Abstract
The hepatitis C virus (HCV) NS3/4A serine protease has been explored as a target for the inhibition of viral replication in preclinical models and in HCV-infected patients. TMC435350 is a highly specific and potent inhibitor of NS3/4A protease selected from a series of novel macrocyclic inhibitors. In biochemical assays using NS3/4A proteases of genotypes 1a and 1b, inhibition constants of 0.5 and 0.4 nM, respectively, were determined. TMC435350 inhibited HCV replication in a cellular assay (subgenomic 1b replicon) with a half-maximal effective concentration (EC50) of 8 nM and a selectivity index of 5,875. The compound was synergistic with alpha interferon and an NS5B inhibitor in the replicon model and additive with ribavirin. In rats, TMC435350 was extensively distributed to the liver and intestinal tract (tissue/plasma area under the concentration-time curve ratios of >35), and the absolute bioavailability was 44% after a single oral administration. Compound concentrations detected in both plasma and liver at 8 h postdosing were above the EC99 value measured in the replicon. In conclusion, given the selective and potent in vitro anti-HCV activity, the potential for combination with other anti-HCV agents, and the favorable pharmacokinetic profile, TMC435350 has been selected for clinical development.
Hepatitis C virus (HCV) was discovered in 1989 as the etiological agent of non-A, non-B hepatitis (7, 20). The subsequent development of diagnostics and blood screening has considerably reduced the rate of new infections (43). However, the chronic nature of the disease and possible long-term liver damage have led to the current global health burden with an estimated 120 to 170 million persons infected (9, 13).
Hepatitis C is mainly transmitted by blood contact, and estimates of the frequency at which exposure results in chronic infection range from 50 to 80% (33). Over decades, a considerable number of infected persons develop fibrosis, cirrhosis, and hepatocellular carcinoma, with chronic HCV infection being the leading cause of liver transplantation (12). There are six major HCV genotypes and multiple subtypes. Genotype 1 is predominant in Europe, North America, Japan, and China, while genotypes 2 and 3 are present in the Mediterranean countries, Far East, and Europe (41). The current standard of care consists of a combination therapy of weekly pegylated alpha interferon (IFN-α) and twice-daily ribavirin and is able to cure ∼80% of patients infected with genotype 2 or 3 but only 40 to 50% of patients infected with genotype 1 (42). Apart from the low success rate in genotype 1 patients, treatment is also associated with a range of side effects, including flu-like symptoms, anemia, and depression (32). Novel, safer, and more potent drugs that target viral replication are needed to improve the treatment outcomes for patients infected with HCV. There are a number of such investigational drugs currently undergoing clinical development.
HCV is a member of the Flaviviridae family of viruses in the Hepacivirus genus and is encoded by a 9.6-kb positive-sense, single-stranded RNA genome. After entry into the cell, the viral genome is released, translated, and processed by host and viral proteases in a series of co- and posttranslational cleavage events. The genome encodes four structural proteins including the core protein, envelope glycoproteins E1 and E2, and p7. The nonstructural (NS) proteins are liberated from the polypeptide chain by autoproteolysis. The processing of the polyprotein at the NS2/NS3 site is mediated in cis by the cysteine-like protease NS2, followed by cleavage at the remaining four cleavage sites mediated by NS3 (28). Full protease activity of NS3 requires the short NS4A peptide as a cofactor for enzymatic function, stability, and anchoring to the endoplasmic reticulum (47).
Increasing molecular knowledge of HCV replication has facilitated the development of specific targeted antivirals. Examples include both nucleoside and nonnucleoside inhibitors of the NS5B polymerase and peptidomimetic substrate/product-based inhibitors of NS3/4A protease (15). The impressive viral load reduction observed during clinical trials with protease inhibitors such as ciluprevir (BILN-2061), boceprevir (SCH503034), and telaprevir (VX-950) and the improved sustained virological response rates shown with telaprevir and boceprevir hold promise for the future therapy of HCV (21, 22, 32, 38, 40).
Here we present the biological profile of TMC435350, a novel inhibitor of the HCV NS3/4A serine protease, using enzymatic, cellular, and rat pharmacokinetic experiments.
MATERIALS AND METHODS
Materials.
TMC435350 was prepared by a method described previously (36). Human recombinant IFN-α was purchased from Calbiochem (La Jolla, CA). Ribavirin was purchased from Sigma Aldrich (St. Louis, MO). The investigational NS5B nucleoside inhibitor NM-107 was purchased from Toronto Research Chemicals (North York, Canada). The NS5B palm domain II inhibitor HCV-796 was synthesized as described earlier (6).
Cells.
Huh7, HepG2, HEK-293T, HT-1080, MT-4, MRC-5, and SAOS-2 cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum and 2 mM l-glutamine. Huh7-Luc cells contained the genotype 1b (con1 based) bicistronic subgenomic replicon (clone ET) encoding a firefly luciferase reporter with cell culture adaptive mutations E1202G, T1280I, and K1846T in the HCV genome. These cells were kindly provided by R. Bartenschlager (adapted from Lohmann et al. [30, 31]). Huh7-SGcon1b cells (i.e., a genotype 1b [con1 based] subgenomic replicon clone with cell culture adaptive mutation S2204I) were obtained from Apath LLC (30, 31). Huh7-SG1a H77 cells (i.e., a genotype 1a [H77 based] subgenomic replicon clone with cell culture adaptive mutations P1496L and S2204I [5]) were obtained from Apath. Replicon cells were maintained in medium as described above with the addition of 0.75 mg/ml G418 (Invitrogen, Carlsbad, CA). Human peripheral blood mononuclear cells (PBMC) were isolated from donor buffy coats by density gradient centrifugation (Ficoll isolation kits; Pharmacia Biotech, Uppsala, Sweden) for the evaluation of cytostatic and cytotoxic activity.
Bacterial expression and purification of recombinant NS3.
The cDNA encoding NS3 amino acids 1 to 191 (H77; genotype 1a) and amino acids 1 to 181 (con1b; genotype 1b) were cloned into pET21d+ and pET16b+, respectively (Novagen, Madison, WI). The NS3 expression constructs were then transformed into Escherichia coli BL21(DE3). Bacterial NS3 expression was induced with IPTG (isopropyl β-d-1-thiogalactopyranoside; 0.4 mM). Three hours after induction, cells were harvested by centrifugation and lysed. Cell debris was removed through ultracentrifugation at 20,000 × g, and clarified cell lysate was further purified by affinity chromatography (Histrap FF crude column; GE Healthcare, Chicago, IL). Fractions containing NS3 protein were then further purified and desalted on prepacked 26/10 HiPrep desalting columns (GE Healthcare) and SP-Sepharose columns (GE Healthcare). Protein purity was assessed on Nu PAGE precast gels (Invitrogen). Purified NS3 samples were concentrated using Centri-Prep concentrators (Millipore, Billerica, MA), and protein concentrations were determined by Bradford assay (Pierce, Rockford, IL).
