Abstract
Rifampin (rifampicin), an important antibiotic agent and a major drug used for the treatment of tuberculosis, exerts immunomodulatory effects. Previous studies have found that rifampin increases inducible nitric oxide (NO) synthase (iNOS) expression and NO production. The present study investigated the potential mechanism(s) underlying these actions. The incubation of human lung epithelial A549 cells with a cytokine mix (interleukin-1β, tumor necrosis factor alpha, and gamma interferon) induced the expression of iNOS mRNA. The addition of rifampin increased the iNOS level by 1.9 ± 0.3-fold at a dose of 10 μg/ml (P < 0.01) and by 4.0 ± 0.3-fold at a dose of 50 μg/ml (P < 0.001). Rifampin treatment also affected the transcription factors that regulate iNOS mRNA: there was an increased and prolonged degradation of the inhibitory subunit of NF-κB, a corresponding increase in the level of cytokine-induced DNA binding of NF-κB (2.1 ± 0.2-fold), and a decrease in the level of expression of peroxisome proliferator-activated receptor gamma (PPARγ). Specifically, the level of PPARγ expression dropped by 15% in response to cytokine stimulation and by an additional 40% when rifampin was added (P < 0.001). Rifampin had no effect on the activation of mitogen-activated protein kinases or the signal transducer and transcription activator (STAT-1). In conclusion, rifampin augments NO production by upregulating iNOS mRNA. It also increases the level of NF-κB activation and decreases the level of PPARγ expression. The increases in the levels of NF-κB activation and NO production probably contribute to the therapeutic effects of rifampin. However, given the role of NF-κB in upregulating many inflammatory genes and the roles of PPARγ in downregulating inflammatory genes and in lipid and glucose metabolism, these findings have implications for potential adverse effects of rifampin in patients with chronic inflammatory diseases and glucose or lipid disorders.
It is now generally recognized that antibiotics act not only on bacteria but also on eukaryotic cells. Evidence is accumulating that diverse antibacterial agents modulate the production of host mediators and may thereby have profound effects on the immune system and on physiological functions (22).
Rifampin (rifampicin) is a widely used antibiotic agent and a major drug for the treatment of mycobacterial infections (3). Its antibacterial activity is mediated by the inhibition of bacterial RNA polymerase. In addition, rifampin exerts immunomodulatory effects. It has been found to suppress macrophage phagocytosis, neutrophil chemotaxis, and delayed-type hypersensitivity and to alter cytokine production in human monocytes and mouse macrophages (17, 20, 21, 28, 37). Recently, in a study of human alveolar epithelial A549 cells, we reported that rifampin increased the levels of cytokine-induced inducible nitric oxide (NO) synthase (iNOS) expression and NO production but decreased the levels of interleukin 1β (IL-1β)-induced arachidonic acid release and prostaglandin E2 (PGE2) production (33, 34).
NO serves as a mediator of three major functions in the body: host defense, vascular homeostasis, and neurotransmission. Its production is controlled by a constitutive isoform and an inducible isoform of NOS. During infection, NO is produced by iNOS in response to bacterial components or a combination of proinflammatory cytokines, such as tumor necrosis factor alpha (TNF-α), IL-1β, and gamma interferon (IFN-γ) (16). In the immune system, NO is involved in diverse immunostimulatory as well as immunosuppressive processes. In some infections, such as those caused by Mycobacterium tuberculosis and Salmonella enterica serovar Typhimurium, the production of NO is crucial for disease control; in others, such as Streptococcus pneumoniae infections, a high level of NO is detrimental to the host (4).
The regulation of the iNOS response to cytokines occurs predominantly at the transcriptional level (7). The transcription factors and their signaling pathways are cell type and species specific; they may also vary according to the inducer (2). In most cells, iNOS transcription requires the activation of the transcription factor nuclear factor-kappa B (NF-κB) by TNF-α and IL-1β and activation of signal transducer and transcription activator 1 (STAT-1) by IFN-γ (7, 11). The signal transduction cascade elicited by TNF-α and IL-1β involves several mitogen-activated protein kinases (MAPKs): p38, extracellular signal-regulated kinase (ERK), and Jun N-terminal protein kinase (JNK)/stress-activated protein kinase (11, 15, 31).
