Abstract
A study was designed to investigate the molecular epidemiological characteristics of multidrug-resistant outbreak-related Pseudomonas aeruginosa isolates collected in a university hospital in northern Greece. Of 29 nonreplicate P. aeruginosa isolates resistant to carbapenems and ceftazidime, 14 were positive for metallo-β-lactamase production. PCR analyses with primers specific for blaVIM and blaIMP revealed that 13 isolates carried a novel blaVIM-2 gene variant, designated blaVIM-17, and only 1 isolate carried blaVIM-2, a gene predominant among P. aeruginosa strains in Greek hospitals. Pulsed-field gel electrophoresis of XbaI-digested genomic DNAs showed a close genetic relationship for 12 of 13 blaVIM-17-carrying outbreak-related isolates, which were of the O11 serotype; the clonally unrelated isolate carrying blaVIM-17 was of the O12 serotype. PCR mapping strategies for the detection of class 1 integrons and sequencing approaches revealed the presence of integrons containing one blaVIM cassette flanked by two aacA29 cassettes. These integrons were similar but not identical to In59 (GenBank accession number AF263519) initially described in France. All isolates carrying blaVIM-17, regardless of their genetic profile, had an identical integron, named In59.3, indicating that although the hospital outbreak was mainly due to clonal dissemination, the horizontal transmission of the blaVIM-17-containing integron among P. aeruginosa isolates should also have occurred. An outbreak-related isolate and a control strain, both of which carried the blaVIM-2 gene but which were clonally distinct, had an identical integron, named In59.2, which differed only at the level of the blaVIM gene from In59.3 integrons, suggesting a common ancestry. The spread of the blaVIM-17-containing integron in clonally unrelated P. aeruginosa isolates without any evidence of plasmid carriage is probably associated with a transposon.
Pseudomonas aeruginosa is an important pathogen able to cause severe infections which are difficult to treat because of the resistance to multiple antimicrobial agents (1, 26, 28). Carbapenems are considered potent agents for the treatment of infections caused by multiresistant strains, mainly because of their stability against most β-lactamases. However, the increasing use of these compounds has resulted in the emergence of carbapenem-resistant P. aeruginosa strains (7, 39). Carbapenem resistance has been associated with impaired outer membrane permeability; upregulation of the efflux system; hyperproduction of a chromosomal AmpC-type cephalosporinase; and the production of enzymes that hydrolyze carbapenems, such as the metallo-β-lactamases (MBLs) and a KPC-type β-lactamase (14, 17, 35, 36, 37).
MBLs or class B β-lactamases are zinc-dependent enzymes characterized by broad hydrolytic activity against all β-lactams except aztreonam (21). Genes encoding MBLs are located as cassettes in integrons that provide them with the potential for expression and dissemination (9, 21, 37). To date, five MBL types, namely, the IMP, VIM, SPM, GIM, and SIM types of MBLs, have been identified; however, the IMP and VIM types are the most commonly detected MBLs worldwide (21, 37).
VIM-type MBLs are predominant in Europe, particularly in the Mediterranean region (21, 37). At present, 18 VIM-type variants have been described (http://www.ncbi.nlm.nih.gov), and another 4 have been assigned to the database at http://www.lahey.org/studies (updated 10 January 2009). In this report, we describe an outbreak caused by multidrug-resistant P. aeruginosa isolates carrying the new variant blaVIM-17 MBL gene in a university hospital in northern Greece. The genetic context of blaVIM-17 and the clonal relationship of the MBL-producing isolates were also determined.
MATERIALS AND METHODS
Bacterial isolates.
The study included 29 nonreplicate P. aeruginosa isolates obtained from clinical specimens between November 2004 and December 2005 from patients with hospital-acquired infections at the Hippokration General Hospital. Bacterial identification was performed with the Vitek 2 and the API 20NE systems (bioMérieux). All isolates, which were selected on the basis of their MICs determined by the broth microdilution method with the Vitek 2 system, presented simultaneous resistance to ceftazidime and carbapenems, which is the criterion for possible MBL production (37). In addition, four clinical P. aeruginosa isolates collected during 2003 were included in this study as blaVIM-2-positive control strains.
