Abstract
The related lipo(depsi)peptide antibiotics daptomycin and friulimicin B show great potential in the treatment of multiply resistant gram-positive pathogens. Applying genome-wide in-depth expression profiling, we compared the respective stress responses of Bacillus subtilis. Both antibiotics target envelope integrity, based on the strong induction of extracytoplasmic function σ factor-dependent gene expression. The cell envelope stress-sensing two-component system LiaRS is exclusively and strongly induced by daptomycin, indicative of different mechanisms of action in the two compounds.
Staphylococcus aureus is a leading cause of nosocomial infections, especially in mechanically ventilated patients. Its remarkable potential to acquire and accumulate high-level resistance against most of the classical antibiotics (including vancomycin) used for the treatment of gram-positive infections is one of the reasons for the ongoing mortality caused by hospital-acquired S. aureus infections (7, 17).
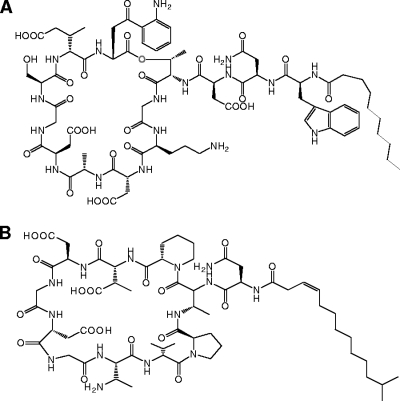
Daptomycin is the first of a new class of cyclic lipodepsipeptide antibiotics (Fig. 1A) with strong bactericidal activities against gram-positive pathogens (2). It interferes with cell envelope integrity, and cell death occurs presumably by either membrane depolarization or membrane perforation (19, 20). Friulimicin B, an acidic, cyclic lipopeptide produced by Actinoplanes friuliensis, shows structural similarities to daptomycin (Fig. 1B) and is also active against multidrug-resistant gram-positive bacteria (1, 22).
FIG. 1.
Chemical structures of the lipodepsipeptide antibiotic daptomycin (A) and the lipopeptide antibiotic friulimicin B (B).
As part of a coordinated effort to study and characterize its mode of action, we have performed comparative in-depth expression profiling for both antibiotics. This technique is a powerful approach to elucidate the inhibitory mechanisms of novel antimicrobial compounds (4, 9) and has been successfully applied to characterize and differentiate antimicrobial actions, often using Bacillus subtilis as a model organism (3, 10). B. subtilis is particularly well suited for studying cell wall antibiotics, since the regulatory network orchestrating its cell envelope stress response (CESR) is well characterized. It consists of four two-component systems and at least four extracytoplasmic function (ECF) σ factors (11).
Here, we present results from an in-depth analysis of the expression signature provoked by the treatment of B. subtilis with sublethal amounts of daptomycin and friulimicin B. Our data show that both antibiotics specifically target cell envelope integrity. But significant differences in the corresponding CESRs, as clearly documented by transcriptomics, proteomics, and detailed gene expression profiling, strongly suggest different modes of action of the two structurally related antibiotics.
(This study was presented in part at the 47th International Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 17 to 20 September 2007 [25]).
Transcriptomics and proteomics.
For microarray experiments, midlogarithmic cultures of B. subtilis were challenged with 1 μg/ml (sublethal amounts) of either daptomycin or friulimicin B. The cells were harvested 10 min postinduction, and cell pellets were directly snap-frozen in liquid nitrogen. RNA preparation and microarray experiments were performed essentially as described previously (13, 23). To validate the gene expression profiles, we also performed two-dimensional gel electrophoresis of the cytoplasmic proteome of B. subtilis cells, quantifying de novo protein synthesis after the addition of daptomycin or friulimicin B by incubating the cultures in the presence of l-[35S]methionine, as described previously (3). The results are summarized in Table 1 and Fig. 2. The complete microarray data sets can be found in the supplemental material and, together with additional supporting information, at http://microbial-stress.iab.kit.edu/87.php.
TABLE 1.