The transcription of iNOS is also subject to negative control. The peroxisome proliferator-activated receptor gamma (PPARγ) is a ligand-dependent transcription factor that belongs to the nuclear receptor superfamily, the members of which control genes crucial for lipid and glucose metabolism (for a review, see reference 14). PPARγ has also been implicated in the suppression of inflammatory genes, including the gene encoding iNOS (5, 9, 19, 26, 27, 30). The ligands for PPARγ consist of both naturally occurring compounds, such as the prostaglandin D2 (PGD2) metabolite 15-deoxy-12,14-prostaglandin J2 (15dPGJ2), and synthetic compounds, such as thiazolidinedione antidiabetic agents. Upon ligand activation, PPARγ is heterodimerized with retinoid X receptor (RXR) and binds to specific peroxisome proliferator (PPARγ) response elements (PPREs) in the promoter region of target genes. Recent studies found that TNF-α and IL-1β downregulated PPARγ expression in rat and human cells (1, 10, 35) and that PPARγ activation blocked cytokine-induced iNOS expression in human lung epithelial cells (5, 24, 30).
The lung epithelial cell line A549 is used as a model of innate immunity in tuberculosis. The cells were shown to produce NO in response to M. tuberculosis infection. Their release of NO was increased by simultaneous stimulation with a mixture of TNF-α, IL-1β-, and IFN-γ, resulting in mycobactericidal activity (25).
Given the important role of NO in the host defense against tuberculosis and the extensive use of rifampin for the treatment of mycobacterial infections, we investigated the mechanism(s) underlying the augmentation of iNOS expression and NO production by rifampin in human alveolar epithelial A549 cells.
(This study was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy/IDSA Meeting, Washington, DC, October 2008.)
MATERIALS AND METHODS
Reagents.
The cell culture medium and its supplements were obtained from Biological Industries (Beit HaEmek, Israel). Recombinant human IL-1β, IFN-γ, and TNF-α were purchased from ProSpect-Tany TechnoGene Ltd. (Rehovot, Israel). Rifampin was obtained from Sigma Chemical (St. Louis, MO). A stock solution of rifampin (50 mg/ml) was prepared in dimethyl sulfoxide and was diluted at least 1:1,000 in culture medium prior to use.
Cell culture and exposure to cytokines.
Human alveolar epithelial A549 cells were obtained from the American Type Culture Collection (ATCC) and were maintained in F-12 medium. Human colon epithelial DLD-1 and HT-29 cells (ATCC collection) were grown in Dulbecco modified Eagle medium. The cells were grown in medium supplemented with 10% heat-inactivated fetal bovine serum, l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 μg/ml), and nystatin (12.5 U/ml) (PSN) at 37°C in a humidified incubator with 5% CO2. The cells were incubated in serum- and PSN-free medium for 24 h before stimulation and were then exposed to cytokines (100 ng/ml each) or cytokines with rifampin (10 to 50 μg/ml) in fresh serum- and PSN-free medium. Cell culture viability was evaluated by the neutral red uptake viability assay, as described previously (33). No differences in viability among the differently treated cells were found (data not shown).
Total RNA isolation and mRNA determination by real-time PCR.
RNA was isolated from cells with an EZ-RNA total isolation kit (Biological Industries), according to the manufacturer's instructions.
Two micrograms of total RNA was reverse transcribed to cDNA with the EZ-First Strand cDNA synthesis kit for reverse transcription-PCR (RT-PCR) (Biological Industries) by using random hexamer primers, according to the manufacturer's instructions. The cDNA samples were examined for iNOS with TaqMan Assay-on-Demand primers by using the TaqMan universal master mix with an ABI 7000 real-time PCR system (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions, and a Prism 7000 sequence detector (Applied Biosystems). The results for each mRNA were normalized to those for β-actin mRNA.
Western blot analysis.
For preparation of cytosolic extracts and Western blot analysis of iNOS, the inhibitory subunit of NF-κB (IκB), ERK, JNK, p38, STAT-1, and PPARγ, A549 cells were seeded in 3-cm dishes (2 × 106 cells/plate), grown for 24 h, and incubated in serum- and antibiotic-free medium for 24 h before exposure to the cytokine mix and rifampin. At the indicated times, the cells were washed twice with cold phosphate-buffered saline, lysed with a buffer (50 mM Tris, pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, 0.1% bromophenol blue, 1.25% 2-mercaptoethanol), and denatured at 95°C. Equal amounts of protein (20 to 30 μg) from total cell extracts, estimated by use of a bicinchoninic acid reagent (Pierce Biotechnology, Rockford, IL), were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Amersham Biosciences, Piscataway, NJ). Nonspecific binding sites were blocked at room temperature with 5% milk in TBST (20 mM Tris, pH 7.8, 150 mM NaCl, 0.1% Tween 20). The membranes were then incubated overnight at 4°C with the following antibodies: rabbit anti-iNOS, mouse anti-PPARγ, rabbit anti-IκB, and rabbit anti-JNK antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-phospho-JNK antibodies (Jackson ImmunoResearch, West Grove, PA); mouse anti-ERK1/2, mouse anti-phospho-ERK1/2, mouse anti-p38, and mouse anti-phospho-p38 antibodies (Sigma); rabbit anti-phospho-STAT-1 antibodies (Cell Signaling Technology, Danvers, MA); and goat anti-actin antibodies (Santa Cruz Biotechnology).