Screening for MBL production and antimicrobial susceptibility testing.
All 29 isolates were screened for MBL production by the double-disk synergy test with EDTA as the inhibitor and two β-lactams (imipenem and ceftazidime) as the substrates (2). The MICs of imipenem (Merck Research Laboratories) and meropenem (Astra-Zeneca Pharmaceuticals) of outbreak-related isolates were determined by Etest (AB Biodisk) and by the broth microdilution method, according to the manufacturers' instruction and the recommendations of the CLSI (formerly NCCLS) (4), respectively. Susceptibility testing results were interpreted according to the criteria of the CLSI. Furthermore, the isolates were tested for their susceptibilities to piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, amikacin, gentamicin, ciprofloxacin, and colistin by the disk diffusion method (4). P. aeruginosa ATCC 27853 was used as a quality control strain.
PCR amplification and molecular analysis of MBL genes.
PCR analyses were performed with whole genomic DNAs extracted (NucleoSpin tissue kit; Macherey-Nagel) from the MBL-producing isolates and by amplification with primers specific for the blaVIM and blaIMP genes, as described previously (38). Sequencing of both strands of the purified amplicons (QIAquick PCR purification kit; Qiagen) was performed with the PCR primers. The identities of the MBLs were initially established by a search of the NCBI database with the BLAST program (http://www.ncbi.nlm.nih.gov), and the MBLs were accurately determined by multiple-sequence alignments by use of the Clustal W program (32). Phylogenetic and molecular evolutionary analyses were conducted with MEGA3 software (11). The sequences of the MBLs whose GenBank accession numbers have been assigned to http://lahey.org/studies were used for comparison.
PCR mapping of blaVIM-containing integrons.
The genetic context of the blaVIM genes was initially assessed by characterization of the variable region of the blaVIM-containing integrons, as described previously (34). PCR mapping was carried out with primers designed on the basis of the 5′ and 3′ conserved segments (CSs) of class 1 integrons, primer INT-F or 5′CS and primer QacR or Sul-R, respectively, in combination with primers VIM-R and VIM-F, designed on the basis of the conserved regions of blaVIM genes (Table 1). The partially overlapping PCR fragments were sequenced with a combination of PCR primers and internally designed primers and were aligned together (see Fig. 2). Amplification reactions were carried out under the following cycling conditions: 5 min at 94°C; 30 cycles of 40 s at 94°C, 40 s at 57°C (for primer pair INT-F and VIM-R and primer pair VIM-F and QacR) or 60°C (for primer pair 5′CS and VIM-R and primer pair VIM-F and Sul-R), and 70 s at 72°C; and finally, 5 min at 72°C. Sequence analysis and comparisons were performed as described above.
TABLE 1.
