Abstract
Mycobacterium abscessus infections tend to respond poorly to macrolide-based chemotherapy, even though the organisms appear to be susceptible to clarithromycin. Circumstantial evidence suggested that at least some M. abscessus isolates might be inducibly resistant to macrolides. Thus, the purpose of this study was to investigate the macrolide phenotype of M. abscessus clinical isolates. Inducible resistance to clarithromycin (MIC > 32 μg/ml) was found for 7 of 10 clinical isolates of M. abscessus previously considered susceptible; the remaining 3 isolates were deemed to be susceptible (MIC ≤ 0.5 μg/ml). Inducible resistance was conferred by a novel erm gene, erm(41), which was present in all 10 isolates and in an isolate of Mycobacterium bolletii (M. abscessus type II). However, the erm(41) alleles were nonfunctional in the three susceptible M. abscessus isolates. No evidence of erm(41) was found in Mycobacterium chelonae, and an isolate of Mycobacterium massiliense appeared to be an erm(41) deletion mutant. Expression of erm(41) in M. abscessus conferred resistance to clarithromycin and erythromycin and the ketolide HMR3004. However, this species was found to be intrinsically resistant, independent of erm(41), to clindamycin, quinupristin (streptogramin B), and telithromycin. The ability to confer resistance to clindamycin and telithromycin, but not quinupristin, was demonstrated by expressing erm(41) in Maycobacterium smegmatis. Exposure of M. abscessus to the macrolide-lincosamide-streptogramin B-ketolide agents increased the levels of erm(41) mRNA 23- to 250-fold within 24 h. The inducible macrolide resistance phenotype of some M. abscessus isolates may explain the lack of efficacy of macrolide-based chemotherapy against this organism.
The rapidly growing mycobacteria (RGM) are significant human pathogens, with approximately 90% of RGM infections being caused by Mycobacterium abscessus, Mycobacterium chelonae, and Mycobacterium fortuitum (11, 12). In particular, M. abscessus is being recognized as an emerging pathogen following trauma and surgery and in patients with cystic fibrosis (15, 35, 36). Although there appears to be a decline in the incidence of infections caused by some species of slowly growing nontuberculous mycobacteria, infections caused by RGM infections may be increasing (15, 43). Many pathogenic RGM species are considered susceptible to the newer macrolides (e.g., clarithromycin and azithromycin), and these agents are important components in the treatment of RGM infections (11, 12, 17, 19, 43). Paradoxically, however, M. abscessus infections of the lung are often intractable to macrolide-based chemotherapy, although high doses of clarithromycin may lead to clinical improvement in some patients (17). The lack of efficacy of macrolides, such as clarithromycin, is puzzling, as pretreatment isolates are usually reported as susceptible to clarithromycin when CLSI procedures are used (34). Intriguingly, Brown et al. (10) reported that the clarithromycin MIC increased for some M. abscessus isolates if the incubations for the susceptibility assays were extended, although these isolates would still be deemed macrolide susceptible if an incubation period based on CLSI guidelines were used (34). This phenomenon was not observed for M. chelonae isolates (10). An explanation for the instability of clarithromycin MIC is that M. abscessus might have inducible resistance to macrolides. If so, this could explain the lack of efficacy of macrolide-based chemotherapy against M. abscessus infections.
Clinically acquired macrolide resistance in mycobacteria is conferred by mutation in the 23S rRNA gene, leading to a base change at position 2058 or 2059 (Escherichia coli numbering) of the 23S rRNA (28). However, such mutants emerge infrequently during chemotherapy of infections caused by M. abscessus or M. chelonae (45). This finding was particularly significant for M. abscessus, as infections with this organism tend to be chronic and intractable to chemotherapy. In a study of 10 patients with pulmonary M. abscessus infections who were placed on clarithromycin monotherapy, all patients were still infected after 6 months of therapy although clarithromycin resistance had not emerged in any of the cases (18). Recently, intrinsic macrolide resistance was associated with novel ribosome methylases or erm genes in the Mycobacterium tuberculosis complex, the M. fortuitum and Mycobacterium smegmatis groups, Mycobacterium wolinskyi, and Mycobacterium mageritense (3, 13, 24, 25, 31-33). Furthermore, all the mycobacterial erm genes were inducible (3, 31-33). Thus, the aim of this study was to determine whether M. abscessus clinical isolates are inducibly resistant to macrolides and to characterize whether these organisms carry a novel erm gene.
MATERIALS AND METHODS
Bacteria and susceptibility testing.
Thirteen clinical isolates of M. abscessus (formerly PCR restriction enzyme analysis type I), eight clinical isolates of M. chelonae, and one clinical isolate each of Mycobacterium bolletii and Mycobacterium massiliense (the last two species were previously grouped as M. abscessus type II) (21) were chosen from organisms submitted for susceptibility testing and/or identification to the Mycobacteria/Nocardia Laboratory at the University of Texas Health Science Center. They were identified to the species level on the basis of growth rate, growth and colony morphology on Middlebrook 7H10 agar, pigment production, and PCR restriction enzyme analysis of the 441-bp Telenti fragment of the hsp65 gene as previously described (9, 39, 41, 44), with modification for separation of M. bolletii and M. massiliense from M. abscessus (21). Partial gene sequencing of the hsp65 gene was used to confirm the identification of M. bolletii and M. massiliense. Ten of the M. abscessus isolates, both M. bolletii and M. massiliense isolates, and all the M. chelonae isolates were deemed macrolide susceptible (clarithromycin MIC ≤ 0.5 μg/ml) on the basis of a broth microdilution assay that conformed to CSLI guidelines (34). However, five of the susceptible M. abscessus isolates (MC719, MC720, MC723, MC724, and MC725) were previously reported as showing trailing endpoints on extended susceptibility testing (10). Three isolates (MC879-2, MC1082-2, and MC1448) were macrolide resistant (clarithromycin MIC > 128 μg/ml), having an A2058→C or an A2058→G 23S rRNA gene mutation (45). These highly resistant mutants were associated with relapse (45), and for MC879-2 and MC1082-2, the pretreatment macrolide-susceptible isolates were available, i.e., MC879 and MC1082. Mycobacterium abscessus strain MAB30 was a pretreatment isolate cultured from a pediatric patient at Childrens Hospital Los Angeles, and M. chelonae ATCC 35752T was obtained from the ATCC (Manassas, VA). Mycobacterium smegmatis variant ermKO4 is a macrolide-hypersusceptible variant of strain mc2155 and is described elsewhere (31). As well as a surrogate host, M. smegmatis ermKO4 was included as an additional reference organism for susceptibility testing against lincosamide and streptogramin B agents (for MICs, see references 31 and 33) and the ketolides telithromycin (HMR3467) and HMR3004 (MICs of 2 μg/ml and 0.5 μg/ml, respectively).