The membranes were washed and incubated with a secondary antibody (Santa Cruz Biotechnology), as appropriate, namely, horseradish peroxidase-linked anti-rabbit, anti-mouse, or anti-goat immunoglobulin antibody, at room temperature for 1 h and were washed three times with TBST. Bound antibodies were detected by the enhanced chemiluminescence method. Densitometry was performed with the VersaDoc imaging system (Bio-Rad Laboratories, Hercules, CA).
EMSA.
Nuclear extracts from A549 cells were harvested with an NE-PER extraction kit (Pierce). A biotin end-labeled probe (Sigma) containing the NF-κB consensus sequence (5′-AGTTGAGGGGACTTTCCCAGGC) was incubated with a nuclear extract (2 μl) by using a light-shift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce). The reaction mixture was then subjected to gel electrophoresis on a 5% native polyacrylamide gel with triborate EDTA (TBE) buffer and was transferred to a nylon membrane (Pierce). The biotin end-labeled DNA was detected by using the streptavidin-horseradish peroxidase conjugate and the chemiluminescent substrate (EMSA kit; Pierce). The specificity of the bands was confirmed with cold NF-κB oligonucleotide.
Statistical analysis.
The results are presented as the means ± standard errors of the means. The unpaired t test was used to compare the results for cells treated with cytokines or cytokines plus rifampin with those for cells treated with cytokines and medium alone (control).
RESULTS
Effect of rifampin on iNOS mRNA expression.
To investigate the mechanism underlying the enhancement of iNOS protein expression and NO production by rifampin, we first examined the effect of rifampin on iNOS mRNA expression. A549 cells were incubated with a cytokine mix (IL-1β, TFNα, and IFN-γ at 100 ng/ml each) in the presence or absence of rifampin. After 19 h, the cells were lysed and the RNA was purified. RT-PCR experiments were performed to analyze mRNA expression. For normalization, the expression of β-actin was determined in a parallel reaction with the same RNA sample.
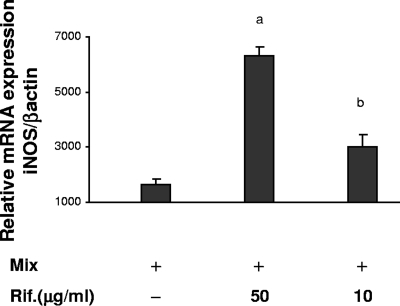
The unstimulated A549 cells did not express mRNA for iNOS. Incubation of the cells with cytokines resulted in a marked increase in the level of iNOS mRNA expression. The subsequent addition of rifampin caused a further dose-dependent increase in the level of iNOS mRNA: 1.9 ± 0.3-fold at a dose of 10 μg/ml (P < 0.01) and 4.0 ± 0.3-fold at a dose of 50 μg/ml (P < 0.001) (Fig. 1). Rifampin alone did not induce iNOS mRNA expression (data not shown).
FIG. 1.
Effect of rifampin (Rif.) on cytokine-induced iNOS mRNA expression. RNA was extracted 19 h after stimulation with IL-1ß, TNF-α, and IFN-γ (Mix; 100 ng/ml each) in the presence or the absence of rifampin, and iNOS mRNA was quantified by a TaqMan real-time RT-PCR. The amount of mRNA in the control cells, which were treated with medium alone, was defined as 1. The values represent the change compared with the results for the control cells. Data are presented as the means ± standard errors of the means from four separate experiments. The P values for the results for the mix versus the results for the mix plus rifampin were P < 0.001 (a) and P < 0.01 (b).
Effect of rifampin on NF-κB activation.
NF-κB binding to the iNOS promoter is an essential step in iNOS transcription. Thus, we sought to determine if rifampin affects NF-κB activation.
In the resting state, NF-κB exists in the cytoplasm as a complex bound to an inhibitor protein, IκB. Upon stimulation, IκB is degraded and the NF-κB released is translocated to the nucleus and activates target genes. We used immunoblotting to study the degradation of IκB in the cytoplasm and the DNA binding of the nuclear extracts with a consensus NF-κB probe in the presence of rifampin.