NS3/4A protease assay.
In vitro inhibition of NS3/4A activity was determined using a fluorescence resonance energy transfer cleavage assay with the RetS1 peptide substrate (Anaspec, San Jose, CA), derived from the genotype 1a NS4A-4B junction, and bacterially expressed full-length NS3 protease domain, supplemented with an NS4A peptide (Anaspec). Briefly, NS3/4A was preincubated in the presence of TMC435350 for 10 min, and then the RetS1 substrate was added and fluorescence was continuously measured for 20 min (excitation, 355 nm; emission, 500 nm). Cleavage of the substrate was expressed as a percentage of the cleavage seen with the vehicle control.
Calculation of Ki.
All inhibitors were analyzed assuming competitive inhibition: Ki = IC50/(1 + S/Km). The Michaelis-Menten constant, Km, was determined graphically with initial velocity measurements obtained at various substrate concentrations (S). The half-maximal inhibitory concentration (IC50) value was calculated from the inhibition values of a series of TMC435350 concentrations using Grafit (Erithacus Software, London, United Kingdom) and Graphpad (GraphPad Software, San Diego, CA).
Host cellular protease assays.
Inhibition of human cathepsins S, L, B, E, and D was measured using fluorescence-based assays as previously described (3, 44). Human cathepsins B, D, and L were obtained from Calbiochem (La Jolla, CA); human cathepsins S and E were expressed in the baculovirus system (3).
Inhibition of other proteases was determined by measuring hydrolysis of the following specific para-nitroaniline-tagged peptides according to the manufacturer's instructions (American Diagnostica, Greenwich, CT, unless described otherwise): Spectrozyme CATG for chymase, cathepsin G, and chymotrypsin; Spectrozyme HLE for human leukocyte elastase; Spectrozyme PKal for kallikrein; S-2765 (DiaPharma, West Chester, OH) for factor Xa; Spectrozyme tPA for tissue plasminogen activator; Spectrozyme PL for plasmin; Spectrozyme TH for thrombin and tryptase; S-2765 (DiaPharma) for trypsin; Spectrozyme FV11a for factor VIIa; L1335 (BACHEM, Bubendorf, Switzerland) for proteinase 3; Spectrozyme UK for urokinase; and Spectrozyme PL for streptokinase.
Cathepsin S activity was also measured in a cellular invariant chain degradation assay using the human JY B-cell line as previously described (44).
HCV replicon inhibition assays.
For the luciferase reporter assay, Huh7-Luc cells were seeded at a density of 2,500 cells/well in a 384-well plate in Dulbecco's modified Eagle's medium plus 10% fetal calf serum and incubated with a range of concentrations of serially diluted TMC435350, in a final dimethyl sulfoxide (DMSO) concentration of 0.5% in the absence of G418. After 72 h of incubation, Steady Lite reagent (PerkinElmer, Waltham, MA) was added in a 1:1 ratio to the medium, and luciferase signal was measured using a ViewLux reader (PerkinElmer).
For RNA detection (quantitative reverse transcriptase PCR [qRT-PCR]), replicon-containing cells were seeded at a density of 3,000 for Huh7-Luc cells and 6,000 cells for con1b and SG1a in 96-well plates and incubated for 72 h, with a compound dilution range containing 0.5% DMSO and no G418. Subsequently, cells were lysed with Cell-to-cDNA lysis buffer (Ambion, Austin, TX) and lysate was submitted to a standard reverse transcription procedure for 30 min at 42°C using the Expand RT-PCR kit (Roche, Basel, Switzerland) and an HCV 5′-untranscribed region (UTR)-specific primer (HCV-270R; 5′-GACCCAACACTACTCGGCTA-3′) and a primer specific for the single cellular transcript of the ribosomal subunit RPL13 gene as a cellular RNA control (RPL13A-Rev; 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′). Following reverse transcription, the cDNA was added to an RT-PCR master mix (Applied Biosystems, Foster City, CA) containing primers and probes for the HCV 5′-UTR (reverse primer, HCV-270R; forward primer, HCV-40F [5′-CTCCCCTGTGAGGAACTAACTG-3′]; probe, FAM-BHQ1 [5′-TGGCGTTAGTATGAGTGTCGTGCAG-3′]) and cellular RPL13A (reverse primer, RPL13A-Rev; forward primer, RPL13A-Fw [5′-CCTGGAGGAGAAGAGGAAAGAGA-3′]; probe, YakimaYellow-BHQ1 [5′-TCCACTACCGGAAGAAGAAACAGCT-3′]). PCRs were run on an ABI PRISM 7900 (Applied Biosystems) using the following program: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The difference in the threshold cycle number between treated and mock wells was used to calculate percent inhibition. The 50% effective concentration (EC50) and 50% cytotoxic concentration (CC50) values were determined directly from dose-response curves from cell-based experiments.
The 9-day replicon assay was performed essentially as described previously (25, 26). Briefly, replicon cells (500 cells/well in 96-well plates) were treated for 9 days with a range of concentrations of TMC435350 in the absence of G418. For combination studies, TMC435350 was used alone or in combination with recombinant IFN-α (at 1 × EC50). Medium containing compounds was refreshed every 3 days, and cells were harvested on days 3, 6, and 9 of incubation at 37°C in a humidified 5% CO2 atmosphere. HCV RNA and cellular RPL13A transcript levels were quantified as described above, and HCV RNA levels were normalized to RPL13A.
Immunoblotting.
Replicon cells were seeded at a density of 60,000 cells per well in a 24-well plate and treated for 72 h with a range of concentrations of TMC435350 as described above. Cells were lysed using M-Per (mammalian protein extraction reagent; Pierce, Rockford, IL), and Western blotting was done following standard procedures (iBlot system; Invitrogen). Proteins were electroblotted onto nitrocellulose, which was subsequently blocked and further incubated with HCV NS5B-specific antisera (raised in rabbits by immunization with bacterially expressed NS5B from HCV strain J4), in phosphate-buffered saline containing 5% milk and 0.1% Tween 20, for 1 h at room temperature. The detection system consisted of horseradish peroxidase-labeled anti-rabbit immunoglobulin G (IgG; Amersham, Buckinghamshire, United Kingdom) and Super Signal system (Pierce). Peroxidase signal was detected using LAS3000 (Fuji, Tokyo, Japan). Subsequently, antibodies were stripped from the membrane using Re-Blot Plus (Chemicon, Billerica, MA) for 30 min at room temperature and reprobed with mouse monoclonal antitubulin antibody (Sigma-Aldrich, St. Louis, MO). Staining with horseradish peroxidase-labeled antimouse IgG (Amersham) and detection were performed as described above.
