Abstract
Daptomycin is the first of a new class of cyclic lipopeptide antibiotics used against multidrug-resistant, gram-positive pathogens. The proposed mechanism of action involves disruption of the functional integrity of the bacterial membrane in a Ca2+-dependent manner. We have used transcriptional profiling to demonstrate that treatment of Bacillus subtilis with daptomycin strongly induces the lia operon including the autoregulatory LiaRS two-component system (homologous to Staphylococcus aureus VraSR). The lia operon protects against daptomycin, and deletion of liaH, encoding a phage-shock protein A (PspA)-like protein, leads to threefold increased susceptibility. Since daptomycin interacts with the membrane, we tested mutants with altered membrane composition for effects on susceptibility. Deletion mutations of mprF (lacking lysyl-phosphatidylglycerol) or des (lipid desaturase) increased daptomycin susceptibility, whereas overexpression of MprF decreased susceptibility. Conversely, depletion of the cell for the anionic lipid phosphatidylglycerol led to increased resistance. Fluorescently labeled daptomycin localized to the septa and in a helical pattern around the cell envelope and was delocalized upon the depletion of phosphatidylglycerol. Together, these results indicate that the daptomycin-Ca2+ complex interacts preferentially with regions enriched in anionic phospholipids and leads to membrane stresses that can be ameliorated by PspA family proteins.
Within the soil microenvironment, bacteria compete for limited nutrients by the production of antibiotics that serve to inhibit the growth of their competitors. Indeed, the majority of antibacterial compounds in clinical use are natural products of soil-dwelling organisms and, in many cases, are produced by Streptomyces spp. and related members of the Actinomycetales (16). Many of these compounds target the synthesis of the peptidoglycan (PG) cell wall layer or disrupt membrane function. Those compounds in clinical use owe their selectivity to the fact that eukaryotic cells lack PG and have a different membrane lipid composition from those of most bacteria.
Antibiotics have also proven to be useful tools for microbial cell biology and have allowed the visualization of the subcellular location of cell envelope biosynthetic processes. For example, fluorescently labeled vancomycin and ramoplanin, antibiotics that bind specifically to the un-cross-linked PG precursors and/or lipid II, have served to confirm the helical arrangement of the lateral cell wall biosynthetic complexes (14, 58), which was also shown in studies using green fluorescent protein-tagged envelope proteins (53).
We have used Bacillus subtilis as a model system to investigate the genetic and physiological responses to both antibiotics and coculture with antibiotic-producing strains. To date, our studies have focused on cell envelope antibiotics (including vancomycin, bacitracin, and fosfomycin) and bacteriocins, such as nisin, duramycin, and sublancin (17, 43). Exposure to these compounds activates distinct cell envelope stress responses controlled by extracytoplasmic function (ECF) σ factors and two-component regulatory systems (TCS) (34). In most cases, the activity of ECF σ factors and TCS is controlled by transmembrane sensors (anti-σ factors or membrane-bound histidine protein kinases, respectively) which thereby allow gene expression to be regulated in response to changes in the cell envelope. The identification of genes induced by a certain antibiotic stress provides insights into the nature of the antibiotic's target(s) and also aids in the identification of resistance functions, many of which are inducible by their cognate antibiotic (30).
Daptomycin, a cyclic lipopeptide antibiotic originally purified from Streptomyces roseosporus (46), is notable for its activity against methicillin-resistant Staphylococcus aureus and certain streptococci and enterococci. The mechanism of action of daptomycin has been controversial. Initial studies suggested that daptomycin inhibited lipoteichoic acid synthesis (6). However, these findings could not be verified (40). The current proposed mechanism of action involves the insertion of its decanoyl side chain into the cytoplasmic membrane in a Ca2+-dependent manner. Subsequent oligomerization, followed by depolarization of the membrane potential and efflux of potassium ions, leads to the arrest of protein, RNA, and DNA synthesis (56). It has been suggested that daptomycin approaches the bacterial membrane in the form of micelles composed of 14 to 16 daptomycin molecules and an equal number of Ca2+ ions, which are proposed to help mask the negative charge of daptomycin. After insertion into the membrane, daptomycin dissipates the membrane potential and leads to cessation of macromolecule synthesis (24, 54, 56). Daptomycin treatment does not result in cell lysis or in daptomycin entering the cytoplasm (6, 13).
Here, we have investigated the genetic and physiological responses of B. subtilis to daptomycin. Using transcriptional profiling, we demonstrate that daptomycin strongly activates the LiaRS TCS, which regulates the liaIHGFSR operon. Mutants defective for liaH, which encodes a phage-shock protein A (PspA)-like membrane stress protein (35), were threefold more susceptible to daptomycin. This susceptibility was further exacerbated in cells additionally lacking the paralogous gene pspA. Fluorescence microscopy studies using Bodipy FL-labeled daptomycin (daptomycin-BDP) together with strains having altered membrane lipid composition support a model in which the daptomycin-Ca2+ complex interacts preferentially with regions enriched in anionic lipids (primarily phosphatidylglycerol [PhG] in B. subtilis) and is localized at new cell division septa and in a helical pattern along the long axis of the cell.
(Part of this work was presented at the 47th Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, September 2007.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and primers used in this study are listed in Tables 1 and 2. Deletion mutants were obtained by replacing genes with antibiotic resistance cassettes using long-flanking homology PCR as described previously (43, 60) in the wild-type W168 (trpC2) or CU1065 (W168 trpC2 attSPβ). Unless otherwise noted, bacteria were cultured in Mueller-Hinton broth supplemented with 50 mg/liter Ca2+ at 37°C with vigorous shaking. The following antibiotics were used for selection when necessary: 100 μg/ml spectinomycin, 10 μg/ml kanamycin, 10 μg/ml chloramphenicol, 20 μg/ml tetracycline, and 1 μg/ml erythromycin with 25 μg/ml lincomycin (macrolide-lincomycin-streptogramin B resistance). Daptomycin and daptomycin-BDP were provided by Cubist Pharmaceuticals (Lexington, MA). For microarray analyses (Table 3), cells were grown to mid-log phase from a 1:1,000 dilution of overnight cultures. To determine the MIC, overnight cultures were diluted 1:100, grown to mid-log phase, and rediluted to 5 × 105 CFU/ml in Mueller-Hinton broth supplemented with 50 mg/liter Ca2+. At least 10 appropriate antibiotic concentrations close to the predicted MIC were added to the cultures at the beginning of the growth curve (including a control without antibiotics) in a total inoculum of 200 μl. Growth was measured spectrophotometrically (optical density at 600 nm [OD600]) using a Bioscreen incubator (Growth Curves USA, Piscataway, NJ) at 37°C with vigorous shaking by monitoring the absorbance every 20 min for 24 h. Inhibition was defined as a final OD600 of <0.05 (at the 24 h time point). The mode of the MIC of a minimum of triplicate experiments is shown in Table 4.
TABLE 1.