To test for inducible resistance, organisms were preincubated in medium alone or in clarithromycin or erythromycin at concentrations between 0.01 and 1 μg/ml prior to determination of the clarithromycin MIC. Preliminary experiments with isolates MC879 and MC958 demonstrated that phenotypic induction of macrolide resistance occurred after 1 day preincubation in 0.1 μg/ml clarithromycin; however, after 3 days preincubation, the phenotypic effects were more pronounced. Consequently, for subsequent experiments, a 3-day preincubation in macrolide was used to test for inducible resistance. Since acquisition of a macrolide-resistant phenotype may require a cell to replace its ribosomes (methylases act on rRNA during ribosome assembly), pretreatment conditions were set to allow cultures to go through at least two generations (based on increase in turbidity).
Isolation of the putative resistance element.
Genomic DNA was isolated from mycobacteria by the method of Belisle and Sonnenberg (7). Five micrograms of DNA isolated from M. abscessus MC719 was digested for 2 h with 10 to 20 units of BamHI. The restricted DNA was size selected to be between 1 kbp and 20 kbp by preparative agarose electrophoresis. The DNA fragments were ligated to the vector pMV261, which is a kanamycin-selectable Escherichia coli mycobacterial shuttle vector (40). Isolation of plasmids conferring macrolide resistance was done by complementation of M. smegmatis ermKO4 as described elsewhere (33). Plasmids that conferred macrolide resistance were analyzed by restriction mapping using the restriction enzymes AseI and PstI, and all were found to have the same pattern of DNA fragments. One plasmid, pMVMC8, was chosen for DNA sequencing and further study.
PCR amplification and cloning of the resistance gene from clinical isolates of M. abscessus.
Crude DNA preparations suitable for PCR were obtained by heating suspensions of mycobacteria in Tris-EDTA buffer at 100°C for 15 to 20 min. Debris was removed from the preparations by centrifugation at 16,000 rpm for 3 min. The basic 50-μl PCR mixture comprised 2 μl DNA, 1 μl Herculase II fusion DNA polymerase (Stratagene, La Jolla, California), 200 μM deoxynucleoside triphosphate, 1 μM each primer, 5% dimethyl sulfoxide, 1× Herculase II PCR buffer (Stratagene). Reactions were run for 2 min at 95°C, followed by 30 to 35 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. For cloning of the resistance element, M. abscessus DNA was amplified with primers MC8-22bam (ACGTTGGATCCGAGCGCCGTCACAAGATGCACA) and MC8-23hind (GCGAGAAGCTTGACTTCCCCGCACCGATTCCAC). The resulting amplimer was digested with the restriction enzyme BamHI and ligated into the promoter trap vector pFPV27 (5), which had been digested with restriction enzymes BamHI and EcoRV. The resulting constructs were transformed into M. smegmatis ermKO4. In addition to MC8-22bam and MC8-23hind, the following consensus primers were used for screening for the resistance element: ERM41-3 (CGAGCCCGCCCTACCAAGTCAC), ERM41-4 (CCGGCCCGTAGCGTCCAATG), MC8-29 (GGGGCTGGTATCCGCTCACTGATG), MC8-27 (GCGCCGCCTGATCACCAGCAC), MC8-22 (GAGCGCCGTCACAAGATGCACA), and MC8-23 (GACTTCCCCGCACCGATTCCAC).
RACE analysis.
The erm(41) transcript was mapped using the 5′ system for rapid amplification of cDNA ends (RACE), version 2.0 (Invitrogen Corp., Carlsbad, CA), following the manufacturer's instructions for GC-rich sequences. The gene-specific nested primers used (outermost to innermost) were as follows: ERM41-4 (see above), ERM41-2 (GGATTCCGGCGTCAAGAGACTC), and ERM41-6 (GCCGTGTCCTGCGCCCAGATCCAC). The resulting product was sequenced using primer ERM41-5 (ACTCCCCTGAGCGAACAC).
Expression analysis.
Mycobacterial RNA in broth cultures of M. abscessus was stabilized by the addition of 2 volumes of RNAProtect bacterial reagent (Qiagen). The RNA was purified using a Qiagen RNeasy mini kit, incorporating a mechanical disruption step with lysing matrix B (QBiogene, Carlsbad, CA) in a FastPrep FP120A instrument (at speed setting 6.5 for 45 s). Residual DNA was removed from the RNA preparations by using a TurboDNase kit (Ambion) as described by the manufacturer. Expression analysis was done by quantitative reverse transcriptase PCR (RT-PCR), using a MyiQ iCycler real-time PCR machine (Bio-Rad, Hercules, CA) and a QuantiTect SYBR green RT-PCR kit (Qiagen) with solution Q PCR additive (Qiagen). The basic reaction conditions were 50°C for 30 min and then 95°C for 15 min (to inactivate reverse transcriptase and to activate the Taq DNA polymerase), followed by 40 cycles at 94°C for 15 s, 60°C for 15 s, and 72°C for 20 s. Approximately 50 to 100 ng of RNA was added per reaction. The primers used were ERM41-3 and ERM41-4, targeting the erm gene (sequences are given above), and COAE-3 (GCACCGCGCCACTCTCAATG) and COAE-4 (CGAGTTTCGATGTCCGCGTGAA), targeting the downstream coaE gene. The results for each RNA preparation were normalized to 23S rRNA by using primers MS23S-1 (CGAATGGCGTAACGACTTCTCA) and MS23S-3 (GTAGTGAAGGTCCCGGGGTC) and to the rpsA gene by using primers RPSA-22 (CAAGTAGCCGTCAACGACA) and RPSA-23a (GACACCTTCGGTCTTGTAAC). Normalization was based on algorithms outlined by Vandesompele et al. (42).
DNA sequencing and analysis.
Plasmid DNA and PCR products were sequenced using the BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) and run on an ABI PRISM 3100 genetic analyzer. BLAST searching (2) of the DNA and protein databases (including those for unfinished prokaryote genomes) was done through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). The computer software program MacVector, versions 9.5 and 10 (MacVector, Inc., Cary, NC), was used for general sequence analysis (e.g., restriction site and open reading frame [ORF] analysis) and ClustalW polypeptide and DNA alignment. Analysis of DNA and RNA folding was done through the web server at http://www.bioinfo.rpi.edu/zukerm/cgi-bin/rna-index.cgi (27, 48).
RESULTS
Inducible resistance in M. abscessus.
To determine if M. abscessus carries an inducible macrolide resistance element, 10 macrolide-susceptible isolates and 2 macrolide-resistant isolates (with A2058→G/C 23S rRNA gene mutations) were preincubated either in medium alone or in medium containing clarithromycin or erythromycin prior to determination of the clarithromycin MIC (Table 1). The 10 susceptible isolates had clarithromycin MICs of ≤0.5 μg/ml when the broth susceptibility assays were read on day 3, irrespective of preincubation conditions. However, two clearly distinct patterns of growth emerged for these isolates on extended incubation (5 to 14 days). For isolates MAB30, MC1028, and MC1082, the clarithromycin MICs remained at ≤0.5 μg/ml during the observation period and were not affected by preincubation with macrolide, i.e., these isolates appeared to be clarithromycin susceptible. For the remaining seven susceptible isolates (MC879, MC958, MC719, MC720, MC723, MC724, and MC725), growth in clarithromycin concentrations of >0.5 μg/ml increased from day 5, and the clarithromycin MICs reached >32 μg/ml by day 9. Furthermore, preincubation in macrolide enhanced the growth of these isolates in clarithromycin (as illustrated by the differential clarithromycin MICs on days 5 and 7), and even at day 9, there was more growth in the higher concentrations of clarithromycin (>8 μg/ml) for organisms preincubated in macrolide than for organisms preincubated in medium alone (data not shown).