Marker genes induced by daptomycin and/or friulimicin B
| Gene(s)a | Induction by:b
|
Regulator(s)c | Localization (putative)d | Homology, function, remarkse | |
|---|---|---|---|---|---|
| DAP | FRI | ||||
| ywaC | 4.5 ± 4.3 | 8.7 ± 3.8 | σV, σM, σW | C | Putative GTP-pyrophosphokinase |
| mreBH | 3.9 ± 1.9 | 3.1 ± 1.2 | C | Control of cell shape; membrane-associated | |
| ydaH | 3.3 ± 0.3 | 9.1 ± 2.4 | σM | M | Conserved membrane protein |
| yqjL | 3.3 ± 0.3 | 8.9 ± 1.6 | σV, σM | C | Putative hydrolase |
| bcrC | 3.3 ± 1.0 | 8.2 ± 2.8 | σV, σM, σW, σX | M | Undecaprenyl pyrophosphate phosphatase |
| yrhH | 3.1 ± 1.3 | 8.5 ± 3.4 | σV, σM, σW | C | Putative methyltransferase |
| liaIH(GFSR) | 429 ± 53 | − | LiaRS | M, S | Conserved membrane protein; phage-shock protein A homolog (three-component regulatory system) |
| gerAAABAC | 15 ± 2.9 | − | (LiaRS) | M, S | Downstream lia operon, known polar effect from PliaI |
| ybeF | 4.6 ± 0.9 | − | M | Conserved membrane protein | |
| sigM-yhdLK | − | 7.4 ± 4.0 | σM | C, M | ECF σ factor |
| yjbC-spx | − | 7.2 ± 1.7 | σV, σM, σW | C | Glutaredoxin family; transcriptional regulator Spx |
| sms-yacKL | − | 7.1 ± 0.5 | σM | C, C, M | DNA repair/binding proteins; membrane protein |
| radC | − | 6.9 ± 2.1 | σM | C | DNA repair protein |
| ypuA | − | 6.5 ± 2.3 | σM | S | Conserved hypothetical |
| ypbG | − | 6.4 ± 1.0 | σM | S | Putative phosphoesterase |
| ypuD | − | 6.2 ± 0.7 | σM | S | Unknown |
| ycgRQ | − | 5.9 ± 0.6 | σV, σM | M | Conserved membrane protein; permease |
| yrhIJ | − | 5.7 ± 0.8 | σM | C, | Cytochrome P450; transcriptional repressor BscR |
| sigV-yrhM | − | 5.1 ± 2.0 | σV | C, M | ECF σ factor |
| yfnI | − | 4.7 ± 2.0 | σM | M (S)f | Similar to phosphoglycerol transferases |
| yebC | − | 4.1 ± 0.6 | σM | M | Unknown |
| yppC | − | 4.1 ± 0.4 | C | Conserved hypothetical | |
| ywnJ | − | 4.1 ± 0.1 | σM, σX | M | Unknown |
| ywtF | − | 3.9 ± 0.6 | σM | C (S)f | Putative transcriptional regulator |
| pbpI | − | 3.8 ± 1.3 | M | Class B penicillin-binding protein | |
| rodA | − | 3.8 ± 0.9 | σM | M | Control of cell shape and elongation |
| ylxW | − | 3.5 ± 0.3 | σM | M | Unknown |
| yoxD | − | 3.7 ± 0.2 | C | Putative 3-oxoacyl-acyl-carrier protein | |
| yqiG | − | 3.4 ± 0.4 | C | Putative NADH-dependent flavin oxidoreductase | |
| yjbQ | − | 3.4 ± 0.2 | M | Putative Na+/H+ antiporter | |
Only genes that were induced ≥threefold in three independent experiments by daptomycin and/or friulimicin B are shown. The proteins corresponding to the underlined genes were also significantly upregulated in the cytoplasmic proteome (Fig. 2).
Average induction ratio of the highest value for each locus (usually the first gene in an operon) and the corresponding standard deviation are given. DAP, daptomycin; FRI, friulimicin B; −, no significant induction.
Assignment of regulators is based on the corresponding regulon papers: LiaRS (12), σM (8), σV (24), σW (6), and σX (5).
Localization of the corresponding proteins is based on the presence of transmembrane regions (membrane proteins) and signal peptides (secreted proteins) detected with SMART. C, cytoplasmic proteins; M, membrane proteins; S, secreted proteins.
Putative function is derived from BSORF/Subtilist entries (at http://bacillus.genome.ad.jp/ and http://genolist.pasteur.fr/SubtiList/genome.cgi, respectively), NCBI blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi), or SMART (http://smart.embl-heidelberg.de/) analysis.
YfnI and YwtF are assigned to secreted proteins based on experimental evidence (21).