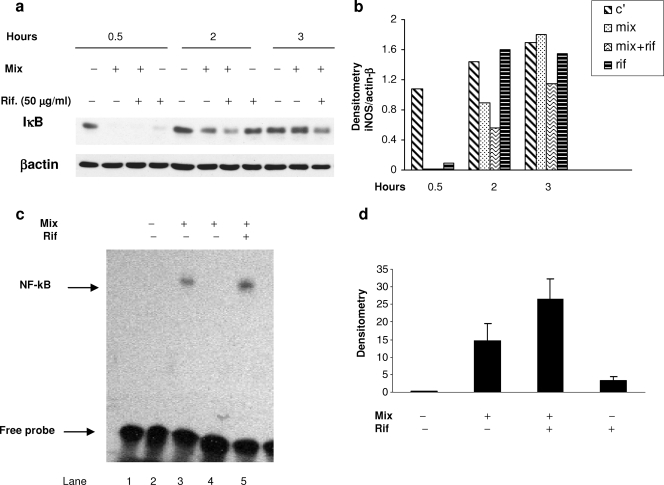
Incubation of the A549 cells with the cytokine mix caused the complete degradation of IκB at 30 min. Normalization to unstimulated levels occurred at 2 h. In cells cotreated with rifampin, the degradation of IκB was increased and prolonged (it was still evident at 3 h) (Fig. 2a). Treatment with rifampin alone also induced the degradation of IκB at 30 min, followed by a rapid return to the basal, unstimulated level.
FIG. 2.
Effect of rifampin (Rif.) on NF-κB activation. (a) Effect of rifampin on IκB degradation by Western blot analysis of IκB expression in A549 cells stimulated with IL-1ß, TNF-α, and IFN-γ (Mix; 100 ng/ml each) in the presence or absence of rifampin (50 μg/ml). Cytosolic extracts were prepared at the indicated times. Similar results were obtained in three similar experiments. (b) Densitometric analysis of the blot presented in panel a. C′, control. (c) EMSA for NF-κB activation. Equivalent nuclear extracts prepared from cells 2 h after stimulation with IL-1ß, TNF-α, and IFN-γ (Mix) with or without rifampin (50 μg/ml) were assayed for binding activity with an oligonucleotide-containing the NF-κB consensus sequence. Specific DNA binding complexes were identified by competition with unlabeled probe (lane 4). Lane 1, labeled probe alone. (d) Results of densitometric analysis from three experiments.
On EMSA, we found that exposure to the cytokine mix induced the DNA binding of NF-κB. The addition of rifampin (50 μg/ml) increased the level of DNA binding of NF-κB 2.1 ± 0.22-fold, but this result did not reach statistical significance (P = 0.07) (Fig. 2d). Rifampin alone also induced low-level NF-κB DNA binding. The specificity of the binding was confirmed by competition with an excess of unlabeled probe.
Effect of rifampin plus IFN-γ on iNOS expression.
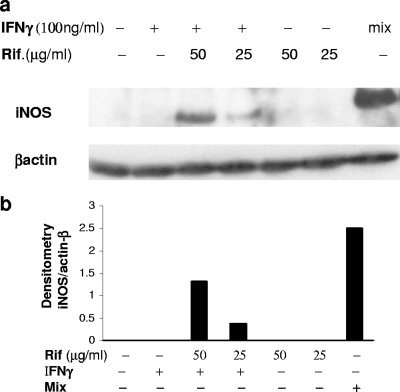
The transcription of iNOS in A549 cells requires the activation of NF-κB by TNF-α or IL-1β and the activation of STAT-1 by IFN-γ (7). To further confirm the contribution of rifampin to NF-κB activation and iNOS induction, we examined the expression of the iNOS protein in A549 cells incubated with rifampin and IFN-γ. Total cell extracts were prepared 20 h after stimulation and were tested by Western blot analysis. As shown in Fig. 3, neither rifampin nor IFN-γ alone induced iNOS expression, whereas incubation of the cells with rifampin together with IFN-γ induced iNOS, albeit by a much smaller amount than the amount induced by the cytokines (Fig. 3).
FIG. 3.
Effect of rifampin (Rif.) together with IFN-γ on the expression of iNOS. (a) Western blot analysis of the effect of IFN-γ and rifampin (Rif) on iNOS expression in A549 cells at 24 h after stimulation with IFN-γ (100 ng/ml) alone, rifampin alone, or IFN-γ plus rifampin compared to the level of β-actin expression. (b) Densitometric analysis of the blot presented in panel a. Similar results were obtained in three separate experiments.
Effect of rifampin on cytokine-induced MAPK and STAT-1 phosphorylation.