TABLE 1.
Effects of macrolide preincubation and extended screening on susceptibility of M. abscessus to clarithromycin
| Isolates | Preincubation (3 days)
|
Clarithromycin MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| Agenta | Concn (μg/ml) | Day 3 | Day 5 | Day 7 | Day 9 | Day 14 | |
| MC879, MC958, MC719, | None | ≤0.5 | 1-2 | 8-32 | >32 | >32 | |
| MC720, MC723, MC724 | ERY | 0.1 | ≤0.5 | 4-8 | >32 | >32 | >32 |
| CLR | 0.1 | ≤0.5 | 4-8 | >32 | >32 | >32 | |
| MAB30, MC1028, MC1082 | None | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| ERY | 0.1 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| CLR | 0.1 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| MC879-2, MC1448 | None | >32 | >32 | >32 | >32 | >32 | |
| ERY | 0.1 | >32 | >32 | >32 | >32 | >32 | |
| CLR | 0.1 | >32 | >32 | >32 | >32 | >32 | |
ERY, erythromycin; CLR, clarithromycin.
These results could have been explained by the selection of a subpopulation of constitutively resistant organisms. To investigate this, cultures of isolates MC879 and MC958 growing in 32 μg/ml clarithromycin were harvested, washed in medium (to remove clarithromycin), and plated on nutrient agar. The susceptibilities of the derived organisms to clarithromycin were retested and found to be equivalent to those of the parental cultures (data not shown), i.e., there was no constitutive shift in phenotype following growth in macrolide, either in the preincubation or in the susceptibility assay. Thus, 7 of the 10 “susceptible” M. abscessus isolates were inducibly resistant to clarithromycin.
As expected, the clarithromycin MICs were high (>32 μg/ml) for the two 23S rRNA gene mutants MC879-2 and MC1448 when scored on day 3; preincubation in macrolide did not affect growth in clarithromycin-containing wells during the observation period (data not shown).
In order to investigate whether the inducible resistance extended to other macrolide-lincosamide-streptogramin B-ketolide (MLSK) agents, the MICs for clarithromycin, clindamycin, quinupristin (streptogramin B), and the ketolides telithromycin (HMR3467) and HMR3004 were assessed on days 4, 7, 10, and 14 for isolates MC879, MC958, and MC1028, with the last being noninducible and clarithromycin susceptible. For all three isolates, the MICs for clindamycin and quinupristin were 64 to 128 μg/ml on day 4 and >256 μg/ml by day 7, and the MICs for telithromycin were 32 to 64 on day 4 and >128 μg/ml by day 7. Thus, the M. abscessus isolates expressed high-level intrinsic resistance to these agents, even in the clarithromycin-susceptible isolate MC1028. The MICs for clarithromycin and HMR3004 against isolates MC879 and MC958 increased from ≤2 μg/ml on day 4 to 32 to 64 μg/ml on day 10 and on day 14. In contrast, the MICs for these agents against isolate MC1028 remained at ≤2 μg/ml throughout the screening period. Thus, macrolide-resistant isolates of M. abscessus were also resistant to the ketolide HMR3004.
Identification of the resistance element.
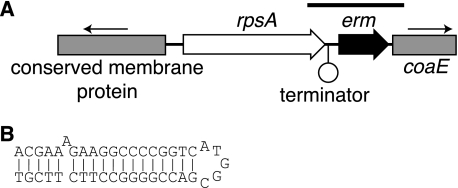
A 4.0-kbp region of the M. abscessus genome (Fig. 1A), cloned in plasmid pMVMC8, was found to confer inducible macrolide resistance to M. smegmatis ermKO4, with induced clarithromycin and erythromycin MICs of 32 and >128 μg/ml, respectively. Within the cloned region (GenBank accession no. EU177504), the only complete known gene was rpsA, which encodes the S1 ribosomal protein. However, on more-detailed analysis, a small ORF was identified immediately downstream of rpsA, and this ORF encoded a 173-amino-acid polypeptide chain with conserved domains (rADc and RrnaAD) characteristic of adenine rRNA methylases. Between rpsA and the putative methylase genes was a DNA hairpin (ΔG; −20.6 kcal/mol) consistent with a transcriptional terminator (Fig. 1B). This suggested that the methylase gene was most likely expressed from its own promoter rather than as part of an operon with the upstream rpsA gene. This was a significant consideration since the two genes were only 123 bp apart. In fact, the first possible initiation codon of the methylase gene was only 68 bp downstream from the terminator, suggesting that the untranslated leader region was very short.
FIG. 1.
(A) Gene organization of the 4.0-kbp region of M. abscessus isolate MC719 DNA that complemented the macrolide-susceptible phenotype of M. smegmatis ermKO4. The lines with arrows show the transcriptional orientations of the partially cloned genes, coaE, and the putative conserved membrane protein. The bar above the genes indicates the region cloned by PCR using primers MC8-22bam and MC8-23hind. (B) Possible transcription terminator downstream from the rpsA gene; the computed ΔG for this structure was −20.6 kcal/mol.
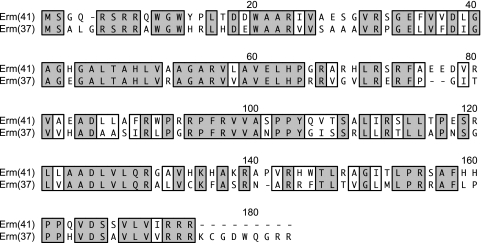
The amino acid sequence of the M. abscessus putative methylase was 73 to 93 amino acids smaller than, and only 30 to 34% identical to, the Erm methylases of other RGM, e.g., Erm(38) of Mycobacterium smegmatis (GenBank accession no. AAN86837), Erm(39) of Mycobacterium fortuitum (GenBank accession no. AAR92235), and Erm(40) of M. mageritense (GenBank accession no. AAS76623). Surprisingly, the best alignment of the putative methylase was that against the Erm(37) adenine rRNA methylase of M. tuberculosis (GenBank accession no. NP_216504.1), with 59% amino acid identity (69% similarity), although the former was slightly shorter (Fig. 2).
FIG. 2.
ClustalW alignment of the predicted polypeptides of Erm(41) of M. abscessus isolate MC719 (GenBank accession no. ABW06859) and Erm(37) of M. tuberculosis strain H37Rv (GenBank accession no. NP_216504.1). Identical residues are shown boxed and shaded; functionally similar residues are shown boxed and not shaded.