Cytoproliferation assays.
The effect of TMC435350 on the proliferation of different cell types was analyzed after 3 to 4 days of compound exposure by incubation with resazurin (Sigma-Aldrich, St. Louis, MO) followed by detection in a fluorescence reader. Inhibition curves were prepared, and CC50 values were calculated.
Isolated PBMC were proliferated in the presence of staphylococcal enterotoxin B in the presence of a range of concentrations of compounds and matching vehicle control (DMSO) dilutions to determine cytostatic activity. The incorporation of labeled [3H]thymidine into the DNA of proliferating cells was measured 7 h after addition of compound, using a Topcount (PerkinElmer). To measure the cytotoxic activity, the staphylococcal enterotoxin B-stimulated PBMC were incubated with the same range of TMC435350 concentrations as used for the cytoproliferation toxicity assay. After 72 h of incubation, the tetrazolium salt WST-1 was added and incubation was continued for another 6 h. Conversion of WST-1 to the dark red formazan product was measured using an enzyme-linked immunosorbent assay reader. The viability of the cells was compared to that of the untreated control. The CC50 and cytostatic (CsC50) values were determined from the dose-response curve.
Selectivity indices for the different cell lines were determined by dividing the CC50 values obtained in the respective cell lines by the EC50, as measured in the HCV Huh7-Luc replicon inhibition assay (luciferase signal detection) as described above (8 nM).
Viral inhibition assays.
Antiviral activities against West Nile virus (WNV-NY99/1; Vero cells), bovine viral diarrhea virus (BVDV-NADL; MDCK cells), yellow fever virus (YFV-17D; VeroE6 cells), Sindbis virus (Sin-V-339; Vero cells), influenza virus (influenza A Virginia/88; MDCK cells), vesicular stomatitis virus (MRC-5 cells), and herpes simplex virus type 2 (G strain; E6 Vero cells) were determined with a cytopathic effect inhibition assay (14). Activity against human immunodeficiency virus type 1 (IIIB; MT-4 cells) was assessed using an enhanced green fluorescent protein-based assay (17). Quantitative PCR was used for anti-human hepatitis B virus (HepG2.2.15 cells) activity (19). A tetrazolium-based colorimetric method was used to determine anti-respiratory syncytial virus (LO; HeLa/M cells) activity (1). A known positive control was included in each assay.
Protein binding.
The HCV replicon inhibition assays were performed as described above in the presence of extra protein or serum (α-1 acid glycoprotein; human serum albumin, and human serum) at the concentrations indicated in Table 3.
TABLE 3.
Effect of protein binding on antiviral activity of TMC435350
| Proteina | EC50 (nM)
|
Relative increase in EC50 vs 10% FCS | No. of independent measurements | |
|---|---|---|---|---|
| Median | IQRb | |||
| 10% FCS | 8.1 | 7.1-11.8 | 134 | |
| 10% FCS + 1 mg/ml AAG | 11.4 | 6.2-14.2 | 1.4 | 18 |
| 10% FCS + 40 mg/ml HSA | 14.1 | 9.7-17.8 | 1.7 | 19 |
| 10% FCS+ 1 mg/ml AAG+ 40 mg/ml HSA | 14.6 | 10.4-22.4 | 1.8 | 19 |
| 40% HS | 11.2 | 8.9-15.2 | 1.4 | 8 |
AAG, α-1 acid glycoprotein; HSA, human serum albumin; HS, human serum.
IQR, interquartile range.
Synergy analysis.
Huh7-Luc replicon cells were treated for 3 days with serially diluted TMC435350 (0.15 to 39.3 nM) in combination with serially diluted IFN-α (0.109 to 14 U/ml), ribavirin (0.32 to 83.6 μM), NM-107 (0.11 to 29.2 μM), or HCV-796 (2 to 61 nM) according to the checkerboard method. The inhibition data were analyzed using the Loewe additivity model (29) with CalcuSyn (Biosoft, Ferguson, MO) (8).
Pharmacokinetics and tissue distribution.
Twenty-four male specific-pathogen-free Sprague-Dawley rats, weighing between 200 and 300 g at the time of dosing, were divided into eight groups of three rats each. Seven groups were dosed orally (p.o.) by gastric intubation of a vitamin E acetate-d-α-tocopheryl polyethylene glycol 1000 succinate-polyethylene glycol 400 solution of TMC435350 at 2 ml/kg body weight to provide a dose of 40 mg/kg. One group was dosed intravenously (i.v.) by slow bolus injection in a tail vein of a 20% 2-hydroxypropyl-β-cyclodextrin formulation of TMC435350 (containing TMC435350, 100 mg/ml 2-hydroxypropyl-β-cyclodextrin, 0.1 N NaOH to pH 8.0 ± 0.1, and mannitol- and pyrogen-free water) at 2 ml/kg body weight to provide a dose of 4 mg/kg. Water and food were available ad libitum during the study.
Blood samples were collected from the orbital venous plexus at 30 min and 1, 2, 4, 8, 24, and 31 h after p.o. dose administration and at 7 min, 20 min, and 1, 3, 8, 24, and 31 h after i.v. dose administration. Samples were centrifuged at 5°C at 1,500 × g for 10 min to allow plasma separation. Tissues were dissected from orally dosed animals.
Samples were analyzed individually for TMC435350 by liquid chromatography-tandem mass spectrometry with a lower limit of quantification of 1 ng/ml (plasma) and 5 ng/g (tissue). Mean ± standard deviation (n = 3) concentrations of TMC435350 were calculated per administration route, per dose level, and per sampling time.
Tissue concentration and individual plasma concentration versus time profiles were subjected to a noncompartmental pharmacokinetic analysis using validated WinNonlin software (v 4.0.1a; Pharsight, Mountain View, CA). An estimate of the absolute bioavailability (Fabs) of TMC435350 was obtained by dividing the dose-normalized mean value of the area under the plasma drug concentration-time curve (AUC) after oral dose administration by the dose-normalized mean AUC value after i.v. dose administration.