Strains used in this study
| Strains | Genotype/purpose description | Reference, source, and/or constructiona |
|---|---|---|
| W168 | trpC2 | BGSC no. 1A1 |
| CU1065 | W168 trpC2 attSPß | 59; Lab stock |
| HB5121 | W168 liaIHGFSR::spc | LFH-PCR → W168 |
| HB0934 | CU1065 liaGFSR::kan | 43 |
| HB0920 | CU1065 liaH::kan | 43 |
| HB0935 | CU1065 liaIH::tet | 43 |
| HB0933 | CU1065 liaR::kan | 43 |
| HB5337 | CU1065 mprF::kan | 52 |
| HB5123 | CU1065 liaIH::tet mprF::kan | HB5337 chr DNA → HB0935 |
| HB0919 | CU1065 pspA::cat | 4 |
| HB5124 | CU1065 liaH::kan pspA::cat | HB0919 chr DNA → HB0920 |
| HB5125 | CU1065 liaIH::tet pspA::cat | HB0919 chr DNA → HB0935 |
| HB5126 | CU1065 liaIH::tet mprF::kan pspA::cat | HB0919 chr DNA → HB5123 |
| HB0938 | CU1065 yhcYZ::cat | T. Mascher, unpublished |
| HB5127 | CU1065 liaH::kan yhcYZ::cat | HB0938 chr DNA → HB0920 |
| HB0031 | CU1065 sigM::kan | 12 |
| HB0020 | CU1065 sigW::mls | 7 |
| HB7007 | CU1065 sigX::spc | 27 |
| HB0097 | CU1065 sigX::spc sigM::kan | 9 |
| HB0096 | CU1065 sigW::mls sigM::kan | 11 |
| HB0030 | CU1065 sigX::spc sigW::mls | M. Cao, unpublished |
| HB0982 | CU1065 sigM::kan sigW::mls sigX::spc | 42 |
| HB5128 | CU1065 liaH::kan sigW::mls | HB0020 chr DNA → HB0920 |
| HB5129 | CU1065 liaH::kan pspA::cat sigW::mls | HB0020 chr DNA → HB5124 |
| HB5130 | CU1065 sigX::spc sigM::kan liaIH::tet | HB0935 chr DNA → HB0097 |
| HB5131 | CU1065 sigM::kan sigW::mls sigX::spc liaIH::tet | HB0935 chr DNA → HB0982 |
| HB5132 | CU1065 sigW::mls pspA::cat | HB0919 chr DNA → HB0020 |
| HB5133 | CU1076 sigM::kan sigW::mls sigX::spc pspA::cat | HB0919 chr DNA → HB0982 |
| HB5134 | W168 des::spc | LFH-PCR → W168 |
| HB5135 | W168 des::spc pspA::cat | HB0919 chr DNA → HB5134 |
| HB5346 | CU1065 ugtP::mls | 52 |
| HB5361 | CU1065 pssA::spc | 52 |
| HB5347 | CU1065 ywnE::tet | 52 |
| HB5343 | CU1065 psd::mls | 52 |
| HB5350 | CU1065 mprF::kan ugtP::mls | 52 |
| HB5388 | CU1065 mprF::kan pssA::spc | 52 |
| HB5344 | CU1065 psd::mls mprF::kan | 52 |
| HB5136 | CU1065 mprF::kan pspA::cat | HB0919 chr DNA → HB5337 |
| HB5363 | CU1065 PxylA-mprF at the thrC locus, Spcr | L. I. Salzberg, unpublished |
| BFA2809 | W168 trpC2 pgsA::pMutin4 | 2 |
| HB0950 | CU1065 attSPß2Δ2::PliaI-cat-lacZ | 44 |
| HB0070 | CU1065 sigM::kan SPß (PM-cat-lacZ) | 9 |
| HB7022 | CU1065 SPß7019 (PX-cat-lacZ) | 27 |
| HB0050 | CU1065 SPß::PsigW-cat-lacZ | 28 |
| HB0021 | CU1065 sigW::mls SPß::PsigW-cat-lacZ | 12 |
Long-flanking homology PCR (LFH-PCR) was applied as described previously to construct some of the deletions, using the primers listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| No. | Name | Sequencea |
|---|---|---|
| 2443 | Lia-Do-Fwd(spec) | 5′-CGTTACGTTATTAGCGAGCCAGTCGCGGCTGCTATTTGTTTGCGCC-3′ |
| 2444 | Lia-Do-Rev | 5′-GGAGGGCTCTTCATCTGATCCG-3′ |
| 2445 | Lia-Up-Rev(spec) | 5′-CAATAAACCCTTGCCCTCGCTACGAAAAACGCCATGCACGAGGC-3′ |
| 2446 | Lia-Up-Fwd | 5′-GCTGTCACATTATGCGGCGC-3′ |
| 3197 | Des-Up-Fwd | 5′-CCCACTTCTCAACATTTGCAA-3′ |
| 3198 | Des-Up-Rev(spec) | 5′-CGTTACGTTATTAGCGAGCCAGTCCGACTTGCTTTGTCAGCTGT-3′ |
| 3199 | Des-Do-Fwd(spec) | 5′-CAATAAACCCTTGCCCTCGCTACGGTTTGTGTCATT TGGGCTAT-3′ |
| 3200 | Des-Do-Rev | 5′-CCCAAGCGTATCATGGAGAT-3′ |
Sequences complementary to antibiotic resistance cassettes for LFH-PCR are underlined.
TABLE 3.
Daptomycin stimulona
| Gene | Signal intensityb
|
Fold change | Gene functionc | Regulon (reference) | |
|---|---|---|---|---|---|
| +DAP | −DAP | ||||
| liaH | 78,930 | 59 | 1,336 | PspA homolog, cell membrane protection | LiaRS (35) |
| liaI | 78,935 | 61 | 1,294 | Putative membrane protein | LiaRS (35) |
| liaG | 37,639 | 133 | 283 | Putative membrane-anchored protein | LiaRS (35) |
| liaF | 50,616 | 341 | 149 | Negative regulator of LiaR | LiaRS (35) |
| liaS | 15,435 | 122 | 126 | TCS histidine kinase | LiaRS (35) |
| liaR | 3535 | 47 | 75 | TCS response regulator | LiaRS (35) |
| yhcY | 149 | 45 | 3.3 | TCS histidine kinase | LiaRS (35) |
| yhcZ | 631 | 289 | 2.2 | TCS response regulator | LiaRS (35) |
| yvrI | 2,754 | 4.8 | 574 | σ factor yvrI | YvrI (41) |
| yvrL | 1,390 | 2.6 | 535 | Negative regulator of yvrI | YvrI (41) |
| yqjL | 303 | 39 | 7.8 | Hydrolase; paraquat resistance | σMW (11) |
| ydaH | 306 | 49 | 6.2 | Uncharacterized membrane protein | σM (32) |
| ypbG | 211 | 58 | 3.6 | Marker for inhibition of cell wall biosynthesis | σM (32) |
| yebC | 155 | 43 | 3.6 | Hypothetical conserved | σM (17) |
| ywaC | 124 | 36 | 3.5 | ppGpp synthase | σM (48) |
| bcrC | 358 | 115 | 3.1 | Similar to bacteriocin transport permease | σM (9) |
| rodA | 150 | 56 | 2.7 | Cell division membrane protein | σM (17) |
| yhdL | 208 | 82 | 2.5 | ECF anti-σ factor | σM (17) |
| maf | 605 | 245 | 2.5 | Cell division and shape determination | σM (17) |
| murG | 386 | 163 | 2.4 | Peptidoglycan biosynthesis | σM (17) |
| murB | 710 | 353 | 2.0 | UDP-N-acetylenolpyruvoyl glucosamine reductase | σM (17) |
| sigM | 199 | 93 | 2.1 | ECF σ factor | σM (26) |
| yuaF | 707 | 148 | 4.8 | Hypothetical protein | σW (29) |
| yuaG | 329 | 168 | 2.0 | Flotillin-like protein | σW (29) |
| yxjI | 144 | 68 | 2.1 | Unknown | σW (10) |
| ywrE | 159 | 76 | 2.1 | Unknown | σW (10) |
| ydjP | 328 | 168 | 2.0 | Similar to chloroperoxidase | σW (10) |
| yoeB | 2,855 | 1,332 | 2.1 | Protection against autolysins | YycFG (51) |
| yfiC | 11,780 | 3,349 | 3.5 | Similar to ABC transporter (ATP-binding protein) | |
| fabHA | 1,203 | 521 | 2.3 | Fatty acid biosynthesis | |
| fabHB | 73 | 26 | 2.8 | Fatty acid biosynthesis | |
| yvcB | 1,214 | 45 | 27 | Unknown | |
| yvkN | 302 | 12 | 26 | Unknown | |
| yvzA | 424 | 42 | 10 | Unknown | |
| yvzC | 261 | 10 | 26 | Hypothetical protein | |
| yvpB | 257 | 19 | 14 | Hypothetical protein | |
| sdpI | 2,233 | 252 | 8.9 | Immunity protein for SdpC; signal transduction | SdpR (19) |
| ywdD | 163 | 31 | 5.3 | Hypothetical protein | |
| ywdE | 504 | 51 | 9.9 | Hypothetical protein | |