In order to establish that the putative methylase gene conferred macrolide resistance, the gene was isolated by PCR from eight M. abscessus isolates: four isolates with inducible macrolide resistance (MC719, MC720, MC879, and MC958); one isolate with high-level constitutive resistance, with a clarithromycin MIC of >128 μg/ml (MC879-2); and three macrolide-susceptible isolates (MAB30, MC1028, and MC1082). The amplimers encompassed the last 60 codons of the rpsA gene, the putative erm gene, and the first 14 codons of the downstream coaE gene (Fig. 1A). For seven of the eight isolates, the amplimers were consistent with the expected size of 892 bp; however, the amplimer from isolate MC1082 was approximately 600 bp. These products were cloned into the promoterless mycobacterial shuttle vector pFPV27 and transduced into M. smegmatis ermKO4. Table 2 shows the macrolide phenotypes of the resulting M. smegmatis transformants. The alleles cloned from inducibly resistant M. abscessus isolates conferred to M. smegmatis inducible resistance against the macrolides erythromycin and clarithromycin, the ketolide telithromycin, and the lincosamide clindamycin. In contrast, these erm alleles did not affect susceptibility to the streptogramin B agent quinupristin. The alleles from the three susceptible M. abscessus isolates (MAB30, MC1028, and MC1082) did not confer macrolide, ketolide, or lincosamide resistance to M. smegmatis ermKO4 and thus were nonfunctional.
TABLE 2.
Drug susceptibilities of M. smegmatis isolates transformed with different erm(41) allelesa
| Donor M. abscessus isolate(s)b | Donor phenotype | Preincubation (20-24 h)
|
MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Agent | Concn (μg/ml) | ERY | CLR | CLN | Q | TEL | ||
| MC879, MC958, MC719, MC720 | Inducibly resistant | None | 32 | 2 | 64 | 128 | 8 | |
| ERY | 2 | >128 | 16 | >512 | 128 | >128 | ||
| MC879-2 | Resistantc | None | 32 | 2 | ND | ND | ND | |
| ERY | 2 | >128 | 16 | ND | ND | ND | ||
| MAB30, MC1028, MC1082 | Susceptible | None | 4 | ≤0.5 | 32 | 128 | 4 | |
| ERY | 2 | 4 | ≤0.5 | 32 | 128 | 4 | ||
| Vector control | NA | None | 4 | ≤0.5 | ND | ND | ND | |
| ERY | 2 | 4 | ≤0.5 | ND | ND | ND | ||
ERY, erythromycin; CLR, clarithromycin; CLN, clindamycin; Q, quinupristin; TEL, telithromycin; NA, not applicable; ND, not determined. CLN, Q, and TEL were tested only against M. smegmatis isolates transformed with erm(41) alleles derived from M. abscessus isolates MC879 and MC1028.
Source of the erm(41) allele.
The donor M. abscessus isolate MC879-2 had an A2058→C 23S rRNA gene mutation and was constitutively resistant to clarithromycin (MIC > 32 μg/ml).
The phenotype conferred to M. smegmatis by the putative methylase gene derived from the M. abscessus isolate MC879-2 was identical to that conferred by the allele derived from the parental isolate MC879 and the other inducibly resistant isolates (Table 2). This was significant because M. abscessus isolate MC879-2 had an A2058→C 23S rRNA gene mutation and, consequently, expressed high-level, constitutive resistance to clarithromycin (MIC > 32 μg/ml) (Table 1). Thus, there was no evidence that changes in erm(41) (e.g., derepression) were involved in the acquisition of high-level macrolide resistance conferred by 23S rRNA gene mutation. Furthermore, a functional, and inducible, methylase gene was maintained in M. abscessus even after acquisition of an rRNA mutation conferring high-level macrolide resistance.
Comparison of the results in Table 2 with the M. smegmatis phenotype conferred by the original 4.0-kbp genome fragment isolated from the complementation studies (see above) established that the putative methylase gene did confer inducible macrolide resistance, that expression was driven from its native promoter, and that the proposed transcriptional terminator immediately downstream from the rpsA gene was functional. With the data establishing that the methylase gene conferred an inducible macrolide resistance phenotype, the gene was submitted to the Nomenclature Center for MLS Genes, maintained by Marilyn C. Roberts (37; http://faculty.washington.edu/marilynr/), and was designated erm(41).
In order to investigate the basis for macrolide susceptibility in some M. abscessus isolates, the erm(41) DNA sequences of seven inducibly resistant isolates (MC879, MC958, MC719, MC720, MC723, MC724, and MC725) and the three susceptible isolates (MAB30, MC1028, and MC1082) were compared. This comparison is summarized in Fig. 3A. For the seven inducibly resistant isolates, there were eight base substitutions (i.e., polymorphisms), with only three resulting in a change of amino acid. Overall, these erm(41) alleles were >98.3% identical.
FIG. 3.
(A) Summary of the DNA sequences in the erm(41) regions of 10 M. abscessus isolates: MC719 (GenBank accession no. EU177504), MC720 (GenBank accession no. EU590124), MC723 (GenBank accession no. EU590125), MC724 (GenBank accession no. EU590126), MC725 (GenBank accession no. EU590127), MC1082 (GenBank accession no. EU590128), MAB30 (GenBank accession no. EU590129), MC879 (GenBank accession no. EU872494), MC958 (GenBank accession no. EU872495), and MC1028 (GenBank accession no. EU872496). The reference sequence was that derived from M. abscessus isolate MC719, and the base numbering is from the first base of the erm(41) ORF. The base changes are shown above the sequence, with the resulting amino acid change (when appropriate); the boxes show the two sequence differences of the susceptible isolates MAB30 and MC1028, relative to the reference sequence. The line immediately underneath the sequence represents the deletions in the erm(41) allele of susceptible isolate MC1082. The thick lines above the sequence represent the three most stable predicted RNA helix loop structures in the leader region and the first 100 bases of the erm(41) transcript. Experimentally determined TSS and putative −10 and −35 elements are shown. (B) ClustalW DNA alignment of the promoter, leader region, and first three codons of erm(41) with the equivalent region of the erm(37) gene of M. tuberculosis (derived from GenBank accession no. NC_000962, bases 2231617 to 2231688). The TSS of erm(41) is indicated, and identical bases are shown boxed and shaded; the amino acid residues for the predicted N termini of the Erm(41) and Erm(37) polypeptides are shown above and below the DNA alignment, respectively.
The DNA sequences of the susceptible isolates MAB30 and MC1028 were identical and differed from our reference sequence (isolate MC719) by only two bases: (i) a T-to-C transition at position 28 (T28→C), leading to a Trp→Arg amino acid change at codon 10, and (ii) a neutral A-to-G transition at position 255 (A255→G). Of these differences, A255→G was also present in resistant isolates MC720, MC725, MC879, and MC958. Thus, the T28→C mutation was the most likely explanation for the lack of function of the erm(41) alleles from isolates MAB30 and MC1028. Although the predicted Trp→Arg change in codon 10 caused by the T28→C mutation may explain the loss of function, it is possible that this mutation blocks either transcription (unlikely, since the regulatory regions for all alleles were identical) or translation of the erm(41) gene.
Consistent with the unusually small size of the cloned amplimer (∼600 bp) from isolate MC1082, this allele had two deletions in the erm(41) gene: one of 2 bp and the other of 274 bp (bases 64 to 65 and 159 to 432) (Fig. 3A). These deletions explained the loss of function of this allele. Yet, the total loss of 276 bp, or exactly 92 codons, retained the reading frame at the 3′ end of the gene. This was probably not a coincidental finding, since a frameshift in the erm(41) gene would have led to translation of the aberrant polypeptide into the downstream gene coaE and thus interfered with the expression of this gene. The location of the 2-bp deletion (bases 64 to 65) (Fig. 3) suggested that the initiation codon for erm(41) was upstream from this point; the only conventional candidate (i.e., ATG or GTG) was the initiation codon shown in Fig. 3A.