RESULTS
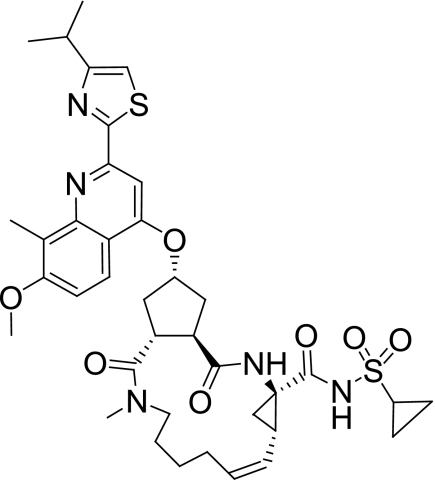
Medicinal chemistry lead optimization was performed on a series of cyclopentane-containing NS3/4A protease inhibitors leading to the discovery of TMC435350 (Fig. 1) (36). Here, we report the biological and pharmacokinetic profile of TMC435350.
FIG. 1.
Structure of TMC435350.
Specific inhibition of HCV NS3/4A enzyme.
The effect of TMC435350 on NS3/4A proteolytic activity was determined in biochemical protease assays using the RetS1 substrate (derived from the genotype 1a NS4A-4B junction) and bacterially expressed genotype 1a (H77) and 1b (con1b) protease domains. Compound inhibition was time dependent, and the overall Kis were estimated to be 0.5 nM for genotype 1a and 0.4 nM for genotype 1b, respectively.
To determine if TMC435350 had additional activities on cellular proteases, a panel of human enzymes was incubated with cognate substrates and compound. TMC435350 was tested initially at a fixed concentration of 10 μM, and if more than 50% inhibition was observed, the activity of TMC435350 against this specific protease was again determined using a dilution range to calculate an IC50. In vitro inhibition values on host proteases were at least over 1,000 times higher than those for HCV NS3/4A protease. TMC435350 only exhibited submicromolar activity on human cathepsin S with an IC50 of 0.8 μM. However, in a relevant cellular assay for cathepsin S activity (p10 fragment accumulation assay), TMC435350 was found to be inactive up to the highest concentrations tested (10 μM), suggesting that the observed inhibitory effect in vitro is not likely to be relevant in vivo.
Selective inhibition of HCV replication.
Studies of HCV replication in vitro have been hampered for many years by the lack of robust cell culture systems for HCV. The development of the HCV replicon system (4, 31) and recently the description of HCV infectious cell culture systems (27, 46, 48) are breakthroughs that have greatly enabled HCV cell culture studies and the discovery of antivirals. The anti-HCV activity of TMC435350 was first determined in cell culture using the Huh7-Luc HCV genotype 1b replicon cell line containing a luciferase reporter (Fig. 2A and Table 1). In this replicon system, the region of the HCV genome coding for structural proteins has been replaced by marker genes. A gene coding for neomycin allows selection and long-term passage of the Huh7 hepatoma cells containing the HCV genome, and a luciferase gene facilitates the monitoring of HCV replication. After 72 h of incubation, percent inhibition of the luciferase signal was determined at each dilution point relative to medium control. The inhibitory effect was dose dependent, and the EC50 and EC90 values determined for TMC435350 were 8 nM and 24 nM, respectively (Table 1). The luciferase reporter data from the replicon assay were corroborated by direct qRT-PCR quantification of replicon RNA (EC50 of 13 nM).
FIG. 2.
Inhibition of HCV replication in genotype 1b replicon cells treated with TMC435350. (A) Huh7-Luc replicon cells were treated for 72 h with a dilution range of TMC435350 concentrations relative to control conditions (no compound). The dose-response curve of % inhibition of luciferase signal is shown; each point represents the mean and standard deviation of four independent experiments. The EC50 is indicated. (B) Dose-dependent reduction of HCV protein expression in TMC435350-treated replicon cells using an immunoblot. Equal amounts of each cell lysate were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent immunoblot analysis with specific antibodies against NS5B and α-tubulin. Molecular masses are given in kDa.
TABLE 1.
Antiviral activity of TMC435350 in genotype 1a and 1b repliconsa
| Replicon cell line (readout) | Subtype | EC50 (nM)
|
EC90 (nM)
|
No. of independent measurements | ||
|---|---|---|---|---|---|---|
| Median | IQRb | Median | IQR | |||
| Huh7-Luc (luciferase) | 1b | 8.1 | 4.5-11.9 | 23.6 | 15.3-28.2 | 134 |
| Huh7-Luc (qRT-PCR) | 1b | 13.0 | 11.3-14.4 | 28.7 | 27.6-57.3 | 21 |
| Huh7-SGcon1b (qRT-PCR) | 1b | 25.2 | 17.9-34.6 | 118.0 | 102.4-144.6 | 17 |
| Huh7-SG1a H77 (qRT-PCR) | 1a | 28.4 | 19.0-39.7 | 120.4 | 104.4-187.0 | 19 |
Inhibition of HCV RNA replication in three different HCV replicon cell lines using luciferase or qRT-PCR readout is shown.
IQR, interquartile range.
To exclude a replicon clone-specific effect and to determine the potency of TMC435350 on a genotype 1a replicon, Huh7-derived replicon cells engineered with either genotype 1a (H77) or genotype 1b (con1b) sequences were treated with TMC435350 and HCV replicon RNA levels were measured (Table 1). EC50s of 28 and 25 nM, respectively, were determined. In addition, no significant effect of TMC435350 on the cellular ribosomal protein RPL13A transcript level was observed in any of the cell lines tested, supporting the specificity of the inhibitor for HCV rather than having an impact on replicon cell viability or division (data not shown).
Anti-HCV activity of TMC435350 in the replicon system was also assessed at the protein level by immunoblot analysis. After a 72-h incubation with TMC435350, a dose-dependent reduction of NS5B expression was shown in replicon cell lysates, while no reduction of the cellular protein α-tubulin was observed (Fig. 2B).
Together, these data suggest potent and specific activity of TMC435350 against HCV genotype 1a and 1b replication.
The effect of TMC435350 on the viability of different human cell lines was assessed using a 3- to 4-day proliferation assay with a resazurin readout. In the Huh7-Luc HCV genotype 1b replicon cell line, the CC50 was found to be 47 μM, indicating specific inhibition of HCV by TMC435350 (selectivity index, CC50/EC50 = 5,875). For hepatoma Huh7 and HepG2 cells and kidney HEK-293T cells, no cytotoxicity was observed with TMC435350 concentrations of up to 42 μM, which translates into a selectivity index (using an EC50 of 8 nM) for HCV of >5,250. In the human T-cell line MT-4 and cells derived from lung (MRC-5), bone (SAOS-2), and connective tissue (HT-1080), CC50 values were slightly lower (16 to 21 μM), which results in a selectivity index of ≥2,000.
TMC435350 was also studied in PBMC isolated from human donors for effects on cell viability. No cytotoxic (CC50 = 33 μM) or cytostatic (CsC50 = 10 μM) effect was detected below 10 μM, resulting in a selectivity index clearly above 1,000 for PBMC. Long-term culture of Huh7 replicon cells for up to 5 weeks with 1 μM TMC435350 had no effect on cellular viability (data not shown).