| ywfB | 299 | 38 | 7.9 | Similar to bacilysin biosynthesis protein | |
| yknU | 2,917 | 372 | 7.8 | Similar to ABC transporter (ATP-binding protein) | |
| hutM | 126 | 40 | 3.2 | Histidine permease | |
| ypeB | 469 | 151 | 3.1 | Sporulation protein | |
| yitI | 630 | 215 | 2.9 | Probable acetyltransferase | |
| rpmB | 892 | 1,631 | 0.5 | Ribosomal protein L28 | |
| degR | 115 | 216 | 0.5 | Activation of degradative enzymes (AprE, NprE, SacB) | |
| yisY | 138 | 264 | 0.5 | Similar to chloride peroxidase | |
| ydgK | 39 | 77 | 0.5 | Similar to bicyclomycin resistance protein | |
| yocH | 759 | 1,513 | 0.5 | Similar to cell wall-binding protein (autolysin) | YycFG (3, 51) |
| yxeQ | 60 | 119 | 0.5 | Putative MmgE/Prp family protein | |
| lmrB | 261 | 534 | 0.5 | Lincomycin resistance | |
| rpsT | 541 | 1,140 | 0.5 | Ribosomal protein S20 | |
| yxeM | 66 | 156 | 0.4 | Similar to amino acid ABC transporter (binding protein) | |
| ywhE | 386 | 926 | 0.4 | pbpG, penicillin-binding protein (sporulation) | |
Genes induced or repressed >2-fold are shown with their respective antibiotic-responsive regulator. Induced genes controlled by regulator(s) known to be responsive to antibiotic-elicited stress are shown together with genes of predicted function or those with unknown function that are strongly induced (>5-fold). Induced and repressed genes were filtered to remove those with low overall expression levels (combined signal intensity < 100). Altogether, daptomycin induced 83 genes at least twofold (234 genes, ≥1.5-fold) and repressed 25 genes at least twofold (181 genes, ≥1.5-fold) (NCBI GEO series accession no. GSE13900).
“+DAP” and “−DAP” correspond to average results of triplicates of the signal intensities of daptomycin-treated or untreated samples, respectively.
Functions were assigned based on the SubtiList database or BSORF entry.
TABLE 4.
MIC of B. subtilis mutants and strains with altered membrane composition, or deletion of transcriptional regulators
| Strain/deletion mutanta | DAP MIC (μg/ml)b |
|---|---|
| W168 | 1.0 |
| CU1065 | 1.0 |
| LiaR-regulated | |
| liaIHGFSR | 0.3 |
| liaH | 0.4 |
| liaIH | 0.4 |
| liar | 0.5 |
| liaF | 1.0 |
| liaGFSR | 0.5 |
| liaIH mprF | 0.2 |
| liaH yhcYZ | 0.2 |
| yhcYZ | 0.8 |
| ECF σ-regulated | |
| sigM | 0.8 |
| sigW | 0.8 |
| sigX | 1.0 |
| sigXM | 0.6 |
| sigWM | 0.7 |
| sigXW | 0.9 |
| sigMWX | 0.6 |
| pspA | 1.0 |
| LiaR/ECF σ-regulated | |
| liaH sigW | 0.3 |
| liaH pspA sigW | 0.2 |
| liaH pspA | 0.2 |
| liaIH pspA | 0.2 |
| liaIH mprF pspA | 0.2 |
| sigXM liaIH | 0.2 |
| sigMWX liaIH | 0.2 |
| sigW pspA | 0.8 |
| sigMWX pspA | 0.5 |
| Membrane alterations | |
| W168 at 25°C | 0.7 |
| des at 25°C | 0.4 |
| des | 0.9 |
| des pspA | 0.9 |
| ugtP | 0.9 |
| pssA | 1.0 |
| ywnE | 1.0 |
| psd | 1.0 |
| mprF | 0.5 |
| mprF ugtP | 0.6 |
| mprF pssA | 0.9 |
| psd mprF | 0.9 |
| mprF pspA | 0.5 |
| mprF overexpression | 1.3 |
| pgsA depletion | 8.0 |
| IPTG-induced pgsA | 1.0 |
liaIHGFSR and des mutants are derived from strain W168. Other mutants are derived from strain CU1065.
MIC was determined by liquid growth inhibition experiments. Data represent the mode of the lowest daptomycin (DAP) concentration that led to complete growth inhibition (minimum of triplicates).
RNA preparation and microarray analyses.
Cultures of W168 were grown to mid-log phase (OD600 of 0.4) and split into two flasks. One sample was treated with 1 μg/ml daptomycin (1× MIC) for 20 min; the other sample was used as a nontreated control. Total RNA was isolated from three different biological replicas with the RNeasy minikit (Qiagen Sciences, MD). After DNase treatment with the Turbo DNA-free kit (Ambion), RNA concentrations were quantified using a NanoDrop spectrophotometer (NanoDrop Tech. Inc., Wilmington, DE). The corresponding cDNA was synthesized from 20 μg total RNA and differentially labeled according to the manufacturer's instructions, using the SuperScript Plus indirect cDNA labeling system (Invitrogen). Before and after indirect labeling with Alexa Fluor 555 or Alexa Fluor 647 (for at least 3 h at room temperature), cDNA was purified using the Qiagen PCR purification kit (Qiagen, MD) and quantified with a NanoDrop spectrophotometer. Both labeled cDNA populations were combined (approximately 100 pmol coupled cDNA each), denatured, and hybridized to a microarray slide overnight at 42°C for 16 to 18 h. After washing, hybridized microarray slides were scanned with a GenePix 4000B array scanner (Axon Instruments, Inc.). Our B. subtilis W168 microarrays, consisting of 4,109 gene-specific antisense oligonucleotides (65-mers; Sigma-Genosys), were printed at the W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University. Each slide contains 8,447 features corresponding to duplicates of each open reading frame-specific oligonucleotide, additional oligonucleotides of control genes, and 50% dimethyl sulfoxide blank controls. Images were processed using the GenePix Pro 4.0 software package, which produces (red/green) fluorescence intensity pairs for each gene. Each expression value is represented by at least six separate measurements (duplicate spots on each of three arrays). Mean values and standard deviations were calculated with Microsoft Excel. The normalized microarray data sets were filtered to remove those genes that were not expressed at levels significantly above background in either condition (sum of mean fluorescence intensity, <20). In addition, the mean and standard deviation of the fluorescence intensities were computed for each gene, and those for which the standard deviation was greater than the mean value were ignored. The induction values were calculated by dividing the signal intensities of daptomycin-treated samples by those of untreated samples. These data are available in Table S1 of the supplemental material. The graph in Fig. 1 represents average results of triplicates.