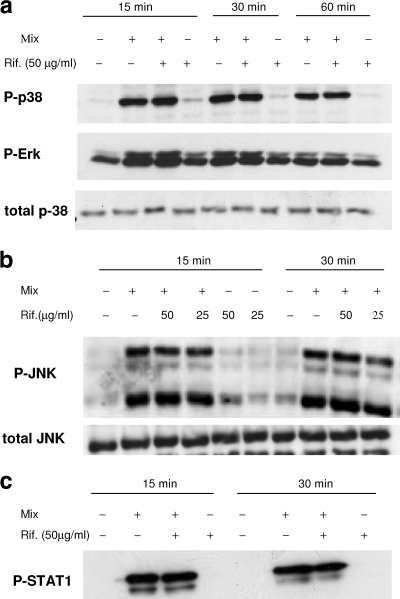
Previous studies have reported that the MAPK signaling proteins are involved in cytokine-induced iNOS transcription in A549 cells (12, 31). Therefore, we investigated whether rifampin upregulates iNOS mRNA via activation of the three major MAPKs, namely, ERK, JNK, and p38. We used immunoblot analysis with specific antibodies against their phosphorylated forms. Incubation of the cells with the cytokine mix induced the rapid and transient phosphorylation of ERK1/2, JNK, and p38 at 15 and 30 min; the phosphorylation started to decline at 60 min. The addition of rifampin (50 μg/ml) did not increase the phosphorylation beyond that induced by the mix (Fig. 4 a and b).
FIG. 4.
Effect of rifampin (Rif.) on the activation of MAPK and STAT-1. Cells were incubated with IL-1ß, TNF-α, and IFN-γ (Mix) in the absence or the presence of the indicated concentration of rifampin (Rif.). Cytoplasmic extracts were prepared at the indicated times and were subjected to Western immunoblotting with antibodies specific for the phosphorylated forms of ERK1/2, JNK, p-38, and STAT-1 (P-Erk, P-JNK, P-p38, and P-STAT-1, respectively). The results were compared to those obtained with ERK1/2, JNK, and p38. Data from one of three experiments with similar results are presented.
The Janus kinase signal transducer and activator of transcription (Jak-STAT-1) pathways, triggered by IFN-γ, is also involved in the activation of the iNOS promoter. As shown in Fig. 4c, the addition of rifampin had no effect on STAT-1 phosphorylation.
Effect of rifampin on PPARγ expression.
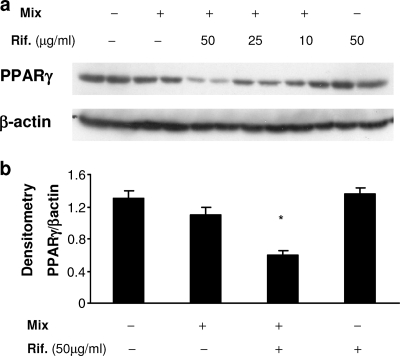
On the basis of earlier findings that the anti-inflammatory nuclear receptor PPARγ is involved in the downregulation of iNOS expression (18, 30) and that TNF-α and IL-1β decrease the levels of PPARγ expression in human liver cells (10), we sought to determine if rifampin modulates NO production via the PPARγ pathway. A549 cells were incubated with the cytokine mix in the presence or absence of rifampin for 24 h, and the PPARγ expression in whole-cell extracts was analyzed by immunoblotting. PPARγ was highly expressed in untreated A549 cells, in agreement with previous findings (23). Following incubation with the cytokines, there was a slight but not significant decrease in the level of PPARγ expression. The addition of rifampin further reduced the PPARγ level in a dose-dependent manner. Exposure to rifampin alone had no effect on the basal (unstimulated) level of expression of PPARγ (Fig. 5).
FIG. 5.
Downregulation of PPARγ expression by rifampin (Rif.). A Western blot analysis of the level of PPARγ expression in A549 cells was performed 24 h after treatment with medium alone; IL-1ß, TNF-α, and IFN-γ (Mix; 100 ng/ml each); and the mix with different doses of rifampin (Rif). The level of PPARγ expression was then normalized to the level of β-actin expression. (a) A representative blot from one of five experiments with similar results is shown. (b) Results of densitometric analysis in five experiments. *, P < 0.001 for the mix versus the mix plus rifampin.
Effect of rifampin on iNOS expression in human colon epithelial cells.