The three susceptible isolates, MAB30, MC1028, and MC1082, were all pretreatment isolates, and except for the last, no follow-up isolates were available. Thus, it was not known whether reversion of the T28→C mutation could occur during chemotherapy. Analysis by PCR of MC1082 with its relapse-associated derivative MC1082-2 demonstrated that the erm(41)-derived amplimers were the same size (∼600 bp). This result was consistent with the erm(41) deletions characterized in MC1082 (see above) being unchanged in isolate MC1082-2.
Analysis of the erm(41) leader region.
With 5′ RACE and sequencing of the amplification product, the transcription start site (TSS) was mapped to the adenine at position −28 relative to the initiation codon (Fig. 3A). Upstream from the TSS were candidates for the −10 (TATCTT) and −35 (TATCGA) elements, with a spacing of 19 bp. The promoter and leader region of erm(41) were 61% identical to the equivalent bases immediately upstream of the erm(37) gene of M. tuberculosis (Fig. 3B). Moreover, the leader region [i.e., between the TSS and the erm(41) initiation codon] and the first two codons showed 70% nucleotide identity between the two genes. Thus, the putative regulatory regions and genes of the erm genes of M. abscessus and M. tuberculosis were highly similar.
In addition to the erm(41) reading frame, a second ORF was identified in the leader region. This ORF had potential initiation codons at positions −28 (i.e., the first three bases of the transcript), −16, −13, and −10 relative to the proposed erm(41) initiation codon and overlapped the erm(41) gene by 50 bp. Although the encoded polypeptide lacked significant identity with other known proteins (BLAST analysis E values of >2), the second ORF might encode a leader peptide involved in the regulation of erm(41). To explore this, the RNA sequences of the leader region and the erm(41) gene were screened for the helix loop (or hairpin) structures typical of gene regulation by translational attenuation. Within the leader region and the first 100 bp of the erm(41) gene, only three helix loop structures with ΔG values more negative than −10 kcal/mol were predicted (Fig. 3A). The ΔG values for helix loops 1 and 2 were −19.9 and −19.5, respectively, and together represented the most stable conformation in this region of RNA. However, if helix loop 1 were destabilized (e.g., by a stalled ribosome), then helix loop 2 would become the most stable structure (ΔG, −26.0 kcal/mol), blocking the formation of helix loop 3. In order for this scenario to regulate expression of erm(41), the initiation codon and ribosome binding site should be in the second half of helix loop 3. However, there was no conventional initiation codon in that region. Furthermore, evidence presented above suggested that the initiation codon of the erm(41) gene was that depicted in Fig. 3A.
The most stable DNA helix loop structure involved bases −5 to 24, depicted in Fig. 3A (similar to helix-loop 1), with a ΔG value of −12.1 kcal/mol, which may have been stable enough to act as a weak transcription terminator. However, the proximity of this structure to the 5′ end of the transcript (20 bp upstream) was inconsistent with transcriptional attenuation as a regulatory mechanism for erm(41).
Analysis of erm(41) expression.
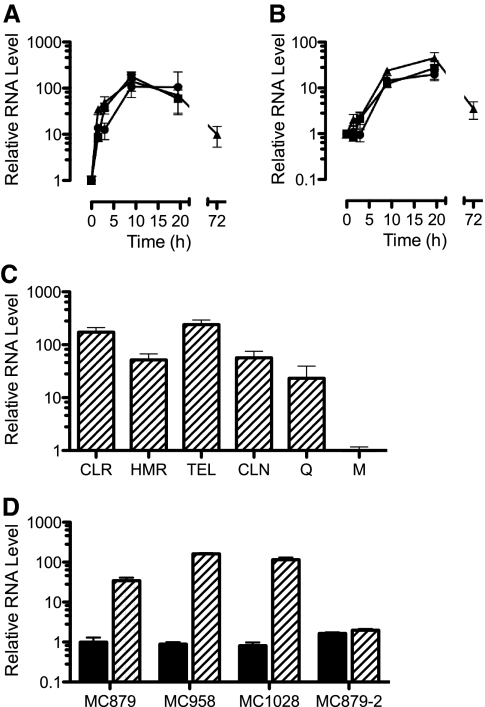
The phenotypic induction of resistance suggested that expression of erm(41) was regulated by exposure to macrolides. Real-time RT-PCR analysis revealed that macrolide exposure did indeed lead to increases in erm(41) RNA levels, reaching peaks of 50- to 100-fold above the baseline (i.e., noninduced) after 9 to 20 h (Fig. 4A). After only 1.5 h of incubation in 0.1 μg/ml clarithromycin, the RNA levels in isolate MC879 had increased 34-fold. The kinetics of erm(41) induction were similar for 0.1 μg/ml clarithromycin and 0.1 μg/ml erythromycin, and although the kinetics for 1 μg/ml clarithromycin were delayed, by 9 to 20 h, erm(41) RNA levels were equivalent for all induction conditions (Fig. 4A). The level of erm(41) RNA decreased ninefold above the baseline after 72 h.
FIG. 4.
RNA levels, relative to the baseline (time zero), for erm(41) (A) and coaE (B) in M. abscessus isolate MC879 after addition of clarithromycin at 0.1 μg/ml (triangle) or 1 μg/ml (circle) or erythromycin at 0.1 μg/ml (square). (C) Relative erm(41) RNA levels for isolate MC879 incubated for 24 h in clarithromycin (CLR; 1 μg/ml), HMR3004 (HMR; 1 μg/ml), telithromycin (TEL; 32 μg/ml), clindamycin (CLN; 128 μg/ml), or quinupristin (Q; 128 μg/ml); RNA levels are relative to those for organisms incubated in medium alone (M). (D) erm(41) RNA levels at the baseline (shaded bars) and after 24 h of incubation with erythromycin at 0.1 μg/ml (hatched bars) for four isolates of M. abscessus. All levels are relative to the baseline erm(41) RNA level of isolate MC879. Isolate MC1028 had a nonfunctional erm(41) allele, and isolate MC879-2 had a functional erm(41) allele but carried a 23S rRNA gene mutation conferring constitutively high-level macrolide resistance.
The 19-bp spacing between the erm(41) and coaE genes suggested that these genes were cotranscribed. Therefore, it was not surprising that the levels of coaE RNA increased in the presence of macrolide, reaching peaks of 17- to 34-fold above the baseline (Fig. 4B). Again, similar to the expression of erm(41), the level of coaE RNA dropped to ∼4-fold above the baseline by 72 h. These results were consistent with erm(41) and coaE being cotranscribed, i.e., part of an operon. However, the peak expression level of erm(41) was 5.1 ± 1.0-fold higher (P < 0.001) than that of coaE (averaged over the three induction conditions).