Selectivity toward different DNA and RNA viruses.
To determine antiviral specificity, the activity of TMC435350 was tested against a panel of DNA and RNA viruses. No antiviral activity was observed up to the highest concentration used for all viruses tested (10 to 100 μM; data not shown).
In vitro combination studies.
The effect of combining TMC435350 with different anti-HCV agents was explored in Huh7-Luc replicon cells. Combinations of TMC435350 with IFN-α, ribavirin, the polymerase nucleoside inhibitor NM-107, or the polymerase nonnucleoside inhibitor HCV-796, were analyzed. Anti-HCV activity was determined using luciferase readout, and the effects of combination were calculated using the Loewe additivity model (Fig. 3 and Table 2).
FIG. 3.
Mutual influence between TMC435350 and IFN-α as analyzed by the Loewe additivity model. Huh7-Luc replicon cells were treated for 72 h with compound combinations. The combination index value for an effective dose of 50%, 75%, or 90% inhibition (ED50, ED75, and ED90, respectively) was calculated. The graph is a representative example of the data in Table 2.
TABLE 2.
Mutual influence between TMC435350 and IFN-α, ribavirin, NM-107, and HCV-796 as analyzed by the Loewe additivity modela
| Combination compound | CI value for:
|
Influenceb | ||
|---|---|---|---|---|
| ED50 | ED75 | ED90 | ||
| IFN-α | 0.77 | 0.57 | 0.43 | Synergistic |
| Ribavirin | 0.98 | 0.94 | 0.92 | Additive |
| NM-107 | 0.58 | 0.64 | 0.64 | Synergistic |
| HCV-796 | 0.86 | 0.88 | 0.92 | Additive to synergistic |
Huh7-Luc replicon cells were treated for 72 h with combinations of the compounds shown, and the combination index (CI) value for ED50, ED75, or ED90 was calculated.
CI values of <0.9, 0.9 to 1.1, and >1.1 indicate synergy, an additive effect, or antagonism, respectively.
No antagonism of anti-HCV activity was found between TMC435350 and any of the other molecules, and no evidence for changes in cytotoxicity (CC50) was observed. With regard to specific combinations, ribavirin showed an additive effect, whereas a clear synergy between TMC435350 and IFN-α was observed. The combination of TMC435350 and the polymerase nucleoside inhibitor NM-107 showed a synergistic effect. An additive to synergistic effect was found when combining TMC435350 with the NS5B palm domain inhibitor HCV-796. To confirm these results, a second set of experiments were performed, and the data were evaluated using the MacSynergy software based on the algorithm of Prichard and Shipman (35). The results confirmed the findings described here (see the supplemental material).
To further explore the effect of combining TMC435350 with IFN-α, Huh7-Luc replicon cells were treated for 9 days with either molecule alone or in combination (Fig. 4). Individual treatment with 5 nM TMC435350 or with 0.5 U/ml IFN-α alone resulted in a 1-log10 reduction of replicon RNA after 9 days. When the two molecules were combined at a concentration of 5 nM for TMC435350 and 0.5 U/ml for IFN-α, a 1.7-log10 drop was observed. Incubation of the cells with 50 nM TMC435350 together with 0.5 U/ml IFN-α reduced HCV RNA levels to −4.5 log10 after 9 days of treatment, which was 1.3 log10 more than that observed with 50 nM TMC435350 alone.
FIG. 4.
Nine-day incubation of genotype 1b replicon cells with TMC435350 alone, IFN-α alone, and TMC435350 in combination with IFN-α. Huh7-Luc replicon cells were harvested after 3, 6, or 9 days of treatment, and RNA levels were assessed by qRT-PCR. The normalized (host RPL13A) HCV RNA levels of treated replicon cells relative to untreated cells are plotted on a log10 scale. □, 0.5 U/ml IFN-α; ▵, 5 nM TMC435350; ▴, 5 nM TMC435350 plus 0.5 U/ml IFN-α; ○, 50 nM TMC435350; •, 50 nM TMC435350 plus 0.5 U/ml IFN-α. Each point represents the mean value of three independent experiments.
Protein binding.
Components of human serum have been shown to bind to and reduce the activity of a number of drugs. Therefore, as part of our preclinical evaluation of TMC435350, we tested the antiviral activity of TMC435350 in the presence of human plasma proteins (α-1 acid glycoprotein and human serum albumin alone or in combination) and human serum (Table 3). Replicon EC50s increased only 1.4- to 1.8-fold relative to the standard conditions.
Pharmacokinetics and tissue distribution.
The plasma pharmacokinetics and oral bioavailability of TMC435350 were studied in male Sprague-Dawley rats after a single p.o. or i.v. administration of 40 or 4 mg/kg, respectively (Table 4).
TABLE 4.
Pharmacokinetic parameters of TMC435350a
| Parameter | Result for TMC435350 dose (route):
|
|
|---|---|---|
| 40 mg/kg (p.o.) | 4 mg/kg (i.v.) | |
| Clearance (liters/h/kg) | 2.3 | |
| V (liters/kg) | 5.3 | |
| Cmax (ng/ml) | 1,430 | |
| tmax (h) | 2.0 | |
| t1/2 8-24 h (h) | 2.6 | 4 |
| AUC0-24 h (ng·h/ml) | 7,740 | 1,740 |
| AUC0-∞ (ng·h/ml) | 7,750 | 1,750 |
| Fabs (%) | 44 | |
Single-dose pharmacokinetic studies were conducted with TMC435350 in Sprague-Dawley rats by both the oral and i.v. routes.
After i.v. administration of an aqueous β-cyclodextrin solution of TMC435350 at a dose of 4 mg/kg, the mean plasma concentration at 0 h (C0) was estimated to be 3,560 ng/ml. Plasma concentrations declined with an average half-life for 8 to 24 h (t1/2 8-24 h) of 4 h, and total plasma clearance amounted to 2.3 liters/h/kg, which is moderate. The apparent volume of distribution at steady-state (V) was 5.3 liters/kg, indicating moderate distribution of the compound to the tissues.
After oral administration of a polyethylene glycol solution of TMC435350 at a dose of 40 mg/kg, the mean peak plasma concentration (Cmax) amounted to 1,430 ng/ml and was observed 2 h postdose (time to peak plasma concentration [tmax]). The absolute bioavailability (Fabs) in rats of TMC435350 (calculated using values for AUC from 0 to infinity [AUC0-∞]) was estimated to be 44%.