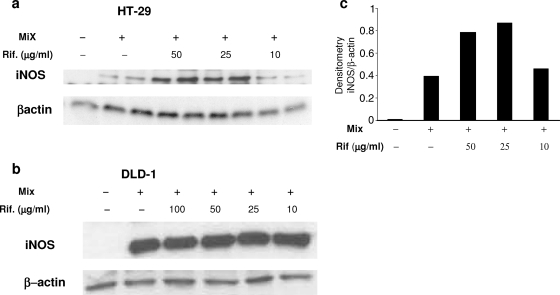
Because iNOS expression is differently controlled in various cells (12), we also examined the effect of rifampin on iNOS expression in two human colon cell lines, HT-29 and DLD-1 cells. As shown in Fig. 6, treatment with the cytokine mix induced iNOS expression in both cell lines. Similar to its effect in A549 cells, rifampin enhanced the level of iNOS expression in HT-29 cells in a dose-dependent manner (Fig. 6a). However, it did not increase the level of iNOS expression in cytokine-stimulated DLD-1 cells (Fig. 6b).
FIG. 6.
Effect of rifampin (Rif.) on iNOS expression in human colon epithelial cells. Western blot analysis of iNOS expression in HT-29 cells (a) and DLD-1 cells (b) was performed 24 h after stimulation with the cytokine mix or the cytokine mix plus rifampin (Rif.), and the level of expression was normalized to the level of β-actin expression. Representative blots of one of two experiments with similar results are shown in panels a and b. (c) Densitometric analysis of the blot presented in panel a.
DISCUSSION
The present study provides evidence that rifampin increases the level of cytokine-induced NF-κB activation, reduces the level of PPARγ expression, and augments cytokine-induced iNOS transcription. The increased transcription of iNOS leads to the increased expression of iNOS protein and NO production, as previously reported by our group (33).
The effect of rifampin on NF-κB in the present study was demonstrated by several approaches. Cotreatment of the A549 cells with rifampin and the cytokine mix was associated with an increased and prolonged degradation of IκB. Notably, rifampin alone also induced IκB degradation for a short time (30 min). The activation of NF-κB by rifampin was also reflected by the expression of the iNOS protein in cells cotreated with rifampin and IFN-γ, although neither of these stimulators alone induced iNOS production. Previous studies with A549 cells have shown that both the NF-κB pathway, triggered by IL-1ß and/or TNF-α, and the Jak-STAT-1 pathway, triggered by IFN-γ, are critical for iNOS transcription (7, 11). Thus, our finding that IFN-γ with rifampin induces iNOS indicates that rifampin not only enhances cytokine-induced NF-κB activation but also functionally triggers the NF-κB pathway sufficiently to induce iNOS transcription, without the presence of TNF-α or IL-1ß.
Since iNOS transcription could be differently regulated in different cells (2), we also investigated the effect of rifampin in colonic epithelial DLD-1 and HT-29 cells. The results supported the involvement of rifampin in NF-κB activation. As in A549 cells, rifampin increased the level of iNOS expression in HT-29 cells in a dose-dependent manner, although it was ineffective in DLD-1 cells. Interestingly, resting DLD-1 cells were reported to contain high basal levels of activated NF-κB. Furthermore, other studies with DLD-1 cells showed that IFN-γ alone induced iNOS expression and that NF-κB did not function as a major effector in the cytokine-mediated induction of iNOS mRNA (13). Thus, the inability of rifampin to enhance iNOS expression in DLD-1 cells is consistent with its role as an effector of NF-κB.
We also examined whether rifampin affects the phosphorylation of the MAPK signaling proteins ERK-1/2, p38, and JNK, which regulate the NF-κB pathway (12, 15, 31). Western blot analysis revealed that the phosphorylation of all three kinases increased rapidly in the presence of the cytokine mix, but the addition of rifampin did not increase it further. Rifampin also had no effect on the cytokine-induced activation of the signaling protein and transcription factor STAT-1. These results indicate that the action of rifampin is not mediated via the direct activation of MAPKs or STAT-1.
The nuclear receptor and transcription factor PPARγ has been implicated in the downregulation of inflammatory genes and specifically of iNOS. Several authors have demonstrated that the activation of PPARγ with endogenous or synthetic ligands blocked cytokine-induced iNOS expression in various human and mouse cells, including human lung epithelial cells (9, 19, 24, 27, 30). To shed further light on the mechanism of action of rifampin, we explored its possible effect on PPARγ expression. We found that resting A549 cells expressed a high level of PPARγ, and stimulation with the cytokine mix did not significantly reduce its level of expression. The addition of rifampin to the mix significantly reduced the level of PPARγ expression. We suspect that rifampin exerts a dual effect on PPARγ. Our earlier studies showed that rifampin is a potent inhibitor of arachidonic acid release and PGE2 production (34). Arachidonic acid is one of the natural ligands of PPARγ (14) and is also a precursor of the PGD2 metabolite 15dPGJ2, another well-recognized ligand of PPARγ. Therefore, a substantial reduction in arachidonic acid release may lead to a drop in the concentration of 15dPGJ2, in the same way in which it caused a drop in the concentration of PGE2, both of which are products of arachidonic acid via the cyclooxygenase pathway. The concomitant reduction in the level of expression of PPARγ and in the concentration of its ligands may amplify the effect of rifampin on PPARγ, leading to increased levels of iNOS transcription.