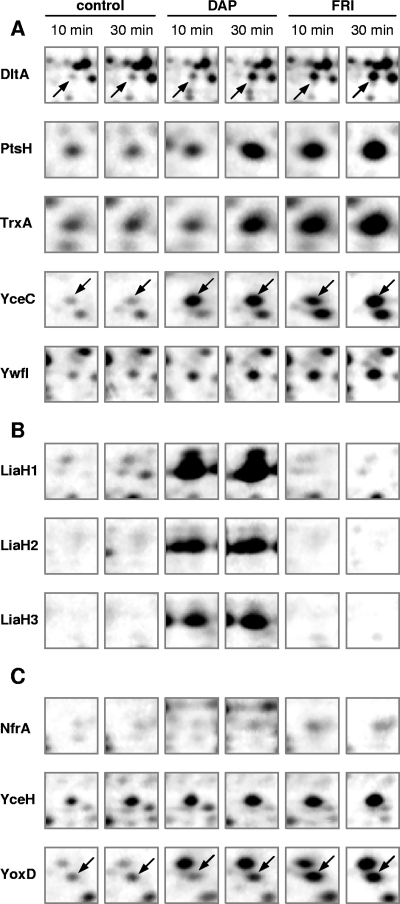
FIG. 2.
Synthesis patterns of marker proteins after induction with daptomycin (DAP) or friulimicin B (FRI) compared to patterns of untreated control cells. Details from the two-dimensional gels of the cytoplasmic proteome (“spot albums” of marker proteins) are shown for two time points postinduction with each of two compounds. (A) Proteins induced by both antibiotics. (B) Daptomycin-specific spots. (C) Friulimicin B-specific spots. See text for details.
Both antibiotics induced a limited number of genes, most of which could be assigned to known CESR regulons. Daptomycin specifically and strongly activated the LiaRS two-component system, with more than 200-fold induction of its primary target genes, liaIH. This induction has also been observed recently in an independent study (9a) and is in good agreement with data from the orthologous VraSR system of S. aureus which was also induced by daptomycin (16). Moreover, a strong LiaH induction was also observed with proteomics analysis, where it was identified in three strong neighboring spots (differing in their isoelectric points), indicative of posttranslational modifications (Fig. 2).
Both compounds induced numerous genes known to be regulated by ECF σ factors. This ECF-dependent response was much stronger for friulimicin B (Table 1). In addition, only seven genes/proteins of unknown regulation were differentially expressed (Table 1 and Fig. 2), including the actin homolog mreBH, which was induced about three- to fourfold by both compounds. Five more genes without known regulator, some of which are potentially involved in cell envelope biogenesis, specifically responded to friulimicin B (Table 1). All genes identified in our analysis have been linked to CESR of B. subtilis previously (data not shown). While no expression signature available so far resembles that of friulimicin B, both the transcriptome and the proteome profile for daptomycin closely resemble those of bacitracin (3, 14).
In-depth gene expression profiling.
The results of our microarray study led to three follow-up analyses on the specificity of the corresponding CESR. (i) We analyzed the induction of all seven ECF σ factors by quantitative real-time reverse transcriptase PCR (RT-PCR), based on the known and highly ECF-specific autoregulation of their own genes, to determine the respective inducer spectrum and strength. The primers used for amplification are listed in Table 2. Both antibiotics activate σM and σV, with friulimicin B provoking a significantly stronger response. In addition, friulimicin B also induced the uncharacterized ECF σ factor σYlaC (Table 3).
TABLE 2.
Strains and oligonucleotides used in this study
| Strain or oligonucleotide | Relevant genotype or fragment amplified | Source, construction, reference, or sequencea |
|---|---|---|
| B. subtilis strains | ||
| W168 | Wild-type strain; trpC2 | Lab stock |
| BFS2470 | W168 liaI::pMUTIN | Zoltan Pragai (Harwood lab); 14, 15 |
| TMB389 | W168 liaIH::Tetr | W168, transformed with chromosomal DNA of strain HB0935; 14 |
| Oligonucleotides | ||
| 822/823 | sigY-RT | Fwd, ACAAGAAGAACAGCGGCTGA; rev, TTCCTGAACAAGTTCCTCGC |
| 824/825 | sigX-RT | Fwd, ACAGAAGACCTTCTTCAAGAG; rev, CGCTGTCTGATTGTTTGCTG |
| 826/827 | sigM-RT | Fwd, GTTTACAGGTTCCTGCTCTC; rev, ATGAAGGCGTTTCGCGCCA |
| 828/829 | sigW-RT | Fwd, TCAGCTTTGCTACCGTATGC; rev, TTGCGAATGCGGTCAATGGT |
| 830/831 | sigV-RT | Fwd, AAGCGTTGCTTGTCACATGC; rev, GTTCCTGACCGTTTCAACTG |
| 832/833 | sigZ-RT | Fwd, GCATCTCCCAAATCTGATCG; rev, TTTTCTTCCTCTGCACTGTCA |
| 834/835 | ylaC-RT | Fwd, TTGAGGACTTGTATCGGCAG; rev, GAGCCATGTTCTGATGGAAG |
| 125/126 | liaH-RT | Fwd, TGAAACAGCACACGATTGCC; rev, GTTTGCCTGTTCATAGGAAGC |
| 156/157 | rpsJ-RT | Fwd, GAAACGGCAAAACGTTCTGG; rev, GTGTTGGGTTCACAATGTCG |
| 158/159 | rpsE-RT | Fwd, GCGTCGTATTGACCCAAGC; rev, TACCAGTACCGAATCCTACG |
Fwd, forward; rev, reverse.