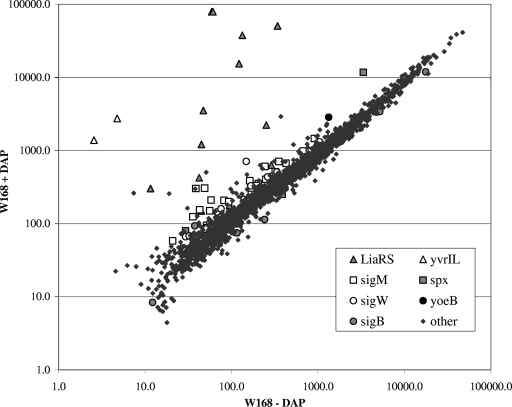
FIG. 1.
Daptomycin stimulon in B. subtilis. The scatter plot represents the average expression levels of treated (+) versus untreated (−) cultures of B. subtilis W168 (daptomycin [DAP] at the MIC of 1 μg/ml; 20 min) from triplicate microarray analyses. The key lists highly expressed genes as grouped by their corresponding transcriptional regulators.
Fluorescence microscopy.
Cells were either treated with the lipophilic membrane dye FM 4-64 (Invitrogen), Bodipy FL-labeled vancomycin (vancomycin-BDP) (Invitrogen), or daptomycin-BDP (Cubist Pharmaceuticals). Daptomycin-BDP is an N-BodipyFL-ornithine derivative of daptomycin. The activity of daptomycin-BDP was confirmed by measuring the MIC of B. subtilis wild type and a set of more-susceptible mutants. In all cases, the MIC corresponded to an ∼10-fold higher MIC than that induced by daptomycin treatment. Furthermore, daptomycin-BDP was able to induce cat-lacZ reporter fusions to liaI independent of the solvent. One hundred microliters of cells from exponential or stationary growth phases was incubated with 4 μl daptomycin-BDP (0.5 mg/ml) in 50% dimethyl sulfoxide for 10 min, with 2 μl vancomycin-BDP (0.1 mg/ml) mixed 1:1 with 0.1 mg/ml vancomycin, or with 1 μl FM 4-64 (0.5 μg/ml) for 20 min. After washing in Mueller-Hinton broth the cells were mounted in the antifade reagent Citifluor (Ted Pella, Inc.) on poly-l-lysine (Sigma-Aldrich)-treated slides. Nomarski optics differential interference contrast (DIC) or fluorescent images were taken with an Olympus BX61 epifluorescence microscope with a UPlanApo 100× (numerical aperture of 1.35) objective. The microscope is equipped with fluorescence filter cubes for viewing DAPI (4′,6-diamidino-2-phenylindole), fluorescein isothiocyanate, and Cy3. Images were acquired using a Cooke SensiCam with a Sony interline chip. Image acquisition and postprocessing were performed using the SlideBook software package (Intelligent Imaging).
Cluster analysis.
Results of whole genome microarray analyses of B. subtilis with a set of antimicrobial compounds from our data and from a study by Hutter et al. (30) were compared by complete linkage clustering (arrangement based on treatment and genetic response similarity) using the Gene Cluster 3.0 software. The resulting cluster was visualized with Treeview 1.60, written by Michael Eisen (18).
Microarray data accession number.
The complete set of raw and normalized data for each of the triplicate DNA microarray experiments involving B. subtilis treated with daptomycin is available at the Gene Expression Omnibus database (http://acbi.alm.nih.gov/geol/) under accession no. GSE13900.
RESULTS AND DISCUSSION
Daptomycin strongly activates the LiaRS regulon in B. subtilis.
Determination of antibiotic stimulons reveals the transcriptional responses to the imposed stress and often provides clues to both the relevant mode of action and the induction of possible defense mechanisms (30). Here, we have used DNA-based microarray analyses to investigate the initial transcriptional responses to treatment of B. subtilis with daptomycin at the MIC of 1.0 μg/ml. As monitored after 20 min of exposure, daptomycin induced ∼83 genes at least twofold, and many of these are members of known antibiotic-responsive regulons (Fig. 1 and Table 3) (17, 44).
The strongest response to daptomycin treatment was the induction of the autoregulated liaIHGFSR operon which encodes the antibiotic-responsive LiaRS TCS (Table 3). Lia was named in reference to its strong induction by lipid-II-interacting antibiotics (35, 44), although it is now clear that there is an imperfect correlation between lipid II binding and induction (61). The Staphylococcus aureus VraSR system is orthologous to the LiaRS TCS (35). It has been implicated in mediating antibiotic resistance (22, 38) and was also strongly induced by daptomycin, as deduced by microarray analyses (47).
In addition to the liaI operon itself, daptomycin induced several other genes in a LiaR-dependent manner. These included the yhcYZ operon, which was previously shown to be a direct target of LiaR activation (35), and the yvrI and yvrL genes. The yvrI gene has recently been shown to be a divergent member of the σ70 family of transcriptional regulators (41) and activates expression of oxalate decarboxylase (OxdC), a major cell wall protein (1). It is not yet clear if yvrI is a direct or indirect target of LiaR activation.
In B. subtilis, a number of genes involved in antibiotic resistance and cell wall metabolism are regulated by the ECF σ factors σW and σM. Daptomycin weakly induced six genes of the σW regulon (10, 12) and 19 genes of the σM regulon (17, 32) (Table 3). To monitor the transcriptional responses of the liaI, sigW, and sigM genes to daptomycin treatment, we used transcriptional (cat-lacZ) reporter fusions and found that all were induced by daptomycin (data not shown).
Phage-shock protein A homologs LiaH and PspA protect against daptomycin.
Of the liaIHGFSR genes, liaFSR are well conserved within gram-positive bacteria with a low G+C content. Only the bacilli also contain the phage-shock protein A homolog LiaH. S. aureus harbors the liaFSR orthologs SA1702 and vraSR (35). When testing for daptomycin susceptibility, Muthaiyan et al. observed an increased susceptibility of a vraSR mutant strain to daptomycin (0.78 μg/ml versus 1.0 μg/ml for the wild type) (47). Here, we have used growth inhibition studies to examine whether the daptomycin-induced liaI operon affects susceptibility to daptomycin. A deletion of the entire liaI operon significantly increased susceptibility from an MIC of 1.0 μg/ml for the wild type to an MIC of 0.3 μg/ml (Table 4). A null mutant of the negative regulator liaF, which overexpresses LiaIH (34, 35), did not appear to increase resistance.
To test for possible functional redundancy between liaH and its paralog pspA (which was not strongly induced by daptomycin treatment despite being part of the σW regulon) we tested single and double mutants of these loci. A pspA deletion produced almost no change in MIC compared to that of the wild type, but the liaH pspA double mutation further decreased the MIC by twofold relative to that of the liaH single mutant. These results indicate that the PspA (phage-shock protein A) homologs, LiaH and PspA, both contribute to decreased daptomycin susceptibility and that the induction of LiaH by daptomycin is adaptive.