Besides profoundly suppressing the expression of PPARγ and its ligand, rifampin may also inhibit the activity of PPARγ. Rifampin binds to the nuclear receptor PXR, which creates a heterodimer with RXR, itself a partner of PPARγ in the hetrodimerization and activation of genes (29). Therefore, it is possible that rifampin competes with PPARγ for their mutual nuclear receptor, RXR.
The exact mechanism involved in the inhibition of iNOS transcription by PPARγ is not completely understood. It was shown that PPARγ competes with NF-κB and STAT-1 for the limited amounts of coactivator proteins, thereby reducing their activities (18). Hence, the rifampin-induced activation of NF-κB may be associated with the decrease in the level of PPARγ expression. Recently, a novel PPRE was identified in the murine iNOS promoter that binds to PPARγ (6). The PPRE contributed to negative iNOS expression in response to inflammatory stimuli. The same authors reported that rat and human promoters of iNOS contain sequences with significant homology to the mouse PPRE promoter. Therefore, it is possible that PPARγ acts directly on the iNOS promoter in A549 cells as well.
Rifampin has long been regarded as an anti-inflammatory drug. Whether it also serves as a ligand of the corticosteroid receptor remains controversial (8, 32). Our study shows that rifampin is a proinflammatory agent and acts via the suppression of PPARγ expression and the activation of NF-κB, which most probably leads to the augmentation of cytokine-induced iNOS transcription and NO production. However, rifampin may have a compound effect in inflammation, since it also inhibits arachidonic acid release and PGE2 production (34). In this aspect, it rather resembles corticosteroids.
The increase in iNOS transcription was achieved with pharmacologically relevant concentrations of rifampin, since its peak concentration in the serum following administration of the usual 600-mg dose is 15 μg/ml, whereas in other tissue and fluids, rifampin reaches higher levels (21, 36).
In conclusion, rifampin augments NO production by upregulating iNOS mRNA transcription possibly through two mechanisms: increasing the level of NF-κB activation and decreasing the level of PPARγ expression. The increases in the levels of NF-κB activation and NO production most probably contribute to the therapeutic effects of rifampin. However, NF-κB is involved in the upregulation and PPARγ is involved in the downregulation of many inflammatory genes, and PPARγ is also important in lipid and glucose metabolism. Others have reported that treatment with a PPARγ agonist improves insulin resistance and yields beneficial effects in controlling atherosclerosis (6). Therefore, our study suggests that rifampin administration may have unfavorable effects in patients with chronic inflammatory diseases and glucose or lipid metabolism disorders.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Afif, H., M. Benderdour, L. Mfuna-Endam, J. Martel-Pelletier, J. P. Pelletier, N. Duval, and H. Fahmi. 2007. Peroxisome proliferator-activated receptor gamma 1 expression is diminished in human osteoarthritic cartilage and is downregulated by interleukin-1beta in articular chondrocytes. Arthritis Res. Ther. 9:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aktan, F. 2004. iNOS-mediated nitric oxide production and its regulation. Life Sci. 75:639-653. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard, J. S. 1998. The ying and yang of rifampicin. Nat. Med. 4:92-96. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan, C. 2001. Nitric oxide and the immune response. Nat. Immunol. 2:907-914. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. W., Y. H. Chang, C. J. Tsi, and W. W. Lin. 2003. Inhibition of IFN-gamma-mediated inducible nitric oxide synthase induction by the peroxisome proliferator-activated receptor gamma agonist, 15-deoxy-delta 12,14-prostaglandin J2, involves inhibition of the upstream Janus kinase/STAT1 signaling pathway. J. Immunol. 171:979-988. [DOI] [PubMed] [Google Scholar]