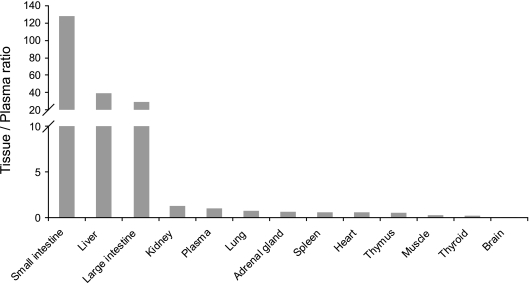
The tissue distribution of TMC435350 was evaluated in male Sprague-Dawley rats by comparing tissue and plasma exposures at several time points up to 31 h after a single oral dose of 40 mg/kg (Fig. 5). The highest tissue/plasma AUC ratios were observed in the small intestine (ratio of 128), large intestine (ratio of 29), and liver (ratio of 39). In other tissues, concentrations were similar to or below those in plasma, with tissue/plasma ratios ranging between 0.5 and 1.3 in kidneys, adrenal glands, lung, spleen, heart, and thymus. Distribution of TMC435350 to muscle, thyroid gland, and brain was limited.
FIG. 5.
Tissue distribution of TMC435350 after a single oral administration of 40 mg/kg in Sprague-Dawley rats. AUC0-31 h (except for muscle, AUC0-8 h) tissue/plasma ratios of TMC435350 are shown. Each bar represents the mean value for three rats.
Tissue concentrations of TMC435350 reached maximum values within 4 h postdose in all tissues sampled, except the large intestine (tmax of 8 h) (data not shown). The TMC435350 concentration in liver remained above the EC99 up to 31 h postdosing, while the concentrations in plasma were above the EC99 at 8 h and around the EC50 at 24 h postdosing (Fig. 6).
FIG. 6.
Time-dependent exposure after single oral administration of 40 mg/kg TMC435350 in plasma and liver tissue of Sprague-Dawley rats. Plasma (⧫) and liver (▴) tissue concentrations of TMC435350 are plotted over time, and EC50, EC90, and EC99 values as determined for TMC435350 in Huh7-Luc replicon cells are indicated. Each point represents the mean value for three rats.
DISCUSSION
The treatment prospects for HCV-infected patients have improved considerably with the introduction of pegylated IFN-α-ribavirin combination therapy. However, further improvements in sustained virological response rates, tolerability, and treatment duration are still needed, especially for genotype 1 patients. It is generally expected that antiviral drugs that specifically inhibit the function of viral proteins essential for virus replication, such as NS3/4A protease and NS5B polymerase, will increase cure rates and shorten treatment duration.
There are essentially two chemically distinct classes of NS3 protease inhibitors: the α-ketoamide electrophilic trap-containing inhibitors (telaprevir and boceprevir) and the macrocyclic class (ciluprevir, ITMN-191/R-7227, and TMC435350) (32), with reported activity in patients infected with HCV. Telaprevir and boceprevir are the most advanced specifically targeted antivirals in clinical development for HCV, and results from phase 2b studies have been reported recently (21, 23).
The first protease inhibitor showing an impressive reduction in viral load was the macrocyclic inhibitor ciluprevir, which reduced HCV RNA by 2 to 3 log10 after 2 days of dosing. Further development of this molecule was stopped, due to concerns of cardiotoxicity observed in rhesus monkeys after administration of supratherapeutic doses (16, 22). Although the exact mechanism was not reported, the cardiovascular toxicity observed with this pioneer drug is assumed to be compound specific.
An extensive medicinal chemistry exploration of novel macrocyclic molecules was initiated (2, 36), using biochemical protease assays and the cellular replicon system to identify active molecules. In addition, a cascade of early ADME (absorption, distribution, metabolism, and excretion) assays and pharmacokinetic analysis in rats were employed to identify potent inhibitors with good oral bioavailability. Initial lead compounds with good activity still had a rather poor pharmacokinetic profile, providing the biggest challenge during lead optimization, as described elsewhere (36). More than 1,000 compounds were synthesized. Based on potent and specific anti-HCV activity as well as favorable preclinical pharmacokinetic properties, TMC435350 was selected for further characterization and ultimately for clinical development.
TMC435350 displayed potent inhibition of HCV NS3/4A protease of genotypes 1a and 1b, with Kis of 0.5 and 0.4 nM respectively. The compound is over 1,000 times less active against 20 human proteases, including human leukocyte elastase, trypsin, and chymotrypsin, indicating specificity of TMC435350 for the HCV NS3/4A protease.
Different genotype 1a and 1b replicons were inhibited with EC50s ranging from 8 nM to 28 nM, confirming the potency and selectivity of TMC435350. The anti-HCV effect of TMC435350 was also analyzed in a recent study using a different set of genotype 1 replicon cell lines (24). The EC50s of 2.8, 0.27, and 4.6 nM in Con1-, HCV-N, and H77-based replicon cell lines, respectively, obtained in that study confirmed the potent anti-HCV activity of TMC435350. The difference from the EC50s observed by us is not fully understood but could be due to the respective experimental conditions (such as replicon cell number, growth rate, and replication level). Such differences have been reported before for other HCV inhibitors (34). Interestingly, the TMC435350 inhibition in the replicon model also correlates well with the inhibition of replication of the HCV genotype 1a H77 strain in an infectious system (24). Using this system in an infectivity reduction assay, TMC435350 remained fully active, with an EC50 of 6.2 nM.
The cytotoxic and cytostatic concentrations of TMC435350 (CC50 and CsC50) obtained in a panel of cell lines derived from different tissues and primary PBMC were greater than 10 μM, resulting in a selectivity index of >1,000. In addition, TMC435350 showed no antiviral effect against different DNA and RNA viruses, including closely related Flaviviridae family members such as bovine viral diarrhea virus and yellow fever virus, further supporting the specificity of the drug candidate.
It is likely that future therapies will consist of specifically targeted antiviral drugs in combination with IFN-α and ribavirin in order to enhance the antiviral effect and to raise the threshold for resistance development, ultimately improving sustained virological response rates (32). To analyze the potential of combining TMC435 with other HCV inhibitors, studies with different classes of agents were performed in the replicon system using the Loewe additivity model for analysis. The combination of TMC435350 with IFN-α was synergistic, whereas the combination with ribavirin was additive, suggesting that TMC435350 could be useful in combination with IFN-α and ribavirin. The combination with the NS5B inhibitors NM-107 and HCV-796 also resulted in a synergistic effect in this assay. In addition, the combination of TMC435350 with IFN-α significantly improved the antiviral effect in a 9-day replicon assay, reducing HCV replicon RNA by 4.5 log10 in combination compared to 3.2 log10 for TMC435350 alone. A similar increase of anti-HCV activity in a 9-day replicon assay after combination with a low dose of IFN has been described for telaprevir (25). More importantly, the addition of IFN-α to telaprevir also increased the viral load reduction in patients from 4.4 to 5.5 log10 IU/ml after 14 days of dosing (10), suggesting that a similar positive effect of combination with IFN-α might be expected with TMC435350 in vivo. Adding to the value of combination therapy, we have also seen that combining TMC435350 with IFN-α reduces the emergence of drug-resistant replicon colonies (data not shown).