The mechanisms by which PspA proteins protect cells against membrane disruption are unclear but are likely to involve direct interactions with the inner surface of the membrane (15). Escherichia coli PspA forms abundant, oligomeric ring-like structures that are speculated to coat the inner surface of the membrane and thereby prevent proton leakage (39). In vitro, E. coli PspA binds preferentially to liposomes containing anionic lipids and suppresses proton leakage (39). LiaH has also been observed to be an abundant oligomeric protein with a similar ultrastructure (T. Mascher, personal communication). Together, these findings are consistent with the notion that daptomycin toxicity results from disruption of the membrane integrity and that the two B. subtilis PspA paralogs can counteract this disruptive effect.
ECF σ factors also contribute to decreased daptomycin susceptibility.
Since several σW and σM regulon members were upregulated upon daptomycin treatment, we tested null mutants of ECF σ factor genes for daptomycin susceptibility. In the singly mutant strains, there was a slight decrease in MIC for sigM and sigW (0.8 μg/ml for both), whereas a sigX mutant was unaffected. Multiply mutant strains displayed even greater susceptibility, with the lowest MIC noted for the sigXM double (0.6 μg/ml) and the sigMWX triple mutants (0.6 μg/ml) (Table 4). Increased daptomycin susceptibility in the multiply mutant strains is consistent with the recent demonstration that these three ECF σ factors have overlapping regulons and multiply mutant strains are often more susceptible to antibiotics than are single mutants (42).
In B. subtilis, several ECF σ factors have been implicated in conferring resistance to cell envelope-active antibiotics. For example, σX regulates the dlt operon and the pssA ybfM psd operon, which reduce the net negative charge of the cell envelope by d alanylation of teichoic acids and insertion of phosphatidylethanolamine into the membrane, respectively. As a result, sigX mutants are more susceptible to cationic antimicrobial peptides (8). The σW regulon includes a large number of genes implicated in resistance against both small molecule inhibitors, such as fosfomycin, and peptide antibiotics, such as sublancin and SdpC (4). Finally, the σM regulon has recently been shown to include many genes known to be important for cell envelope synthesis, and sigM mutants are susceptible to some cell wall antibiotics, such as moenomycin and bacitracin (17, 42). The identity of the ECF σ factor-dependent operons that confer daptomycin protection is not yet clear.
Mutants with altered membrane composition affect daptomycin susceptibility.
Since the decanoyl side chain of daptomycin is predicted to insert into the membrane, we tested whether susceptibility is influenced in strains with altered membrane lipid composition. Daptomycin susceptibility was measured for B. subtilis strains lacking phosphatidylethanolamine (psd, pssA), lysyl-PhG (LPhG) (mprF), glycolipids (ugtP), or cardiolipin (ywnE). Of the null mutants, only the mprF mutant showed a significant difference compared to the wild type (MIC of 0.5 μg/ml versus 1.0 μg/ml [Table 4]). Moreover, overexpression of mprF led to slightly decreased susceptibility (MIC, 1.3 μg/ml). MprF catalyzes the tRNA-dependent modification of PhG with lysine to form the positively charged LPhG (57).
It has been shown earlier that a change in membrane charge due to mprF disruption affects the susceptibility to antimicrobial agents in S. aureus (49). Here, we speculate that the reduction of the net negative charge of the membrane upon increased production of LPhG functions to reduce the affinity of a positively charged daptomycin-Ca2+ complex due to electrostatic repulsion. Indeed, previous studies of S. aureus strains that were selected for increased daptomycin resistance found that point mutations in mprF frequently occurred as an early event during selection (21). However, the effect of these mutations alone was quite modest, and further selection led to additional mutations in the yycFG TCS and RNA polymerase subunit genes rpoBC (21). Since an mprF null mutant is more susceptible to daptomycin, we suggest that these mprF mutations may have been gain-of-function mutations. Independently, Jones et al. found that daptomycin resistance in S. aureus was correlated with the increased translocation of LPhG from the inner to the outer leaflet of the membrane without changing the overall concentration of LPhG (33). An increase of mprF gene expression was not seen upon daptomycin treatment in B. subtilis, but an increase in positive charge through LPhG translocation to the outer leaflet and an additional effect of reduction in the membrane net negative charge by σX could together affect the ability of daptomycin to insert into the membrane.
The physical properties of the membrane are determined by both the membrane head group composition and the length and desaturation of the fatty acyl side chains. In B. subtilis, the fluidity of the membrane is regulated, in large part, by the lipid desaturase Des, which introduces cis double bonds at the fifth position of the fatty acyl chains (Δ5) in response to reduction in temperature. The desaturase is under the control of the DesRK TCS (41a). Deletion of des resulted in increased susceptibility to daptomycin, and this effect was especially notable during growth at low temperatures (MIC at 25°C of 0.4 μg/ml versus 0.7 μg/ml for the wild type). The underlying mechanisms of the effect of des deletion are not entirely clear. The increased rigidity of the membrane in a des mutant might facilitate the membrane disruptive action of daptomycin and impair repair mechanisms by the cell, or the decrease in unsaturated fatty acyl moieties might affect interactions with the decanoyl side chain of daptomycin.
Depletion of PhG greatly decreases daptomycin susceptibility.
The effect of reducing the net negative charge of the cell membrane on daptomycin susceptibility was especially apparent when we studied a strain in which PhG could be depleted from cells by using a conditionally expressed allele of pgsA. PgsA is required for the first step in PhG synthesis from phosphatidic acid (2). Depletion of this essential complex lipid, by transfer of cells to a medium lacking the inducer IPTG (isopropyl-β-d-thiogalactopyranoside), results in cells that lose the characteristic helical staining pattern associated with anionic-lipid-favoring membrane dyes (e.g., FM 4-64). This strain continues to grow for several hours even in the absence of IPTG, as cells gradually become depleted of PhG (2). When PhG-depleted cells were subcultured in a medium lacking IPTG, but containing daptomycin, they were able to grow in the presence of daptomycin concentrations significantly higher than that of the wild type. Conversely, when expression of pgsA was induced by IPTG addition, wild-type levels of daptomycin susceptibility were restored (Table 4). This increased resistance was specific for daptomycin; PhG-depleted cells were unaffected in susceptibility to vancomycin (which targets PG synthesis) and had increased susceptibility to duramycin, which interacts specifically with phosphatidylethanolamine (31). Presumably, in this case, depletion of PhG from the membrane led to an increase in the concentration of phosphatidylethanolamine.
The effects of membrane composition and charge on daptomycin insertion have also been studied by Jung et al. in artificial liposomes (37). By means of fluorescence spectroscopy, differential scanning calorimetry, and 31P nuclear magnetic resonance, they found that daptomycin (with Ca2+) binds to acidic and neutral lipids in different fashions and leads to a change of the structural organization of acidic membranes (induction of nonlamellar lipid phases and membrane fusion) (37). This again emphasizes the influence of membrane lipid composition on daptomycin susceptibility.
Daptomycin preferentially localizes to the cell septum and in a helical pattern along the cell wall.