TABLE 3.
Induction of ECF σ factors and liaH by daptomycin and friulimicin B
| Gene | Induction by:a
|
|
|---|---|---|
| DAP | FRI | |
| sigM | 2.4 ± 0.1 | 8.7 ± 3.8 |
| sigV | 2.4 ± 0.7 | 7.4 ± 2.3 |
| sigW | 1.4 ± 0.3 | 0.9 ± 0.0 |
| sigX | 0.8 ± 0.2 | 0.8 ± 0.0 |
| sigY | 0.9 ± 0.1 | 1.8 ± 0.3 |
| sigZ | 1.0 ± 0.0 | 1.2 ± 0.1 |
| ylaC | 1.0 ± 0.0 | 2.9 ± 0.3 |
| liaH | 1170 ± 426 | 0.9 ± 0.0 |
Levels of change given are the average ± standard deviation of the results of two independent real-time RT-PCR experiments, performed essentially as previously described (23), using an iScript one-step RT-PCR kit with Sybr green (Bio-Rad) according to the manufacturer's recommended procedure. DAP, daptomycin; FRI, friulimicin B.
(ii) The much stronger activation of ECF target genes by friulimicin B was not due to the corresponding lack of liaIH induction, as demonstrated by the induction values of ECF genes in the liaIH mutant strain TMB0389, which were identical to those in the wild type (data not shown). The stronger ECF response to friulimicin B is therefore LiaIH independent and a true antibiotic-specific difference in the corresponding gene induction profiles.
(iii) We also quantified the activity of the LiaR target promoter PliaI as a function of the daptomycin/friulimicin B concentrations over a range of 4 orders of magnitude by performing a β-galactosidase assay (using strain BFS2470 as described previously) (15). PliaI induction was indeed only observed in the presence of daptomycin and in a very narrow window of antibiotic concentrations (between 0.5 and 2 μg/ml) (data not shown). These results strongly suggest different modes of action for daptomycin and friulimicin B.
Conclusions.
Our data clearly allowed the identification of cell envelope integrity as the site of daptomycin and friulimicin B action, but the results strongly suggest mechanistic differences between the two compounds. This assumption is primarily based on the dramatic differences in the LiaRS response. Moreover, friulimicin B activates both σM and σV more strongly than daptomycin and, additionally, induces σYlaC expression (summarized in Fig. 3). The strong similarities of CESR between daptomycin and bacitracin were initially viewed as an indication that daptomycin might interfere with the lipid II cycle of cell wall biosynthesis. But a detailed biochemical mechanism of action study revealed that friulimicin B, like amphomycin but in contrast to the membrane-interfering daptomycin, inhibits cell wall biosynthesis by binding bactoprenol phosphate (18).
FIG. 3.