The potency of TMC435350 was complemented by other desirable preclinical qualities such as a low effect of functional protein binding and a minimal impact on cytoproliferation. The pharmacokinetic parameters in rats after p.o. and i.v. dosing suggest a favorable systemic exposure supportive of once-daily dosing of TMC435350. The elimination half-life in rats was 4 h, and plasma clearance was moderate. After single oral administration of 40 mg/kg to rats, considerably higher TMC435350 concentrations were achieved in the liver relative to blood plasma at all time points tested (liver/plasma ratio of 32:1 to 65:1), which is presumably important in the treatment of a liver disease like HCV infection. While plasma exposure dropped to around the EC50 at 24 h postdosing, the liver concentration remained above the replicon EC99.
HCV has been shown to have multiple interactions with the innate immune system, presumably as a mechanism to control pathways that play an important role in the antiviral immune defense (39). NS3/4A has been shown to cleave IPS and TRIF, two adapter proteins downstream of the immune RNA sensors Rig-I and TLR3, respectively (11). While it appears obvious that an NS3/4A inhibitor could restore this pathway alongside its direct antiviral activity, the drug concentration required to restore activity of these pathways was shown to be 100-fold more than the concentration needed to inhibit the HCV replicon (24). The reasons for this are not clear, and while it remains speculative as to whether the NS3/4A inhibitors have an added therapeutic value via an innate immunity mode of action, there is at least theoretical and in vitro experimental evidence to support this view (18). Our in vitro data are supported by in vivo data in healthy volunteers and patients (37, 45) that exposure to TMC435350 is sufficient to achieve levels in which restoration of the innate immune system is possible, providing a potential advantage of TMC435350 over drugs with lower trough levels.
Further studies to elucidate the biochemical interaction of TMC435350 with HCV protease and inhibition studies on other genotypes, as well as detailed in vitro resistance characterization, are ongoing. The in vitro biological profile of TMC435350 outlined in this study supports the ongoing evaluation of this investigational drug in patients as a once-daily inhibitor of HCV replication.
Supplementary Material
Acknowledgments
Numerous scientists from Tibotec and Medivir have contributed to this project. We thank Rob Thurmond and Lisa Minor (Johnson & Johnson PRD) for help with protease selectivity assays and the Beerse Preclinical Development Group for the pharmacokinetic studies. We thank Luc Geeraert for help with the preparation of the manuscript.
Footnotes
Published ahead of print on 26 January 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andries, K., M. Moeremans, T. Gevers, R. Willebrords, C. Sommen, J. Lacrampe, F. Janssens, and P. R. Wyde. 2003. Substituted benzimidazoles with nanomolar activity against respiratory syncytial virus. Antivir. Res. 60:209-219. [DOI] [PubMed] [Google Scholar]
- 2.Back, M., P.-O. Johansson, F. Wangsell, F. Thorstensson, I. Kvarnstrom, S. Ayesa, H. Wahling, M. Pelcman, K. Jansson, S. Lindstrom, H. Wallberg, B. Classon, C. Rydergard, L. Vrang, E. Hamelink, A. Hallberg, S. Rosenquist, and B. Samuelsson. 2007. Novel potent macrocyclic inhibitors of the hepatitis C virus NS3 protease: use of cyclopentane and cyclopentene P2-motifs. Bioorg. Med. Chem. 15:7184-7202. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. M., L. Karlsson, and R. L. Thurmond. 2003. Cloning, expression, purification, and activity of dog (Canis familiaris) and monkey (Saimiri boliviensis) cathepsin S. Protein Expr. Purif. 28:93-101. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R. 2002. Hepatitis C virus replicons: potential role for drug development. Nat. Rev. Drug Discov. 1:911-916. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, C. J., A. M. Del Vecchio, T. R. Bailey, B. A. Kulkarni, T. H. Faitg, S. R. Sherk, C. W. Blackledge, D. J. Rys, T. A. Lessen, J. Swestock, Y. Deng, T. J. Nitz, J. A. Reinhardt, H. Feng, and A. K. Saha. 31 October 2003. Benzofuran compounds, compositions and methods for treatment and prophylaxis of hepatitis C viral infections and associated diseases. PCT patent WO2004041201.