Fluorescent imaging with daptomycin-BDP was used to study the localization of daptomycin in the cell envelope in B. subtilis. Strikingly, daptomycin-BDP was not distributed evenly throughout the cell membrane but, rather, in a complex, reproducible pattern. The highest concentration was found along the newly formed division septa and in a helical pattern along the long axis of the cell, whereas no fluorescence was detected at the cell poles (Fig. 2A and B). This helical pattern and localization to the septa are reminiscent of localization studies of both the cell wall biosynthetic machinery (58) and anionic phospholipids (including PhG) in B. subtilis (2). Sites of active cell wall biosynthesis have been visualized using vancomycin-BDP (58).
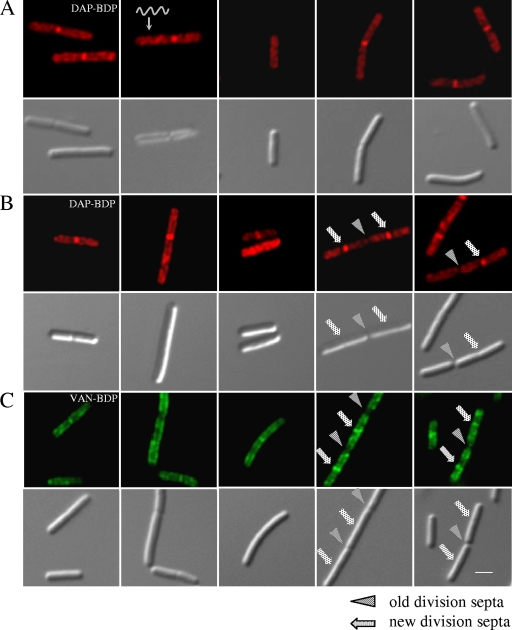
FIG. 2.
Daptomycin-BDP inserts preferentially at new division septa and in a spiral pattern. Fluorescent and DIC micrographs of B. subtilis stained with daptomycin-BDP (DAP-BDP) and vancomycin-BDP (VAN-BDP). (A) B. subtilis W168 treated with daptomycin-BDP at two times the MIC for 10 min (during exponential growth phase). (B) W168 treated with daptomycin-BDP at 10 times the MIC for 10 min. (C) W168 treated with equal amounts of vancomycin and vancomycin-BDP for 20 min. Panels A and B show a spiral localization of daptomycin-BDP and the preferential insertion at newly formed division septa, similar to that of vancomycin-BDP (C). The scale bar represents 2 μm.
We next compared the localization of daptomycin-BDP with vancomycin-BDP which binds to the d-Ala-d-Ala dipeptide of un-cross-linked PG (50). In separate labeling experiments, the daptomycin-BDP and vancomycin-BDP labeling showed very similar patterns (Fig. 2C). Since both antibiotics are bound to the same fluorophore, we were unable to carry out direct colocalization studies. However, pretreatment with vancomycin did not affect labeling by daptomycin-BDP, nor did unlabeled daptomycin interfere with staining with vancomycin-BDP. This suggests that these two antibiotics may have distinct targets, as expected from prior work (56). Recently, Mascio et al. showed that daptomycin also exhibits bactericidal effects on S. aureus cells in stationary growth phases (albeit with higher MIC) (45), which prompted us to test daptomycin-BDP labeling in B. subtilis stationary cells. We observed a similar pattern as that in exponentially growing cells (Fig. 3), albeit with less intense labeling at the division septa.
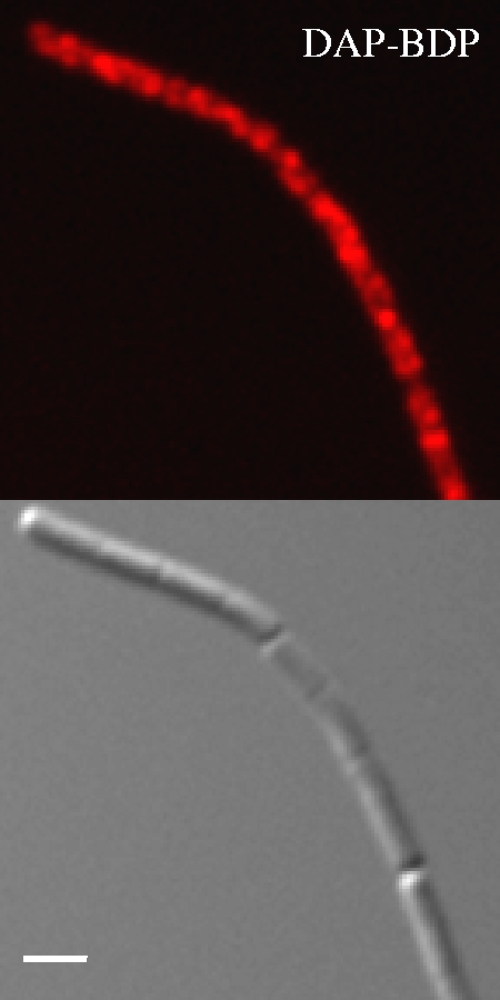
FIG. 3.
Daptomycin-BDP (DAP-BDP) staining of stationary phase cells. Fluorescent and DIC micrographs of stationary growth phase culture of B. subtilis W168 treated with daptomycin-BDP at two times the MIC for 10 min. The staining shows a similar insertion pattern as that of exponential-growth phase cultures. The scale bar represents 2 μm.
In light of the recently reported helical distribution of PhG in B. subtilis membranes (2), we next tested whether PhG depletion affects the observed staining pattern seen with daptomycin-BDP. Remarkably, cells depleted of PhG exhibited a significant loss of staining intensity and failed to exhibit the characteristic helical staining pattern seen in the nondepleted cells (Fig. 4A to C). Very similar effects were noted when the cells were stained instead with the membrane dye FM 4-64 (Fig. 4D and E), which is known to stain regions enriched in PhG (2). Together, these results suggest that the characteristic staining pattern observed with daptomycin-BDP is due to a preferential interaction with the membrane in regions enriched in PhG, consistent with the effects of PhG depletion on daptomycin susceptibility, as noted above.
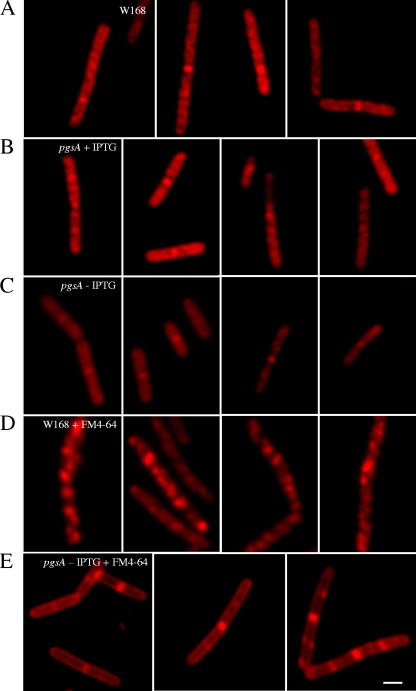
FIG. 4.
Correlation between daptomycin-BDP staining and anionic phospholipid content and distribution. Fluorescent and DIC micrographs of wild-type W168 (A) and pgsA::pMutin grown in the presence (B) or absence (C) of 1 mM IPTG, stained with daptomycin-BDP at two times the MIC for 10 min. Daptomycin-BDP delocalizes when pgsA is not expressed from the IPTG-inducible promoter, suggesting preferential insertion of daptomycin in membrane lipid domains rich in anionic phosphatidylglycerol. W168 and pgsA::pMutin stained with the membrane lipid dye FM 4-64 in the absence of IPTG are shown in panels D and E, respectively. The scale bar represents 2 μm.