Primer pairs and internal primers used for identification of blaVIM-containing integrons
| Primer | Nucleotide sequence (5′ to 3′) | PCR or sequencing targets | Reference or source |
|---|---|---|---|
| INT-F | CGTTCCATACAGAAGCTG | Amplification and sequencing of gene(s) between intI and blaVIM | 16 |
| VIM-R | ATGAAAGTGCGTGGAGAC | 16 | |
| 5′CS | GGCATCCAAGCAGCAAG | Amplification and sequencing of gene(s) between 5′ CS and blaVIM | 13 |
| VIM-R | ATGAAAGTGCGTGGAGAC | ||
| VIM-F | AGTGGTGAGTATCCGACAG | Amplification and sequencing of gene(s) between blaVIM and qacEΔ1 | 16 |
| QacR | CGGATGTTGCGATTACTTCG | 34 | |
| VIM-F | AGTGGTGAGTATCCGACAG | Amplification and sequencing of gene(s) between blaVIM and sulI | |
| Sul-R | CCGACTTCAGCTTTTGAAGG | This study | |
| INT-For | CTTCYARAAAACCGAGGATGC | Sequencing of promoter region and gene between intI and blaVIM | This study |
| VIM2-Rev | CTTACTCAAAAGTTTGAACAT | Sequencing of gene between blaVIM and 5′ CS | This study |
| VIM2-For | CGCTCAGTCGTTGAGTAG | Sequencing of gene between blaVIM and qacEΔ1 | This study |
| aacA29F/Rev | GTCGGCAGCGTCTTGTTCTT | Sequencing of attI1 and promoter region | This study |
| aacA29R/For | GTATCACGCACCGCAAGTT | Sequencing of 3′ CS-qacEΔ1 region | This study |
| QacF | ATCGCAATAGTTGGCGAAGT | Sequencing of 3′ CS-qacEΔ1 and sul1 region | This study |
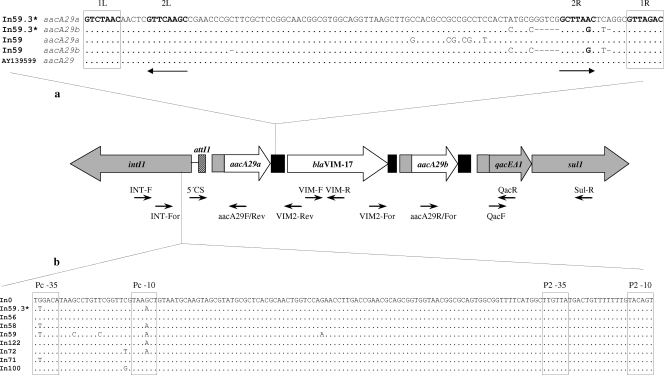
FIG. 2.
Schematic representation of blaVIM-17-containing integron In59.3. Boxes represent coding regions, and the arrowheads on the boxes indicate the corresponding transcriptional orientation; the inserted blaVIM-17 and aacA29 genes are indicated by white boxes, while the CSs at the 5′ and 3′ ends of the integrated gene cassettes, which contain the intI1 gene and the qacEA1 and sul1 genes, respectively, as well as part of the qacE cassette fused upstream of each aacA29 cassette, are indicated by gray boxes. The attI1 and 59-be recombination sites are represented by hatched and black rectangles, respectively. The primers used for PCR mapping and sequencing are shown under the integron structure by thin arrows (Table 1). (a) Comparison of the 59-be recombination sites of the aacA29 gene cassettes. The sequences are shown from the inverse core site to the core site, as they are presented in the circular form of the cassettes. The boldface sequences represent the four conserved regions (29); inverse core and core sites are boxed, and internal 2L and 2R sites are indicated by horizontal arrows. (b) Comparison of the sequence of the promoter region of the blaVIM-17-containing integron (In59.3) with the sequences of the empty In0 integron and some European class 1 integrons. The −35 and −10 sequences of promoters Pc and P2 are boxed. The GenBank accession numbers of the integrons used for comparison are as follows: In0, U49101; In56, AF191564; In58, AF263520; In59, AF263519; In122, AY507153; In72, AF302086; In71, AM180753; In100, AY560837; and In59.3, EU118148. *, sequences identical to those of blaVIM-2-containing integron In59.2 (GenBank accession number EU118149).
Plasmid detection and conjugation experiments.
DNA extracts obtained from the MBL-producing isolates with a NucleoSpin plasmid kit (Macherey-Nagel) and/or by the classic procedure of plasmid DNA preparation (27) were subjected to 0.7% agarose gel electrophoresis, in which Escherichia coli 39R861 and HindIII-digested bacteriophage λ DNA were used as plasmid size markers.
Transfer of imipenem resistance was attempted by plate mating with E. coli 26R793 (Lac− Rifr) and a Rifr P. aeruginosa isolate as recipient strains. Transconjugants were selected on Muller-Hinton agar containing rifampin (rifampicin; 100 μg/ml) and ceftazidime (4 μg/ml) (25).
PFGE typing and serotyping.
Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA (10) was performed in a CHEF-DRIII system (Bio-Rad), and the banding patterns were interpreted according to the criteria established by Tenover et al. (31). Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA) was used to calculate correlation Dice coefficients and to generate a dendrogram by the unweighted pair-group with arithmetic averages clustering method.
Serotyping was performed by the slide agglutination method of the International Antigenic Typing Scheme with commercially available monovalent antisera against the O somatic antigen (Bio-Rad).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study have been assigned to the GenBank database under accession numbers EU118148 and EU118149.
RESULTS AND DISCUSSION
Detection and characterization of MBL genes.
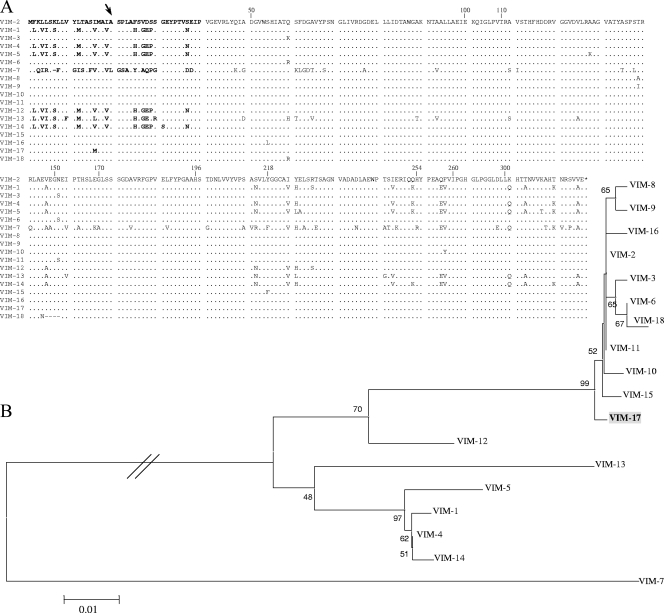
Among the 29 isolates tested, 14 were positive for MBL production by the double-disk synergy test. The presence of MBL genes in all 14 isolates was demonstrated by PCR amplification with the blaVIM-2-specific primers. Sequencing revealed that 13 isolates harbored a new blaVIM-2 variant, designated blaVIM-17, and that 1 isolate harbored blaVIM-2. blaVIM-17 differed from blaVIM-2 by a C48G polymorphism that resulted in an Ile15Met substitution close to the leader peptide cleavage site (Fig. 1A). VIM-17 was clustered among the VIM-2-like MBLs in an evolutionarily separate position and was the only enzyme of this group that presented a substitution in the NH2-terminal region (Fig. 1A and B). Notably, in this region sequence heterogeneity was mostly observed between the VIM-2-like and the VIM-1-like groups (18, 19) (Fig. 1A).
FIG. 1.
Relation of the novel enzyme VIM-17 to other VIM-type MBLs reported to date. The sequences of the GenBank accession numbers of all VIM types assigned to the database at http://lahey.org/studies were used for comparison and phylogenetic analysis. (A) Amino acid sequence comparison of VIM-type MBLs. Dots represent amino acids identical to those in VIM-2. The NH2-terminal region is shown in boldface; the arrow indicates the putative position of the leader peptide cleavage site, as determined by sequencing of the NH2 terminus (3). The numbering is according to the updated BBL scheme (8). (B) Phylogenetic tree based on amino acid sequence analysis of VIM-type MBLs. The tree was constructed by the neighbor-joining method (Poisson correction model). The numbers at the nodes indicate the percentage of 1,000 bootstrap replicates that supported the branch; only bootstrap values of ≥40 are shown on the tree. The novel VIM-17 enzyme identified in this study is in boldface and shaded.
Characterization of genetic context of blaVIM genes.