Although erm(41) conferred resistance to macrolides, ketolides, and lincosamides, it was unclear whether all these agents could induce expression of this gene. To address this issue, the induction of erm(41) expression was assessed following 24 h of incubation in representative MLSK agents (Fig. 4C). The two ketolides telithromycin and HMR3004, as well as clindamycin, led to increased erm(41) RNA levels, ranging from 52-fold (HMR3004) to 250-fold (telithromycin) above those for organisms incubated in medium alone; clarithromycin increased RNA levels 172-fold. In addition, quinupristin (streptogramin B) also increased erm(41) RNA levels (23-fold). Although there was an order-of-magnitude range in RNA levels for the five agents, this may have resulted from differences in induction kinetics rather than differences in peak level of expression. Nevertheless, these results indicated that representatives of all drug classes of the MLSK group acted as inducers of erm(41).
Figure 4D shows a comparison of the inductions of erm(41) in four isolates of M. abscessus following 20 h of incubation with 0.1 μg/ml erythromycin. The levels of erm(41) RNA in noninduced organisms were not significantly different (P = 0.4) for isolates MC879, MC958, and MC1028. Macrolide exposure caused 34- to 160-fold (mean, 104.2 ± 37.4) increases in erm(41) RNA levels for these isolates; compared with the level for isolate MC879, the induced erm(41) RNA levels were significantly higher for isolate MC958 (P < 0.001) and isolate MC1028 (P < 0.005). Macrolide exposure also increased coaE RNA 8- to 31-fold (mean, 18.6 ± 5.3), which was, on average, 5.1 ± 0.4-fold lower than the corresponding erm(41) level for each isolate. This discrepancy between erm(41) and coaE expression levels was consistent with that determined in the time course experiments described above and resulted from a lower absolute level of coaE RNA in macrolide-induced organisms; baseline coaE and erm(41) RNA levels were equivalent.
Thus, expression of erm(41) and coaE was induced by macrolides even in organisms with nonfunctional alleles (i.e., isolate MC1028). This was not unanticipated, as the regulatory regions of all the isolates were identical. These results demonstrated that the disabling event in isolate MC1028 [and MAB30, which was genetically identical in the erm(41) region] was not caused by the T28→C mutation interfering with transcription of the erm(41) allele.
In contrast to what was found for isolates MC879, MC958, and MC1028, exposure of isolate MC879-2 to erythromycin did not significantly (P = 0.6) affect the level of erm(41) RNA (Fig. 4D) or coaE RNA (data not shown). Since isolate MC879-2 had an A2058→C 23S rRNA mutation and was highly resistant to macrolides, these results indicate that regulation of erm(41) expression requires ribosome inhibition by a macrolide.
Species distribution of erm(41).
From the sequences of the analyzed alleles, six consensus primers, spanning the complete erm(41) gene, were identified. These primers were used in three combinations (ERM41-3/ERM41-4, MC8-29/MC8-27, and MC8-22/MC8-23) that amplified 151-bp, 491-bp, and 892-bp products from M. abscessus isolates (except the erm deletion mutants MC1082 and MC1082-2). The binding sites of the ERM41-3, ERM41-4, MC8-29, and MC8-27 primers were internal to the erm(41) gene. The MC8-22 and MC8-23 primers targeted the upstream rpsA gene and the downstream coaE gene, respectively, amplifying across the erm(41) gene. The PCRs were used to screen eight M. chelonae clinical isolates, M. chelonae ATCC 35752T, and one isolate each of M. bolletii and M. massiliense for the presence of the erm(41) gene. The DNA preparations from all tested organisms generated a correctly sized product (113 bp) with the 23S rRNA gene-specific primers MS23-1 and MS23-3 (used for a positive-control PCR). No amplimers were generated with the ERM41-3/ERM41-4 PCR, the MC8-29/MC8-27 PCR, or the MC8-22/MC8-23 PCR against any of the eight M. chelonae clinical isolates or the M. chelonae type strain. These results suggested that M. chelonae lacked the erm(41) gene. The M. bolletii isolate generated amplimers from all the PCRs, i.e., consistent with the presence of the erm(41) gene. Intriguingly, M. massiliense failed to generate an amplimer from the ERM41-3/ERM41-4 PCR but generated amplimers 200 to 250 bp smaller than expected with the MC8-29/MC8-27 and MC8-22/MC8-23 PCRs (∼300 bp and ∼625 bp, compared with the expected sizes of 491 bp and 892 bp, respectively). Thus, although M. massiliense was negative for erm(41), the PCR data suggested that this resulted from a deletion internal to the gene. In fact, the results for M. massiliense were equivalent to those obtained for M. abscessus MC1082 [the erm(41) deletion mutant), although the amplimers from the former were ∼50 bp larger.
DISCUSSION
Mycobacterium abscessus pulmonary infections typically respond poorly to chemotherapy based on macrolides, yet susceptibility testing of the infecting organisms almost always suggests that they are susceptible to clarithromycin. Trailing endpoints and overgrowth of zones of inhibition (for disk diffusion assays) with some isolates of M. abscessus have been reported and have been seen as a complication to the interpretation of susceptibility to macrolides (10, 34). The discovery of a novel inducible erm gene, erm(41), provides an explanation for the lack of efficacy for macrolides against M. abscessus infections and also the equivocal susceptibility testing results. Although the therapeutic implications of erm(41) require further study, it is anticipated that expression of this gene does confer clinically significant resistance to macrolides. The relationship between drug phenotype in susceptibility assays and the presence of a functional erm(41) allele indicates that susceptibility assays for M. abscessus against clarithromycin (and other macrolides) should be implemented to distinguish isolates with inducible resistance from those that are susceptible. This can be achieved by adapting the CSLI guidelines (34), scoring the plates after 3 to 4 days (to identify highly resistant mutants) as well as after 7 to 14 days. Although preincubation in macrolides induces resistance in M. abscessus, adding a preincubation to routine susceptibility assays is unnecessary.
Expression of erm(41) in M. abscessus conferred resistance to the macrolides clarithromycin and erythromycin and the ketolide HMR3004. Furthermore, erm(41) conferred resistance to clindamycin and telithromycin in M. smegmatis even though M. abscessus was intrinsically resistant to these agents [by an erm(41)-independent mechanism]. In contrast, erm(41) did not affect susceptibility to the streptogramin B agent quinupristin. This phenotype is equivalent to that conferred by the erm genes of other RGM (31-33) and erm(37) of M. tuberculosis (13, 24). Most clinically important Erm methylases dimethylate the adenine at position 2058 (A2058) in the 23S rRNA and confer resistance to MLSK agents (23). However, Erm(38) of M. smegmatis and Erm(37) of M. tuberculosis lead to monomethylation of A2058 (24, 25), although a small proportion of ribosomes modified by Erm(38) are dimethylated (25). The monomethylated state of ribosomes modified by the mycobacterial Erm methylases probably accounts for the lack of increased resistance to streptogramin B agents. Consequently, it is likely that the Erm(41) methylase is also a monomethylase.