Cluster analysis of the stimulons of daptomycin and other cell envelope-active antibiotics.
Analyses of the daptomycin stimulon and the effects of membrane mutations on susceptibility, combined with the preceding analysis of daptomycin-BDP localization, are all consistent with a primary mechanism of action involving insertion into the cell membrane in regions enriched in anionic lipids. However, recent studies of the transcriptional response of S. aureus to daptomycin led Muthaiyan et al. to suggest that daptomycin might interfere with both membrane integrity and PG biosynthesis (47). In this organism, daptomycin induced genes characteristic of membrane-disrupting agents (e.g., m-chlorophenylhydrazone) as well as genes induced by cell wall synthesis inhibitors (e.g., vancomycin).
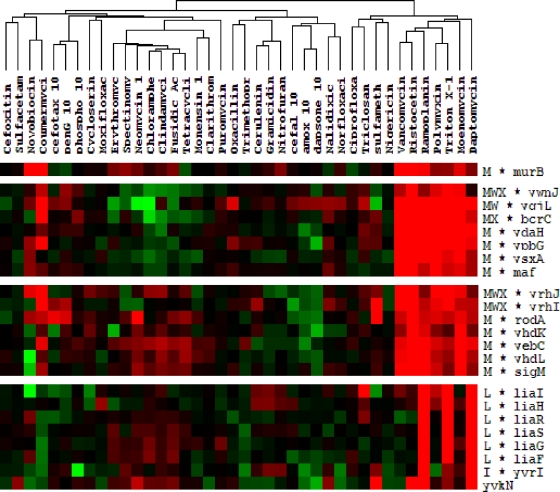
To determine whether the daptomycin stimulon of B. subtilis most closely resembled the stimulons for membrane-perturbing agents or cell wall synthesis inhibitors, we performed a hierarchical clustering analysis using data sets representing the transcriptional responses to daptomycin, vancomycin, moenomycin, and ramoplanin (our results) and 35 other antibiotics from a study by Hutter et al. (30). The daptomycin stimulon is most similar to a cluster of treatment conditions that includes compounds that inhibit PG synthesis (vancomycin, ristocetin, ramoplanin, and moenomycin) and that perturb membrane function (Triton X-114 and polymyxin B). All of these conditions induce the σM regulon, and several compounds are strong inducers of the LiaRS TCS (Fig. 5). These results lead us to suggest that insertion of daptomycin into membrane regions enriched in anionic lipids may have multiple effects, including both disruption of membrane function and perhaps interference with the assembly or function of cell wall biosynthetic complexes.
FIG. 5.
Cluster analysis of B. subtilis microarray studies with 40 different antimicrobial agents. The gene expression patterns of daptomycin-treated B. subtilis are most closely related to those of cells treated with the following cell membrane- and cell wall-active antibiotics: moenomycin, Triton X-114, polymyxin B, ramoplanin, ristocetin, and vancomycin. Cluster analysis was performed on whole genome data sets for each antibiotic (see Materials and Methods), and selected clusters enriched for daptomycin-induced genes are shown. Red indicates induction and green repression after treatment, whereas black corresponds to unchanged gene expression.
Conclusions.
Here, we report a detailed genetic analysis of factors affecting daptomycin susceptibility in B. subtilis. Transcriptome analyses indicate that daptomycin induces sets of genes shown previously to be induced by exposure to membrane-active compounds and cell wall synthesis inhibitors. The LiaRS TCS is strongly induced by daptomycin, and genetic studies indicate that both LiaH and the paralog PspA contribute to decreased daptomycin susceptibility. Analogous to E. coli PspA (36, 39), these proteins are likely to help stabilize the membrane and prevent depolarization. These results are consistent with the expectation that daptomycin acts primarily on the membrane.
Analysis of strains with altered membrane composition suggests that daptomycin interacts preferentially with regions of the membrane enriched in anionic lipids, including PhG, the dominant anionic lipid in B. subtilis. For example, a strain in which PhG is depleted becomes less susceptible to daptomycin, although it retains normal (or even increased) susceptibility to other cell wall- and membrane-active antibiotics. Conversely, strains lacking MprF, which synthesizes the cationic lipid LPhG, are more susceptible to daptomycin. Mutations in mprF have been previously associated with daptomycin resistance (21), although in light of the results here, it seems likely that these are gain-of-function mutations that increase MprF levels or activity. Indeed, overexpression of mprF in B. subtilis decreases daptomycin susceptibility.
Cells depleted of PhG not only display decreased daptomycin susceptibility, they also lose the characteristic helical staining pattern seen in wild-type cells treated with daptomycin-BDP. This may result directly from the reduction in levels of negatively charged membrane lipids, which would thereby decrease the affinity of the positively charged daptomycin-Ca2+ complex for the membrane. Alternatively, or in addition, PhG depletion might result in altered composition or localization of membrane proteins or cell envelope biosynthetic complexes. For instance, Campo et al. reported that reduced PhG led to delocalization of the translocation ATPase SecA in the B. subtilis membrane (5), and Barák et al. observed enrichment of the cell division protein MinD in anionic phospholipid spirals in the membrane (2).
Daptomycin is used clinically as a reserve antibiotic against complicated skin and skin structure infections (23) as well as against S. aureus-induced bacteremia and infective endocarditis (20). To date, reports about resistance to daptomycin in clinical settings have been relatively rare (25, 55). To better understand the evolution of daptomycin resistance, S. aureus strains with increased daptomycin resistance (either selected in the laboratory or arising during clinical treatment) were chosen for DNA sequence analysis. These studies indicate that the evolution of resistance is a multigenic phenomenon. Often, mutations in mprF emerge early, and other contributing mutations occur in the essential yycFG TCS and rpoBC genes (21). To date, there are no documented examples of high-level daptomycin resistance emerging due to a single gene mutation. In light of these findings, it is interesting that cells depleted of PhG display such a dramatic decrease of susceptibility. However, null mutations that confer daptomycin resistance are unlikely to arise in pgsA since, at least in B. subtilis, it is an essential gene.
Supplementary Material
Acknowledgments
We thank Cubist Pharmaceuticals and, in particular, Jared A. Silverman for their generous gift of daptomycin and Bodipy FL-labeled daptomycin, as well as for helpful discussions. We further thank Rebekah Ward for assistance with fluorescence microscopy and Thorsten Mascher for communicating unpublished results and for helpful comments.
This work was supported by grant GM-047446 from NIH.