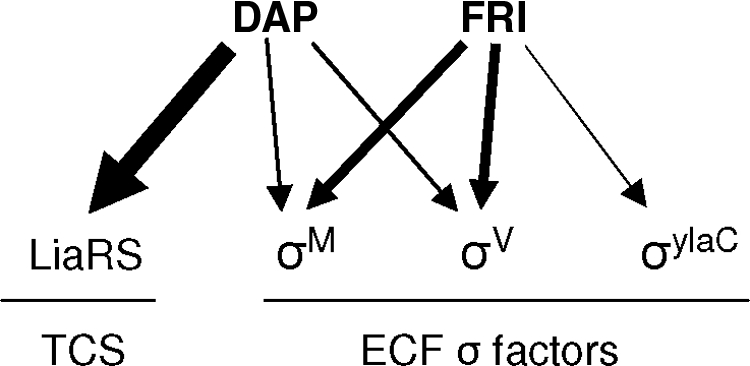
Schematic representation of the regulatory networks orchestrating the daptomycin (DAP) and friulimicin B (FRI) stress responses. The thickness of the arrows corresponds to the strength of induction of the given regulators (see text and Table 1 for details). TCS, two-component system.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to T.M.), the Fonds der Chemischen Industrie (to T.M. and M.H.), the Bundesministerium für Bildung und Forschung (to G.H., U.M., and M.H.; project name, Unternehmen Region-Zentren für Innovationskompetenz; project number from PtJ, 03ZIK012; project running time, June 2005 to May 2010), and the Bildungsministerium of the country Mecklenburg-Vorpommern (to M.H.). Funding for Combinature Biopharm AG (now Merlion Pharmaceuticals GmbH) for friulimicin-related work was granted by the BMBF (project name, BioChancePLUS; project number from PtJ, 0313173; and project running time, April 2004 to March 2007). T.W. was supported by a Chemiefonds Ph.D. scholarship from the Fonds der Chemischen Industrie.
We thank Anja Hoffmann and Susanne Paprotny for excellent technical assistance, Anna-Barbara Hachmann, John D. Helmann, Tanja Schneider, and Hans-Georg Sahl for sharing data prior to publication, and the Decodon GmbH (Greifswald, Germany) for their cooperation.
Footnotes
Published ahead of print on 21 January 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aretz, W., J. Meiwes, G. Seibert, G. Vobis, and J. Wink. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. I. Taxonomic studies of the producing microorganism and fermentation. J. Antibiot. (Tokyo) 53:807-815. [DOI] [PubMed] [Google Scholar]
- 2.Baltz, R. H., V. Miao, and S. K. Wrigley. 2005. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 22:717-741. [DOI] [PubMed] [Google Scholar]
- 3.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazas, M. D., and R. E. Hancock. 2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10:1245-1252. [DOI] [PubMed] [Google Scholar]
- 5.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, runoff transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 7.de Lencastre, H., D. Oliveira, and A. Tomasz. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, H. P., and C. Freiberg. 2007. Applications of transcriptional profiling in antibiotics discovery and development. Prog. Drug Res. 64:23-47. [DOI] [PubMed] [Google Scholar]
- 9a.Hachmann, A.-B., E. R. Angert, and J. D. Helmann. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 12.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jürgen, B., S. Tobisch, M. Wümpelmann, D. Gördes, A. Koch, K. Thurow, D. Albrecht, M. Hecker, and T. Schweder. 2005. Global expression profiling of Bacillus subtilis cells during industrial-close fed-batch fermentations with different nitrogen sources. Biotechnol. Bioeng. 92:277-298. [DOI] [PubMed] [Google Scholar]
- 14.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 15.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordmann, P., T. Naas, N. Fortineau, and L. Poirel. 2007. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr. Opin. Microbiol. 10:436-440. [DOI] [PubMed] [Google Scholar]
- 18.Schneider, T., K. Gries, M. Josten, I. Wiedemann, S. Pelzer, H. Labischinski, and H.-G. Sahl. 2009. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53:XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman, J. A., N. G. Perlmutter, and H. M. Shapiro. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straus, S. K., and R. E. Hancock. 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758:1215-1223. [DOI] [PubMed] [Google Scholar]
- 21.Tjalsma, H., H. Antelmann, J. D. H. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J.-Y. F. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vertesy, L., E. Ehlers, H. Kogler, M. Kurz, J. Meiwes, G. Seibert, M. Vogel, and P. Hammann. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. II. Isolation and structural characterization. J. Antibiot. (Tokyo) 53:816-827. [DOI] [PubMed] [Google Scholar]
- 23.Wecke, T., B. Veith, A. Ehrenreich, and T. Mascher. 2006. Cell envelope stress response in Bacillus licheniformis: integrating comparative genomics, transcriptional profiling, and regulon mining to decipher a complex regulatory network. J. Bacteriol. 188:7500-7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zellmeier, S., C. Hofmann, S. Thomas, T. Wiegert, and W. Schumann. 2005. Identification of σV-dependent genes of Bacillus subtilis. FEMS Microbiol. Lett. 253:221-229. [DOI] [PubMed] [Google Scholar]
- 25.Zühlke, D., B. Voigt, M. Hecker, S. Jordan, T. Mascher, S. Pelzer, and H. Labischinski. 2007. Distinct mode of action of the lipopeptide antibiotic friulimicin B and the lipodepsipeptide daptomycin: a proteomic study, abstr. F1-1641. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.