Mycobacteria are replete with macrolide resistance-associated rRNA methylase genes representing five Erm classes: erm(37) of the M. tuberculosis complex (3, 13, 24), erm(38) of the M. smegmatis group (25, 31, 32), erm(39) of the M. fortuitum group (32, 33), erm(40) of M. mageritense and M. wolinskyi (32), and, now, erm(41) of M. abscessus and M. bolletii. Furthermore, all mycobacterial erm genes are inducible, which raises the possibility of developing macrolide derivatives that do not induce these genes; unfortunately, ketolides induce erm(41) and other mycobacterial erm genes (3, 32). Interestingly, the ketolides emerged as an important group of antibacterial agents, partly because ketolides were believed to be unable to induce erm genes in pathogens such as Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes (8, 47). However, this belief has been questioned by a recent report demonstrating that ketolides induced erm(C) expression in E. coli (4).
Inducible erm genes in many pathogenic bacteria are believed to be regulated primarily by translational attenuation (29, 46), although transcriptional attenuation is involved in at least one case (14, 22). A notable exception to this is erm(37) of M. tuberculosis, which may be regulated by the whiB7 regulatory gene (30). For translational attenuation and transcriptional attenuation, translation of a leader peptide and appropriately positioned helix loop structures in the RNA transcript (for translational attenuation) or in the DNA (for transcriptional attenuation) are required for the regulatory mechanisms to operate (20, 22, 29, 46). Although the erm(41) gene has an a possible leader peptide ORF and several relatively stable helix loop structures, the locations of these elements relative to the TSS and proposed initiation codon for erm(41) do not provide compelling evidence that regulation of this gene involves translational or transcriptional attenuation.
As an alternative to translational or transcriptional attenuation, the increases in erm(41) RNA levels following induction suggest that an inducible transcription factor may be involved. If so, the transcription factor must be conserved, as erm(41) is inducible in M. smegmatis. Since the whiB genes are conserved in mycobacteria (16), it is possible that erm(41) is regulated in a manner similar to that for erm(37) of M. tuberculosis, i.e., by a whiB7 ortholog (30). The similarity of the leader region of erm(41) to the DNA upstream of the erm(37) gene lends support for this possibility, although the comparison between the putative promoters of erm(41) and erm(37) is less compelling. Upstream from the erm(41) TSS are putative −10 (TATCTT) and −35 (TATCGA) elements of the erm(41) promoter. These elements show similarity to other proposed mycobacterial promoters (1, 6), many of which appear to have the consensus sequences TANNNT for −10 elements and WWNCGA for −35 elements. The promoter region for erm(41) does not contain recognition elements of the characterized mycobacterial sigma factors (26, 38), suggesting that an alternative transcription factor is involved in expression of this gene. Clearly, elucidation of the regulatory mechanism of erm(41) requires further study.
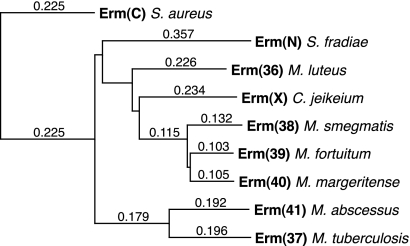
As suggested previously (32), the erm(38), erm(39), and erm(40) methylases of the RGM are closely related to those of corynebacteria and the antibiotic-producing actinomyces (e.g., Streptomyces species). Yet, despite that M. abscessus is also an RGM, erm(41) is more closely related to erm(37) of M. tuberculosis than to any other Erm class. One explanation for this similarity is that M. tuberculosis and M. abscessus acquired their erm genes horizontally from the same ancestral source. In this scenario, the phylogenic analysis (Fig. 5) suggested that M. abscessus acquired erm(41) before the other RGM acquired their erm genes. However, an alternative explanation for the similarity of erm(37) and erm(41) is convergent evolution, i.e., the enzymes have the same function, which restricts the possible gene sequence.
FIG. 5.
Phylogram illustrating the predicted phylogenic relationships between the M. abscessus isolate MC719 Erm(41) methylase (GenBank accession no. ABW06859) and representative Erm methylases from other, closely related bacterial species, including Erm(37) of Mycobacterium tuberculosis (GenBank accession no. NP_216504), Erm(38) of Mycobacterium smegmatis (GenBank accession no. AAN86837), Erm(39) of Mycobacterium fortuitum (GenBank accession no. AAR92235), Erm(40) of M. mageritense (GenBank accession no. AAS76623), Erm(X) of Corynebacterium jeikeium (GenBank accession no. AAK28910), Erm(36) of Micrococcus luteus (GenBank accession no. AAL68827), and Erm(N) of Streptomyces fradiae (GenBank accession no. CAA66307). The phylogeny was rooted on Erm(C) of Staphylococcus aureus plasmid pT48 (GenBank accession no. AAA20192).
Understanding the phylogenic relationships of the mycobacterial erm genes is further complicated by the finding that the position of erm(41) in the M. abscessus chromosome is different from those of erm(37) of M. tuberculosis and other mycobacterial erm genes (although they are all chromosomal genes). The erm(41) gene is adjacent to the rpsA gene in M. abscessus, whereas erm(37) (RV1988) is ∼400,000 bp away from rpsA (Rv1630) in M. tuberculosis, and the erm(38), erm(39), and erm(40) genes of other RGM are all adjacent to the folD gene, which is 2 million bp away from rpsA (based on the M. smegmatis chromosome). These differences in chromosome location, in addition to the sequence divergence, suggest that erm(41) was acquired by M. abscessus independently of the other mycobacterial erm genes.
Mycobacterium abscessus is closely related to M. chelonae; in fact, they were considered subspecies in the past (12). Thus, the apparent absence of erm(41) from M. chelonae is intriguing. Although the biological significance of this difference between the two species is not clear, the lack of erm(41) in M. chelonae may explain why infections with this organism respond better to macrolide-based chemotherapy than infections with M. abscessus. In addition, erm(41) may also be clinically important, as it is a detectable (e.g., by PCR) marker for distinguishing M. abscessus from M. chelonae and other RGM.
In a previous study of erm(39) of M. fortuitum (33), some clinical isolates had nonfunctional alleles. Thus, it was intriguing that the same phenomenon was found for erm(41) of M. abscessus, and M. massiliense may be an erm(41) deletion mutant. This suggested that there were conditions under which loss of methylase function was beneficial. The fact that the nonfunctional alleles were still inducible and probably still led to the synthesis of a polypeptide (at least with M. abscessus) was consistent with the benefit being the loss of methylase function rather than the burden of making the enzyme. Furthermore, since the mycobacterial erm genes are inducible, it also seems unlikely that the selective pressure was the marginal effect of ribosome methylation on growth rate in the absence of macrolide. What drives the maintenance of a nonfunctional erm allele may be an important issue for the management of mycobacterial infections. If the infecting organisms can be artificially pressured to acquire or maintain a nonfunctional erm allele, it may be possible to circumvent the intrinsic macrolide resistance of the erm-positive mycobacteria.
The presence of erm(41) in M. abscessus provides an explanation for the lack of efficacy of macrolide-based treatments of infections with this organism. However, it is not known whether the presence of a nonfunctional erm(41) allele in the infecting isolate is associated with a better outcome. Similarly, it is not known whether susceptible organisms with nonfunctional alleles can acquire reversion mutations during treatment. Clearly, further study is needed to investigate the clinical significance of erm(41) in M. abscessus and M. bolletii.
