bacterial culture and piercing the intrathoracic region of the fly [11]. Fly mortality was then monitored for each population over 48 hours. Survivors from each of the 8 sub-replicates were pooled into a single population to seed the subsequent generation. The control lines were not infected, but were exposed to the same bottleneck in population size as their paired selected lines by randomly selecting a set of individuals to found the next generation. All flies were passaged for a further 5 generations after G10 without selection.
Measurement of antibacterial defense
Survival in selected lines was monitored each generation. A subset of control line flies not used to found subsequent generations were also tested for survival post infection at G6 and G10. After G10, the lines were passaged for another 5 generations without infection followed by an additional assessment of survivorship at G15. Realised heritability of infection survival was calculated for each of the selected lines with sexes pooled as the ratio of the cumulative selection response to the cumulative selection differential [50]. For this calculation, we modelled infection survival as a threshold character following transformation [51].
Life-history traits
Longevity. Virgin female and male flies were kept in separate vials in populations of 20 (5 replicate populations per gender per line) and moved onto fresh food weekly. Fly death was recorded at each food change. Body mass. Flies were placed in vials on the first day of eclosion and aged for a further three days. Flies were then briefly anaesthetized with CO2 and weighed individually on an electronic balance. Traits were measured for both sexes (40 individuals per gender per line). Developmental time and viability. Twelve eggs laid by a female within a 6 hour window were placed onto a vial after mating with a single male (40 replicates per line). The eggs were monitored every 6 hours. The period of time (hours) from the point of oviposition to the recorded time of eclosion was recorded as the development time. Viability was calculated as a percentage of eggs hatched from a possible of twelve. Female Productivity. Pairs of virgin flies were placed together in a vial and males were removed after 24 hours. The mated females (40 replicates per line) were moved onto fresh vials every 5 days to lay eggs. The total number of viable adult offspring produced by each female was recorded as its productivity. Male Mating Success. A selected male and a control male each powdered with micronized dust of distinct colors were placed with a female from the base population for 90 minutes (Variable N, 137 to 215 replicates per line). Female choice was scored by identifying the male that the female had chosen as a mate.
Statistical analysis for life-history traits
Paired T-tests were performed on line means to compare selected lines to controls at the end of the selection regime for all traits. When traits were measured separately in the different sexes, a mixed model analysis of variance was fitted to the line mean data with restricted maximum likelihood:
Both line and treatment×line effects were treated as random whereas sex, treatment and the interaction between them were all treated as fixed.
Mating data were analyzed using a generalized mixed linear mixed model:
in which the effect of the dye, treatment and dye×treatment were treated as fixed and line was random. A binomial error distribution was assumed and a logit link function was used. Generalized mixed linear models were fitted using the GLIMMIX procedure and general mixed linear models were fitted using the MIXED procedure in SAS version 9.1.3 (SAS Institute Inc., Cary NC). Significance testing of all fixed effects used Satterthwaite's approximation for error degrees of freedom.
D. melanogaster microarrays
Microarrays were used to screen for genes demonstrating expression changes in selected lines relative to control lines after bacterial infection in G10. Only male flies were extracted and compared in this analysis. A dual colour reference design paired each selected and control line. Each pair was represented by technical replicates (N = 3) that were then replicated with a dye swap (total N = 6). Microarrays were of the 4×44 K format (Agilent) each containing controls and 3 replicates of each 60 mers feature randomly distributed across the layout. The D. melanogaster genomic sequence (Release 5.4) was obtained from Flybase [28] and was used for construction of oligonucleotides using eArray Version 5.0 (Agilent Technologies Inc., Santa Clara, CA). After removing probes that cross hybridised, a total of 13,875 transcripts which represented 12,041 genes were spotted onto each microarray. Pools of 20 males representing each line were snap frozen in liquid nitrogen and extracted for Total RNA using Trizol (Invitrogen Corp., Carlsbad, CA). RNA was then purified using Qiagen RNeasy kits according to manufacturer's instructions. Further sample preparations and hybridizations were then carried out by the Special Research Centre Microarray Facility at the University of Queensland. Sample quality was examined using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). Fluorescent cDNA was synthesized using Agilent Low RNA Input Linear Amplification Kit with Cyanine 3 or Cyanine 5-CTP.
For each transcript, median signal intensity, background signal intensity, flag and saturation were extracted and analyzed using Genesping v.7.0 (Agilent Technologies Inc., Santa Clara, CA). Probes that were not detected in at least one hybridization were considered uninformative and excluded from further consideration. An intensity dependent (Lowess) normalization (Per Spot and Per Chip) was used to correct for non-linear rates of dye incorporation as well as irregularities in the relative fluorescence intensity between the dyes. Hybridizations from each line were used as replicate data to test for significance of expression changes using the cross-gene error model. The Bonferroni multiple testing correction was used to reduce the occurrence of false positives. All array data have been deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under the accession # E-MEXP-2054.