- 7.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 8.Chou, T. C., and P. Talaly. 1977. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J. Biol. Chem. 252:6438-6442. [PubMed] [Google Scholar]
- 9.Deuffic-Burban, S., T. Poynard, M. S. Sulkowski, and J. B. Wong. 2007. Estimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United States. J. Viral. Hepat. 14:107-115. [DOI] [PubMed] [Google Scholar]
- 10.Forestier, N., H. W. Reesink, C. J. Weegink, L. McNair, T. L. Kieffer, H.-M. Chu, S. Purdy, P. L. M. Jansen, and S. Zeuzem. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640-648. [DOI] [PubMed] [Google Scholar]
- 11.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 12.Ghobrial, R. M., R. Steadman, J. Gornbein, C. Lassman, C. D. Holt, P. Chen, D. G. Farmer, H. Yersiz, N. Danino, E. Collisson, A. Baquarizo, S. S. Han, S. Saab, L. I. Goldstein, J. A. Donovan, K. Esrason, and R. W. Busuttil. 2001. A 10-year experience of liver transplantation for hepatitis C: analysis of factors determining outcome in over 500 patients. Ann. Surg. 234:384-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Global Burden of Hepatitis C Working Group. 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20-29. [DOI] [PubMed] [Google Scholar]
- 14.Gong, E., T. Ivens, C. Van den Eynde, S. Hallenberger, and K. Hertogs. 2008. Development of antiviral assays for profiling compounds against a panel of positive-strand RNA viruses using ATP/luminescence readout. J. Virol. Methods 151:121-125. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, C. P., and P. A. Keller. 2005. Control of hepatitis C: a medicinal chemistry perspective. J. Med. Chem. 48:1-20. [DOI] [PubMed] [Google Scholar]
- 16.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C.-L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 17.Jochmans, D., J. Deval, B. Kesteleyn, H. Van Marck, E. Bettens, I. De Baere, P. Dehertogh, T. Ivens, M. Van Ginderen, B. Van Schoubroeck, M. Ehteshami, P. Wigerinck, M. Götte, and K. Hertogs. 2006. Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 80:12283-12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, C. L., D. M. Owen, and M. Gale, Jr. 2007. Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense. J. Biol. Chem. 282:10792-10803. [DOI] [PubMed] [Google Scholar]
- 19.Korba, B. E., and J. L. Gerin. 1992. Use of a standardized cell culture assay to assess activities of nucleoside analogs against hepatitis B virus replication. Antivir. Res. 19:55-70. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, G., Q. L. Choo, H. J. Alter, G. L. Gitnick, A. G. Redeker, R. H. Purcell, T. Miyamura, J. L. Dienstag, M. J. Alter, C. E. Stevens, et al. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364. [DOI] [PubMed] [Google Scholar]
- 21.Kwo, P., E. Lawitz, J. McCone, E. Schiff, J. Vierling, D. Pound, M. Davis, J. Galati, S. Gordon, N. Ravendhran, L. Rossaro, F. Anderson, I. Jacobson, R. Rubin, P. Mukhopadhyay, E. Chaudhri, L. Pedicone, and J. Albrecht. 2008. Interim results from HCV SPRINT-1: RVR/EVR from phase 2 study of boceprevir plus pegintron (peginterferon alfa-2b)/ribavirin in treatment-naive subjects with genotype-1 CHC. J. Hepatol. 48:S372. [Google Scholar]
- 22.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A.-M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M.-A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C.-L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 23.Lawitz, E., M. Rodriguez-Torres, A. J. Muir, T. L. Kieffer, L. McNair, A. Khunvichai, and J. G. McHutchison. 2008. Antiviral effects and safety of telaprevir, peginterferon alfa-2a, and ribavirin for 28 days in hepatitis C patients. J. Hepatology 49:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Liang, Y., H. Ishida, O. Lenz, T.-I. Lin, O. Nyanguile, K. Simmen, R. B. Pyles, N. Bourne, M. Yi, K. Li, and S. M. Lemon. 2008. Antiviral suppression versus restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology doi: 10.1053/j.gastro.2008.07.023. [DOI] [PubMed]
- 25.Lin, K., A. D. Kwong, and C. Lin. 2004. Combination of a hepatitis C virus NS3-NS4A protease inhibitor and alpha interferon synergistically inhibits viral RNA replication and facilitates viral RNA clearance in replicon cells. Antimicrob. Agents Chemother. 48:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, K., R. B. Perni, A. D. Kwong, and C. Lin. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 29.Loewe, S., and H. Muischnek. 1926. Effect of combinations: mathematical basis of problem. Arch. Exp. Pathol. Pharmakol. 114:313-326. [Google Scholar]
- 30.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 32.Manns, M. P., G. R. Foster, J. K. Rockstroh, S. Zeuzem, F. Zoulim, and M. Houghton. 2007. The way forward in HCV treatment—finding the right path. Nat. Rev. Drug Discov. 6:991-1000. [DOI] [PubMed] [Google Scholar]
- 33.Micallef, J. M., J. M. Kaldor, and G. J. Dore. 2006. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J. Viral Hepat. 13:34-41. [DOI] [PubMed] [Google Scholar]
- 34.Paeshuyse, J., I. Vliegen, L. Coelmont, P. Leyssen, O. Tabarrini, P. Herdewijn, H. Mittendorfer, J. Easmon, V. Cecchetti, R. Bartenschlager, G. Puerstinger, and J. Neyts. 2008. Comparative in vitro anti-hepatitis C virus activity of a selected series of polymerase, protease, and helicase inhibitors. Antimicrob. Agents Chemother. 52:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 36.Raboisson, P., H. de Kock, Å. Rosenquist, M. Nilsson, L. Salvador-Oden, T.-I. Lin, N. Roue, V. Ivanov, H. Wähling, K. Wickström, E. Hamelink, M. Edlund, L. Vrang, S. Vendeville, W. Van de Vreken, D. McGowan, A. Tahri, L. Hu, C. Boutton, O. Lenz, F. Delouvroy, G. Pille, D. Surleraux, P. Wigerinck, B. Samuelsson, and K. Simmen. 2008. Structure-activity relationship study on a novel series of cyclopentane-containing macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of TMC435350. Bioorg. Med. Chem. Lett. 18:4853-4858. [DOI] [PubMed] [Google Scholar]
- 37.Reesink, H., R. Verloes, K. Abou Farha, A. Van Vliet, C. Weegink, G. van't Klooster, F. Aharchi, K. Marien, P. Van Remoortere, H. de Kock, F. Broeckaert, G. Fanning, P. Meyvisch, E. Van Beirendonck, and K. Simmen. 2008. Safety of the HCV protease inhibitor TMC435350 in healthy volunteers and safety and activity in chronic hepatitis C infected individuals: a phase I study. J. Hepatol. 48:S28-S29. [Google Scholar]
- 38.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. M. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 39.Saito, T., and M. Gale, Jr. 2008. Regulation of innate immunity against hepatitis C virus infection. Hepatol. Res. 38:115-122. [DOI] [PubMed] [Google Scholar]
- 40.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 41.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 42.Strader, D. B., T. Wright, D. L. Thomas, L. B. Seeff, and American Association for the Study of Liver Diseases. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147-1171. [DOI] [PubMed] [Google Scholar]
- 43.Takano, S., K. Nakamura, S. Kawai, O. Yokosuka, Y. Satomura, and M. Omata. 1996. Prospective assessment of donor blood screening for antibody to hepatitis C virus by first- and second-generation assays as a means of preventing posttransfusion hepatitis. Hepatology 23:708-712. [DOI] [PubMed] [Google Scholar]
- 44.Thurmond, R. L., S. Sun, C. A. Sehon, S. M. Baker, H. Cai, Y. Gu, W. Jiang, J. P. Riley, K. N. Williams, J. P. Edwards, and L. Karlsson. 2004. Identification of a potent and selective noncovalent cathepsin S inhibitor. J. Pharmacol. Exp. Ther. 308:268-276. [DOI] [PubMed] [Google Scholar]
- 45.van't Klooster, G. A. E., I. Vanwelkenhuysen, R. Hooijmaijers, K. Bol, M. Voets, J. Van Houdt, R. A. L. Verloes, F. Aharchi, K. Marien, P. Van Remoortere, F. Broeckaert, H. de Kock, and K. A. Simmen. 2008. Once-daily regimens of the HCV NS3/4A-protease inhibitor TMC435350 are predicted to provide therapeutic exposure in plasma and liver. J. Hepatol. 48:S321. [Google Scholar]
- 46.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H.-G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wölk, B., D. Sansonno, H.-G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.