Acknowledgments
We thank Linda Bridge Mann, Maria McGlasson, and Steven McNulty (Department of Microbiology, University of Texas Health Science Center, Tyler, TX) for their technical assistance.
Funding for this study was provided by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant RO1-AI052291; the Department of Pathology and Laboratory Medicine, Childrens Hospital Los Angeles; and the Department of Microbiology, University of Texas Health Science Center, Tyler, TX.
Footnotes
Published ahead of print on 26 January 2009.
REFERENCES
- 1.Agarwal, N., and A. K. Tyagi. 2006. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res. 34:4245-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andini, N., and K. A. Nash. 2006. Intrinsic macrolide resistance of the Mycobacterium tuberculosis complex is inducible. Antimicrob. Agents Chemother. 50:2560-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, M., T. Chettiath, and A. S. Mankin. 2008. Induction of erm(C) expression by noninducing antibiotics. Antimicrob. Agents Chemother. 52:866-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, L. P., D. M. Brooks, and P. L. Small. 1998. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol. Microbiol. 29:1167-1177. [DOI] [PubMed] [Google Scholar]
- 6.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 101:31-44. [DOI] [PubMed] [Google Scholar]
- 8.Bonnefoy, A., A. M. Girard, C. Agouridas, and J. F. Chantot. 1997. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 40:85-90. [DOI] [PubMed] [Google Scholar]
- 9.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, Jr., S. H. Chiu, G. O. Onyi, E. C. Bottger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 10.Brown, B. A., R. J. Wallace, Jr., G. O. Onyi, V. De Rosas, and R. J. Wallace III. 1992. Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae-like organisms. Antimicrob. Agents Chemother. 36:180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown-Elliott, B. A., D. E. Griffith, and R. J. Wallace, Jr. 2002. Newly described or emerging human species of nontuberculous mycobacteria. Infect. Dis. Clin. N. Am. 16:187-220. [DOI] [PubMed] [Google Scholar]
- 12.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buriánková, K., F. Doucet-Populaire, O. Dorson, A. Gondran, J. C. Ghnassia, J. Weiser, and J. L. Pernodet. 2004. Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, S. S., S. K. Kim, T. G. Oh, and E. C. Choi. 1997. Role of mRNA termination in regulation of ermK. J. Bacteriol. 179:2065-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Groote, M. A., and G. Huitt. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 42:1756-1763. [DOI] [PubMed] [Google Scholar]
- 16.Geiman, D. E., T. R. Raghunand, N. Agarwal, and W. R. Bishai. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith, D. E., T. Aksamit, B. A. Brown-Elliott, A. Catanzaro, C. Daley, F. Gordin, S. M. Holland, R. Horsburgh, G. Huitt, M. F. Iademarco, M. Iseman, K. Olivier, S. Ruoss, C. F. von Reyn, R. J. Wallace, Jr., and K. Winthrop. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367-416. [DOI] [PubMed] [Google Scholar]
- 18.Griffith, D. E., B. A. Brown-Elliot, R. J. Blinkhorn, and R. J. Wallace, Jr. 1993. Treatment of Mycobacterium abscessus lung disease with clarithromycin. Am. Rev. Respir. Dis. 147:A917. [Google Scholar]
- 19.Griffith, D. E., and R. J. Wallace, Jr. 1996. New developments in the treatment of nontuberculous mycobacterial (NTM) disease. Semin. Respir. Infect. 11:301-310. [PubMed] [Google Scholar]
- 20.Gryczan, T. J., G. Grandi, J. Hahn, R. Grandi, and D. Dubnau. 1980. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 8:6081-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, H. Y., Y. Kook, Y. J. Yun, C. G. Park, N. Y. Lee, T. S. Shim, B. J. Kim, and Y. H. Kook. 2008. Proportion of Mycobacterium massiliense and Mycobacterium bolletii in Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J. Clin. Microbiol. 46:3384-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak, J. H., E. C. Choi, and B. Weisblum. 1991. Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J. Bacteriol. 173:4725-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, M., and S. Douthwaite. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen, C. T., L. Jakobsen, K. Buriankova, F. Doucet-Populaire, J. L. Pernodet, and S. Douthwaite. 2005. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis. J. Biol. Chem. 280:38942-38947. [DOI] [PubMed] [Google Scholar]
- 25.Madsen, C. T., L. Jakobsen, and S. Douthwaite. 2005. Mycobacterium smegmatis Erm(38) is a reluctant dimethyltransferase. Antimicrob. Agents Chemother. 49:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 28.Meier, A., L. Heifets, R. J. Wallace, Jr., Y. Zhang, B. A. Brown, P. Sander, and E. C. Böttger. 1996. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J. Infect. Dis. 174:354-360. [DOI] [PubMed] [Google Scholar]
- 29.Min, Y.-H., A.-R. Kwon, E.-J. Yoon, M.-J. Shim, and E.-C. Choi. 2008. Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob. Agents Chemother. 52:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, R. P., L. Nguyen, J. Gatfield, K. Visconti, K. Nguyen, D. Schnappinger, S. Ehrt, Y. Liu, L. Heifets, J. Pieters, G. Schoolnik, and C. J. Thompson. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 102:12200-12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nash, K. A. 2003. Intrinsic macrolide resistance in Mycobacterium smegmatis is conferred by a novel erm gene, erm(38). Antimicrob. Agents Chemother. 47:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nash, K. A., N. Andini, Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace, Jr. 2006. Intrinsic macrolide resistance in rapidly growing mycobacteria. Antimicrob. Agents Chemother. 50:3476-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nash, K. A., Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace, Jr. 2005. Molecular basis of intrinsic macrolide resistance in clinical isolates of Mycobacterium fortuitum. J. Antimicrob. Chemother. 55:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes, 1st ed. Approved Standard M24-A, 3. NCCLS, Wayne, PA. [PubMed]
- 35.Olivier, K. N., D. J. Weber, R. J. Wallace, Jr., A. R. Faiz, J.-H. Lee, Y. Zhang, B. A. Brown-Elliott, A. Handler, R. W. Wilson, M. S. Schechter, L. J. Edwards, S. Chakraborti, M. R. Knowles, et al. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:828-834. [DOI] [PubMed] [Google Scholar]
- 36.Petrini, B. 2006. Mycobacterium abscessus: an emerging rapid-growing potential pathogen. APMIS 114:319-328. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigue, S., J. Brodeur, P. E. Jacques, A. L. Gervais, R. Brzezinski, and L. Gaudreau. 2007. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 41.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1-research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, D., and L. S. Young. 2004. Nontuberculous mycobacterial infections: a clinical review. Infection 32:257-270. [DOI] [PubMed] [Google Scholar]
- 44.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., A. Meier, B. A. Brown, Y. Zhang, P. Sander, G. O. Onyi, and E. C. Böttger. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhanel, G. G., M. Walters, A. Noreddin, L. M. Vercaigne, A. Wierzbowski, J. M. Embil, A. S. Gin, S. Douthwaite, and D. J. Hoban. 2002. The ketolides: a critical review. Drugs 62:1771-1804. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]