Real-time quantitative PCR for data validation
Quantitative real-time PCR (RT–PCR) was used to validate the expression of a subset of 6 immune genes showing increased expression across all three selected lines on the arrays (Table 3) and that represented some of the major functional categories of the immune response. RNA was extracted as above and then treated with 2 µl of DNase I (Roche) for 30 minutes at 37°C to eliminate genomic DNA. Approximately 0.5 µg of total RNA was reverse transcribed using random primers and SuperScript III reverse transcriptase (Invitrogen) according to manufacturer's protocols. Quantitative PCR (qPCR) was performed on Rotor-gene 6000 (Corbett Life Science, Sydney, NSW) using Platinum®SYBR®Green (Invitrogen Inc, Carlsbad, CA) according to manufacturer's instructions. For each sample a mastermix of 2 µl RNase-free water, 5 µl of SYBR Supermix and 0.5 µl of each primer (10 µM) was added to 2 µl of cDNA. Three replicates were run for each sample. The cycling protocol was as follows; 1 cycle UDG incubation at 50 °C for 2 minutes, 1 cycle Taq activation at 95°C for 2 minutes, 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 5 s, extension at 72°C for 15 s, fluorescence acquisition 78 °C, and 1 cycle of melt curve analysis from 68–95°C in 1°C steps. The raw output data of Cycle Threshold (CT) was normalized by taking into consideration the differences in amplification efficiency of target and the reference genes using Q-gene software [52]. The linear normalized expression (NE) was analyzed using Statistica 8.0 (StatSoft, Inc.). D. melanogaster ribosomal protein rpS17 was used as the reference gene (Table 3).
Table 3. Primers for quantitative real-time PCR.
| Gene name | FlyBase ID | Forward Primer (′5′-3′) | Reverse Primer (′5′-3′) | Product size |
| rpS17 | FBgn0005533 | CACTCCCAGGTGCGTGGTAT | GGAGACGGCCGGGACGTAGT | 81 |
| PGRP-SD | FBtr0076807 | ATGACTTGGATCGGTTTGCT | GCTGGGAGCATGTAACATCA | 198 |
| Thiolester containing protein II | FBtr0079510 | CTGACCTACAAGCACGACGA | CGCCACTCTCCTTCTGTTTC | 184 |
| eater | FBtr0085134 | GCCCTACTGCAAGGGATGTA | GGTGGTTGGATTCAGCTTGT | 190 |
| Black cells | FBtr0086819 | GCACGAATAACCGCACCTAT | AGGATATCGATGCCACGAAC | 196 |
| Drosomycin-4 | FBtr0073061 | GTCCTAATGGTGGCCAACTC | AGCACTTCAGACTGGCACTG | 151 |
| Scavenger receptor C, type I | FBtr0077467 | CTCGGCCTCCAATATAACCA | CTTGTTGATGTGACCGTTGG | 171 |
Supporting Information
Immune gene expression and qPCR validation.
(0.21 MB DOC)
Acknowledgments
The authors would like to thank Karyn Johnson, Scott O'Neill, and three anonymous reviewers for helpful comments on the manuscript.
EM would like to dedicate this manuscript to her mentor, Thomas S. Whittam (1954–2008), Hannah Distinguished Professor, Michigan State University.
Footnotes
The authors have declared that no competing interests exist.