Four outbreak-related isolates carrying blaVIM-17 (isolates D1841, D2816, A2993, and E96) and one isolate carrying blaVIM-2 (isolate D1802), as well as two control strains (strains D2655 and A7914), were selected from among almost all the PFGE clonal types for characterization of the blaVIM-containing integrons. The PCR mapping and sequencing approach revealed class 1 integrons containing one blaVIM cassette (blaVIM-2 or blaVIM-17) flanked by two aacA29 cassettes (Fig. 2) in all five outbreak-related isolates and one control strain (strain A7914). These integrons were similar to In59 (GenBank accession number AF263519), initially detected in a P. aeruginosa isolate in France (20). The 59-base element (59-be) of the blaVIM cassette was complete (72 bp) and identical in both blaVIM-2 and blaVIM-17 cassettes and was also identical to that in blaVIM-2 cassettes found in other integrons, including In59 (6, 12, 18, 20, 22). However, the 59-be's of the aacA29 cassettes differed from those of In59 by six substitutions and by 1 bp in size in aacA29a and aacA29b, respectively (Fig. 2a), with the left and right regions being conserved according to the consensus sequence (29). Notably, the aacA29a 59-be presented 100% identity with an aacA29 59-be detected in an environmental bacterium (GenBank accession number AY139599) (Fig. 2a), indicating that even environmental bacteria could be an important reservoir of resistance gene cassettes (30). At the genetic level, the similarity between members of each lineage did not appear to be limited to the coding sequences but extended to the recombination sites of the genes, suggesting a common ancestry for these aacA29 cassettes. In the promoter region, Pc and P2 promoters were in the strong active and inactive forms (9), respectively (Fig. 2b). Compared to the sequence of In59 (GenBank accession number AF263519), the intI1 gene and the promoter region differed by nine substitutions, two of which were located between the −35 and the −10 hexamers of the Pc promoter.
On the basis of the similarity of their sequences with the sequence of In59, the blaVIM-2-containing integron was named In59.2 and the blaVIM-17-containing integron was named In59.3 (Fig. 2). The integron of the second control strain (strain D2655) differed from the In59.2 and In59.3 integrons, as it contained different cassettes and a different blaVIM-2 cassette (data not shown).
Several class 1 integrons have been found on plasmids with strong dissemination potentials (15, 18, 24, 37). In our study, despite repeated attempts, plasmid DNA was not detectable in any of the outbreak-related isolates and carbapenem resistance was not transferable to recipient strains, implying that the blaVIM-containing integrons of these isolates are most likely located in the chromosome.
Phenotypic, genotypic, and epidemiological features of outbreak-related isolates.
The MICs of imipenem and meropenem for 14 outbreak-related isolates were ≥128 μg/ml and 32 to 128 μg/ml, respectively. All isolates exhibited resistance to the β-lactams tested (except for two isolates found to be susceptible to aztreonam and to cefepime, respectively), ciprofloxacin, and both amikacin and gentamicin. Interestingly, when they were cloned, the aacA29 genes conferred resistance to amikacin but not to gentamicin (20). Given also that only one isolate was susceptible to aztreonam, an antibiotic stable to the hydrolytic activity of MBLs, concomitant resistance mechanisms in addition to the presence of MBLs and aminoglycoside acetyltransferases seem to be involved. Finally, no resistance to colistin was observed, offering a potential therapeutic option.
The emergence of MBL-producing isolates has been associated with the use of broad-spectrum β-lactams, including carbapenems, and the use of aminoglycosides and quinolones (7). Most of our patients (n = 11 [78.6%]) had previously received aminoglycosides (amikacin and/or gentamicin) and only 5 (35.7%) had previously received imipenem, suggesting that the clinical use of aminoglycosides should have contributed to the coselection of MBL-producing P. aeruginosa isolates (5).