Footnotes
Published ahead of print on 21 January 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Antelmann, H., S. Towe, D. Albrecht, and M. Hecker. 2007. The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J. Proteome Res. 6:897-903. [DOI] [PubMed] [Google Scholar]
- 2.Barák, I., K. Muchova, A. J. Wilkinson, P. J. O'Toole, and N. Pavlendova. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 68:1315-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisicchia, P., D. Noone, E. Lioliou, A. Howell, S. Quigley, T. Jensen, H. Jarmer, and K. M. Devine. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180-200. [DOI] [PubMed] [Google Scholar]
- 4.Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60:765-782. [DOI] [PubMed] [Google Scholar]
- 5.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583-1599. [DOI] [PubMed] [Google Scholar]
- 6.Canepari, P., and M. Boaretti. 1996. Lipoteichoic acid as a target for antimicrobial action. Microb. Drug Resist. 2:85-89. [DOI] [PubMed] [Google Scholar]
- 7.Cao, M., B. A. Bernat, Z. Wang, R. N. Armstrong, and J. D. Helmann. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of σW, an extracytoplasmic-function σ factor in Bacillus subtilis. J. Bacteriol. 183:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, M., and J. D. Helmann. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function σ factors. J. Bacteriol. 184:6123-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 11.Cao, M., C. M. Moore, and J. D. Helmann. 2005. Bacillus subtilis paraquat resistance is directed by σM, an extracytoplasmic function σ factor, and is conferred by YqjL and BcrC. J. Bacteriol. 187:2948-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 13.Cotroneo, N., R. Harris, N. Perlmutter, T. Beveridge, and J. A. Silverman. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 15.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 16.D'Costa, V. M., K. M. McGrann, D. W. Hughes, and G. D. Wright. 2006. Sampling the antibiotic resistome. Science 311:374-377. [DOI] [PubMed] [Google Scholar]
- 17.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellermeier, C. D., E. C. Hobbs, J. E. Gonzalez-Pastor, and R. Losick. 2006. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124:549-559. [DOI] [PubMed] [Google Scholar]
- 20.Falagas, M. E., K. P. Giannopoulou, F. Ntziora, and K. Z. Vardakas. 2007. Daptomycin for endocarditis and/or bacteraemia: a systematic review of the experimental and clinical evidence. J. Antimicrob. Chemother. 60:7-19. [DOI] [PubMed] [Google Scholar]
- 21.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hair, P. I., and S. J. Keam. 2007. Daptomycin: a review of its use in the management of complicated skin and soft-tissue infections and Staphylococcus aureus bacteraemia. Drugs 67:1483-1512. [DOI] [PubMed] [Google Scholar]
- 24.Ho, S. W., D. Jung, J. R. Calhoun, J. D. Lear, M. Okon, W. R. Scott, R. E. Hancock, and S. K. Straus. 2008. Effect of divalent cations on the structure of the antibiotic daptomycin. Eur. Biophys. J. 37:421-433. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, R. L., and J. H. Jorgensen. 2008. Inhibitory activities of 11 antimicrobial agents and bactericidal activities of vancomycin and daptomycin against invasive methicillin-resistant Staphylococcus aureus isolates obtained from 1999 through 2006. Antimicrob. Agents Chemother. 52:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horsburgh, M. J., and A. Moir. 1999. σM, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32:41-50. [DOI] [PubMed] [Google Scholar]
- 27.Huang, X., A. Decatur, A. Sorokin, and J. D. Helmann. 1997. The Bacillus subtilis σX protein is an extracytoplasmic function σ factor contributing to survival at high temperature. J. Bacteriol. 179:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 30.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwamoto, K., T. Hayakawa, M. Murate, A. Makino, K. Ito, T. Fujisawa, and T. Kobayashi. 2007. Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys. J. 93:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jervis, A. J., P. D. Thackray, C. W. Houston, M. J. Horsburgh, and A. Moir. 2007. σM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 189:4534-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, T., M. R. Yeaman, G. Sakoulas, S.-J. Yang, R. A. Proctor, H.-G. Sahl, J. Schrenzel, Y. Q. Xiong, and A. S. Bayer. 2008. Staphylococcus aureus clinical treatment failures with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 35.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jovanovic, G., L. J. Lloyd, M. P. Stumpf, A. J. Mayhew, and M. Buck. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281:21147-21161. [DOI] [PubMed] [Google Scholar]
- 37.Jung, D., J. P. Powers, S. K. Straus, and R. E. Hancock. 2008. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem. Phys. Lipids 154:120-128. [DOI] [PubMed] [Google Scholar]
- 38.Kato, Y., T. Suzuki, T. Ida, K. Maebashi, M. Sakurai, J. Shiotani, and I. Hayashi. 2008. Microbiological and clinical study of methicillin-resistant Staphylococcus aureus (MRSA) carrying VraS mutation: changes in susceptibility to glycopeptides and clinical significance. Int. J. Antimicrob. Agents 31:64-70. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi, R., T. Suzuki, and M. Yoshida. 2007. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66:100-109. [DOI] [PubMed] [Google Scholar]
- 40.Laganas, V., J. Alder, and J. A. Silverman. 2003. In vitro bactericidal activities of daptomycin against Staphylococcus aureus and Enterococcus faecalis are not mediated by inhibition of lipoteichoic acid biosynthesis. Antimicrob. Agents Chemother. 47:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLellan, S., T. Wecke, and J. Helmann. 2008. A previously unidentified σ factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis. Mol. Microbiol. 69:954-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Mansilla, M. C., and D. de Mendoza. 2005. The Bacillus subtilis desaturase: a model to understand phospholipid modification and temperature sensing. Arch. Microbiol. 183:229-235. [DOI] [PubMed] [Google Scholar]
- 42.Mascher, T., A. B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function σ factors. J. Bacteriol. 189:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 44.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mascio, C. T., J. D. Alder, and J. A. Silverman. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob. Agents Chemother. 51:4255-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao, V., M. F. Coeffet-Legal, P. Brian, R. Brost, J. Penn, A. Whiting, S. Martin, R. Ford, I. Parr, M. Bouchard, C. J. Silva, S. K. Wrigley, and R. H. Baltz. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507-1523. [DOI] [PubMed] [Google Scholar]
- 47.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nanamiya, H., K. Kasai, A. Nozawa, C. S. Yun, T. Narisawa, K. Murakami, Y. Natori, F. Kawamura, and Y. Tozawa. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67:291-304. [DOI] [PubMed] [Google Scholar]
- 49.Nishi, H., H. Komatsuzawa, T. Fujiwara, N. McCallum, and M. Sugai. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4800-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds, P. E., and E. A. Somner. 1990. Comparison of the target sites and mechanisms of action of glycopeptide and lipoglycodepsipeptide antibiotics. Drugs Exp. Clin. Res. 16:385-389. [PubMed] [Google Scholar]
- 51.Salzberg, L. I., and J. D. Helmann. 2007. An antibiotic-inducible cell wall-associated protein that protects Bacillus subtilis from autolysis. J. Bacteriol. 189:4671-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salzberg, L. I., and J. D. Helmann. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190:7797-7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheffers, D. J., L. J. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749-764. [DOI] [PubMed] [Google Scholar]
- 54.Scott, W. R., S. B. Baek, D. Jung, R. E. Hancock, and S. K. Straus. 2007. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim. Biophys. Acta 1768:3116-3126. [DOI] [PubMed] [Google Scholar]
- 55.Silverman, J. A., N. Oliver, T. Andrew, and T. Li. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverman, J. A., N. G. Perlmutter, and H. M. Shapiro. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staubitz, P., H. Neumann, T. Schneider, I. Wiedemann, and A. Peschel. 2004. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231:67-71. [DOI] [PubMed] [Google Scholar]
- 58.Tiyanont, K., T. Doan, M. B. Lazarus, X. Fang, D. Z. Rudner, and S. Walker. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. USA 103:11033-11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vander Horn, P. B., and S. A. Zahler. 1992. Cloning and nucleotide sequence of the leucyl-tRNA synthetase gene of Bacillus subtilis. J. Bacteriol. 174:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 61.Wecke, T., D. Zühlke, U. Mäder, S. Jordan, B. Voigt, S. Pelzer, H. Labischinski, G. Homuth, M. Hecker, and T. Mascher. 2009. Daptomycin versus friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 53:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.