The work was supported by a UQ Development grant to EM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.McKean KA, Nunney L. Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:7904–7909. doi: 10.1073/pnas.131216398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKean KA, Nunney L. Bateman's principle and immunity: phenotypically plastic reproductive strategies predict changes in immunological sex differences. Evolution. 2005;59:1510–1517. [PubMed] [Google Scholar]
- 3.Evans JD, Pettis JS. Colony-levle impacts of immune responsiveness in noney bees, Apis mellifera. Evolution. 2005;59:2270–2274. doi: 10.1111/j.0014-3820.2005.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 4.Jacot A, Scheuber H, Brinkhof MW. Costs of an induced immune response on sexual display and longevity in field crickets. Evolution. 2004;58:2280–2286. doi: 10.1111/j.0014-3820.2004.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 5.McKean KA, Yourth CP, Lazzaro BP, Clark AG. The evolutionary costs of immunological maintenance and deployment. BMC Evol Biol. 2008;8:76. doi: 10.1186/1471-2148-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazzaro BP. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 2008;11:284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazzaro BP, Sackton TB, Clark AG. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- 9.Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 11.Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host-pathogen interactions. Trends Microbiol. 2004;12:235–242. doi: 10.1016/j.tim.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Schneider DS, Chambers MC. Rogue insect immunity. Science. 2008;322:1199–1200. doi: 10.1126/science.1167450. [DOI] [PubMed] [Google Scholar]
- 13.Leclerc V, Reichhart JM. The immune response of Drosophila melanogaster. Immunol Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- 14.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 15.Haine ER, Moret Y, Siva-Jothy MT, Rolff J. Antimicrobial defense and persistent infection in insects. Science. 2008;322:1257–1259. doi: 10.1126/science.1165265. [DOI] [PubMed] [Google Scholar]
- 16.D'Argenio DA, Gallagher LA, Berg CA, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2001;183:1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, et al. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci U S A. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- 19.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, et al. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 20.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 21.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fauvarque MO, Bergeret E, Chabert J, Dacheux D, Satre M, et al. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb Pathog. 2002;32:287–295. doi: 10.1006/mpat.2002.0504. [DOI] [PubMed] [Google Scholar]
- 23.Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci U S A. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. doi:10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Pearson A, Lux A, Krieger M. Expression cloning of dSR-CI, a class C macrophage-specific scavenger receptor from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1995;92:4056–4060. doi: 10.1073/pnas.92.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 30.Rolff J. Bateman's principle and immunity. Proc Biol Sci. 2002;269:867–872. doi: 10.1098/rspb.2002.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons LW, Roberts B. Bacterial immunity traded for sperm viability in male crickets. Science. 2005;309:2031. doi: 10.1126/science.1114500. [DOI] [PubMed] [Google Scholar]
- 32.Boots M, Begon M. Trade-Offs with Resistance to a Granulosis Virus in the Indian Meal Moth, Examined by a Laboratory Evolution Experiment. Functional Ecology. 1993;7:528–534. [Google Scholar]
- 33.McKean KA, Nunney L. Sexual selection and immune function in Drosophila melanogaster. Evolution. 2008;62:386–400. doi: 10.1111/j.1558-5646.2007.00286.x. [DOI] [PubMed] [Google Scholar]
- 34.Anderson KV. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 35.Leulier F, Lemaitre B. Toll-like receptors–taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 36.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, et al. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. Embo J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ligoxygakis P, Roth S, Reichhart JM. A serpin regulates dorsal-ventral axis formation in the Drosophila embryo. Curr Biol. 2003;13:2097–2102. doi: 10.1016/j.cub.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 38.Reichhart JM. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15:659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Fessler LI, Nelson RE, Fessler JH. Drosophila extracellular matrix. Methods Enzymol. 1994;245:271–294. doi: 10.1016/0076-6879(94)45016-1. [DOI] [PubMed] [Google Scholar]
- 40.Wood W, Jacinto A. Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat Rev Mol Cell Biol. 2007;8:542–551. doi: 10.1038/nrm2202. [DOI] [PubMed] [Google Scholar]
- 41.Braun A, Hoffmann JA, Meister M. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc Natl Acad Sci U S A. 1998;95:14337–14342. doi: 10.1073/pnas.95.24.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elrod-Erickson M, Mishra S, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- 43.Dionne MS, Scheider DS. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis Model Mech. 2008;1:43–49. doi: 10.1242/dmm.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- 46.Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, et al. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun. 2003;71:4059–4066. doi: 10.1128/IAI.71.7.4059-4066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avet-Rochex A, Bergeret E, Attree I, Meister M, Fauvarque MO. Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa. Cell Microbiol. 2005;7:799–810. doi: 10.1111/j.1462-5822.2005.00512.x. [DOI] [PubMed] [Google Scholar]
- 48.Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol Ecol. 2005;14:851–858. doi: 10.1111/j.1365-294X.2005.02445.x. [DOI] [PubMed] [Google Scholar]
- 49.Huston WM, Jennings MP, McEwan AG. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol Microbiol. 2002;45:1741–1750. doi: 10.1046/j.1365-2958.2002.03132.x. [DOI] [PubMed] [Google Scholar]
- 50.Falconer DS, MacKay TFC. Introduction to quantitative genetics. Ed 4. Harlow, Essex, UK: Longmans Green; 1996. [Google Scholar]
- 51.Magnussen S, Kremer A. The beta-binomial model for estimating heritabilities of binary traits. Theo Appl Genet. 1995;91:544–552. doi: 10.1007/BF00222986. [DOI] [PubMed] [Google Scholar]
- 52.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 53.Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL, et al. Proc Natl Acad Sci U S A. 2005;102:2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immune gene expression and qPCR validation.
(0.21 MB DOC)