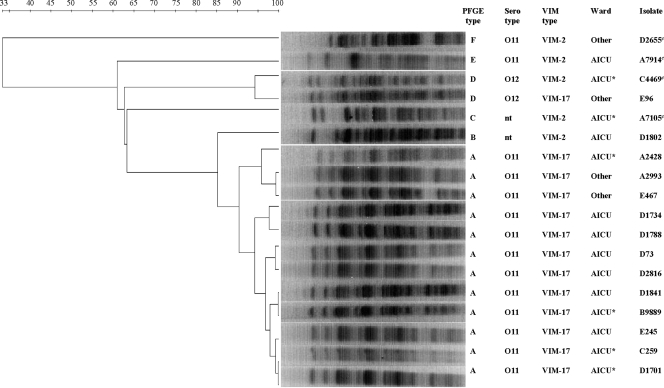
On the basis of the results of PFGE, all the outbreak-related isolates carrying blaVIM-17 except isolate E96 were clustered in the predominant PFGE type, type A (≥90% PFGE similarity) (Fig. 3). Isolate E96 belonged to PFGE type D and was clonally related to one blaVIM-2-positive control strain. Serotyping demonstrated that all strains of type A were serotype O11, in contrast to the isolate of type D, which was serotype O12. blaVIM-2-carrying outbreak-related isolate D1802 belonged to type B and was nontypeable with monovalent antisera. Each of the remaining three blaVIM-2-positive control strains yielded a different restriction profile and belonged to a distinct type (types C, E, and F, respectively) (Fig. 3).
FIG. 3.
Clustering of the outbreak-related P. aeruginosa isolates on the basis of their PFGE profiles after digestion with XbaI. The dendrogram is based on analysis by the unweighted pair-group with arithmetic averages clustering method; percent similarities are shown above the dendrogram. The names and the profiles of the isolates and the hospital wards of the patients are shown on the right of the PFGE gel; control strains are indicated by c. nt, nontypeable; AICU, adult intensive care unit; AICU*, short admission to the AICU; Other, ward other than the AICU.
Ten isolates carrying blaVIM-17 were isolated from patients who were hospitalized in the adult intensive care unit (AICU) or who were transferred to the AICU for a short time during their hospitalization, while three isolates were recovered from patients who had never been admitted to the AICU (Fig. 3). Given the close genetic relationship (type A, serotype O11) observed in all 10 cases associated with the AICU and those two not associated with the AICU, it can be claimed that the most likely origin of the isolates was the AICU, and these isolates further spread to other wards through patient and medical staff traffic. Interestingly, the fact that the clonally unrelated isolate (type D, serotype O12) carrying blaVIM-17, isolate E96, carried the In59.3 integron, identical to that carried by type A isolates, suggests that although the outbreak was mainly due to clonal dissemination, the horizontal transmission of blaVIM-17-containing integron among P. aeruginosa isolates should also have occurred. It is also interesting that the genetically distinguishable (type B) and nontypeable blaVIM-2-carrying isolate recovered from a patient hospitalized in the AICU during the outbreak period carried the In59.2 integron, which was similar to the In59.3 integron carried by type A isolates, suggesting a common ancestry. Furthermore, the In59.2 integron was also found in one clonally distinct blaVIM-2-positive control strain isolated from a patient who had been hospitalized in the AICU 2 years earlier. Another blaVIM-2-positive control strain had a totally different integron with a different blaVIM-2 cassette. These findings suggest that the horizontal spread of the integron itself among P. aeruginosa isolates rather than the spread of the blaVIM cassette among different integrons should have occurred. On the basis of previous reports on other class 1 integrons (6, 9, 23, 33, 37) and given the lack of evidence for plasmid carriage, the spread of blaVIM-containing integrons between genetically unrelated isolates could be associated with a transposon.
Finally, whether the unique substitution characterizing the novel enzyme leads to differences in biochemical characteristics requires further investigation.
Acknowledgments
We are grateful to Spyros Pournaras and his team for providing helpful advice.
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Aloush, V., S. Navon-Venezia, Y. Seigman-Igra, S. Cabili, and Y. Carmeli. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 50:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-beta-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing. 17th informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Cornaglia, G., M. Akova, G. Amicosante, R. Cantón, R. Cauda, J.-D. Docquier, M. Edelstein, J.-M. Frère, M. Fuzi, M. Galleni, H. Giamarellou, M. Gniadkowski, R. Koncan, B. Libisch, F. Luzzaro, V. Miriagou, F. Navarro, P. Nordmann, L. Pagani, L. Peixe, L. Poirel, M. Souli, E. Tacconelli, A. Vatopoulos, and G. M. Rossolini on behalf of the ESCMID Study Group for Antimicrobial Resistance Surveillance (ESGARS). 2007. Metallo-β-lactamases as emerging resistance determinants in gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380-388. [DOI] [PubMed] [Google Scholar]
- 6.Corvec, S., L. Poirel, E. Espaze, C. Giraudeau, H. Drugeon, and P. Nordmann. 2008. Long-term evolution of a nosocomial outbreak of Pseudomonas aeruginosa producing VIM-2 metallo-enzyme. J. Hosp. Infect. 68:73-82. [DOI] [PubMed] [Google Scholar]
- 7.Falagas, M. E., and P. Kopterides. 2006. Risk factors for the isolation of multi-drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: a systematic review of the literature. J. Hosp. Infect. 64:7-15. [DOI] [PubMed] [Google Scholar]
- 8.Garau, G., I. Garcia-Saez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J. M. Frere, and O. Dideberg. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1:109-119. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis. Methods Mol. Med. 15:17-31. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and N. Masatoshi. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Lagatolla, C., E. Edalucci, L. Dolzani, M. L. Riccio, F. De Luca, E. Medessi, G. M. Rossolini, and E. A. Tonin. 2006. Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity J. Clin. Microbiol. 44:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 15.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β -lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miriagou, V., E. Tzelepi, D. Gianneli, and L. S. Tzouvelekis. 2003. Escherichia coli with a self-transferable, multi-resistant plasmid coding for the metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai, H., J. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., T. Lambert, S. Türkoglü, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinteira, S., J. C. Sousa, and L. Peixe. 2005. Characterization of In100, a new integron carrying a metallo-β-lactamase and a carbenicillinase, from Pseudomonas aeruginosa Antimicrob. Agents Chemother. 49:451-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, L. B., and R. A. Bonomo. 2005. Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents, p. 441-508. In V. Lorian (ed.), Antibiotics in laboratory medicine, 5th ed. The Williams & Wilkins Co., Baltimore, MD.
- 26.Rossolini, G. M., and E. Mantengoli. 2005. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin. Microbiol. Infect. 11:17-32. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Preparation of plasmid DNA by alkaline lysis with SDS: minipreparation, section 1.32. In Molecular cloning: a laboratory manual, vol. 1, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Sligl, W., G. Taylor, and P. G. Brindley. 2006. Five years of nosocomial gram-negative bacteremia in a general intensive care unit: epidemiology, antimicrobial susceptibility patterns, and outcomes. Int. J. Infect. Dis. 10:320-325. [DOI] [PubMed] [Google Scholar]
- 29.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 30.Tennstedt, T., R. Szczepanowski, S. Braun, A. Puehler, and A. Schlueter. 2003. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45:239-252. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toleman, M. A., D. Biedenbach, D. Bennett, R. N. Jones, and T. R. Walsh. 2003. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 34.Toleman, M. A., D. Biedenbach, D. M. Bennett, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-beta-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 55:61-70. [DOI] [PubMed] [Google Scholar]
- 35.Villegas, M. V., K. Lolans, A. Correa, J. N. Kattan, J. A. Lopez, J. P. Quinn, and Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 51:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, F., and S. G. Amyes. 2007. Carbapenem resistance in clinical isolates of Pseudomonas aeruginosa. J. Chemother. 19:376-381. [DOI] [PubMed] [Google Scholar]
- 37.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zavascki, A. P., R. P. Cruz, and L. Z. Goldani. 2005. Risk factors for imipenem-resistant Pseudomonas aeruginosa: a comparative analysis of two case-control studies in hospitalized patients. J. Hosp. Infect. 59:96-101. [DOI] [PubMed] [Google Scholar]