- 6.Crosby, M. B., J. Svenson, G. S. Gildeson, and T. K. Nowling. 2005. A novel PPAR response element in the murine iNOS promoter. Mol. Immunol. 42:1303-1310. [DOI] [PubMed] [Google Scholar]
- 7.Ganster, R. W., B. S. Taylor, L. Shao, and D. A. Geller. 2001. Complex regulation of human inducible nitric oxide synthase gene transcription by Stat 1 and NF-κB. Proc. Natl. Acad. Sci. USA 98:8638-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herr, A. S., G. M. Wochnik, M. C. Rosenhagen, F. Holsboer, and T. Rein. 2000. Rifampicin is not an activator of glucocorticoid receptor. Mol. Pharmacol. 57:732-737. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, C., T. Ting, and B. Seed. 1998. PPAR gamma agonists inhibit production of monocytic inflammatory cytokines. Nature 391:82-86. [DOI] [PubMed] [Google Scholar]
- 10.Kim, M. S., T. R. Sweeney, J. K. Shigenaga, L. G. Chui, A. Moser, C. Grunfeld, and K. R. Feingold. 2007. Tumor necrosis factor and interleukin 1 decrease RXRalpha, PPARalpha, PPARgamma, LXRalpha, and the coactivators SRC-1, PGC-1alpha, and PGC-1beta in liver cells. Metabolism 56:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinert, H., A. Pautz, K. Linker, and P. M. Schwarz. 2004. Regulation of the expression of inducible nitric oxide synthase. Eur. J. Pharmacol. 500:255-266. [DOI] [PubMed] [Google Scholar]
- 12.Kleinert, H., P. M. Schwarz, and U. Forstermann. 2003. Regulation of the expression of inducible nitric oxide synthase. Biol. Chem. 384:1343-1364. [DOI] [PubMed] [Google Scholar]
- 13.Kleinert, H., T. Wallerath, G. Fritz, I. Ihrig-Biedert, F. Rodriuez-Pascual, D. A. Geller, and U. Forstermann. 1998. Cytokine induction of NO synthase II in human DLD-1 cells: roles of JAK-STAT, AP-1 and NF-κB-signaling pathways. Br. J. Pharmacol. 125:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kota, B. P., T. H. Huang, and B. D. Roufogalis. 2005. An overview on biological mechanisms of PPARs. Pharmacol. Res. 51:85-94. [DOI] [PubMed] [Google Scholar]
- 15.Kristof, A. S., J. Marks-Konczalik, and J. Moss. 2001. Mitogen-activated protein kinases mediate activator protein-1-dependent human inducible nitric-oxide synthase promoter activation. J. Biol. Chem. 276:8445-8452. [DOI] [PubMed] [Google Scholar]
- 16.Kwon, S., and S. C. George. 1999. Synergistic cytokine-induced nitric oxide production in human alveolar epithelial cells. Nitric Oxide 3:348-357. [DOI] [PubMed] [Google Scholar]
- 17.Labro, M.-T. 2005. Anti-inflammatory activity of ansamycins. Expert Rev. Anti-Infect. Ther. 3:91-103. [DOI] [PubMed] [Google Scholar]
- 18.Li, M., G. Pascual, and C. K. Glass. 2000. Peroxisome proliferator-activated receptor gamma-dependent repression of the inducible nitric oxide synthase gene. Mol. Cell. Biol. 20:4699-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez, M., and M. C. LaPointe. 2003. PPAR γ inhibition of cyclooxygenase-2, PGE2 synthase, and inducible nitric oxide synthase in cardiac myocytes. Hypertension 42:844-850. [DOI] [PubMed] [Google Scholar]
- 20.Mlambo, G., and L. B. Sigola. 2003. Rifampicin and dexamethasone have similar effects on macrophage phagocytosis of zymosan, but differ in their effects on nitrite and TNF-α production. Int. Immunopharmacol. 3:513-522. [DOI] [PubMed] [Google Scholar]
- 21.Pahlevan, A. A., D. J. M. Wright, L. Bradley, C. Smith, and B. M. J. Foxwell. 2002. Potential of rifamides to inhibit TNF-induced NF-κB activation. J. Antimicrob. Chemother. 49:531-534. [DOI] [PubMed] [Google Scholar]
- 22.Pasquale, T. R., and J. S. Tan. 2005. Nonantimicrobial effects of antibacterial agents. Clin. Infect. Dis. 40:127-135. [DOI] [PubMed] [Google Scholar]
- 23.Pawliczak, R., C. Han, X. L. Huang, A. J. Demetris, J. H. Shelhamer, and T. Wu. 2002. 85-kDa cytosolic phospholipase A2 mediates peroxisome proliferator-activated receptor gamma activation in human lung epithelial cells. J. Biol. Chem. 36:33153-33163. [DOI] [PubMed] [Google Scholar]
- 24.Ricote, M., A. C. Li, T. M. Willson, C. J. Kelly, and C. K. Glass. 1998. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 391:79-82. [DOI] [PubMed] [Google Scholar]
- 25.Roy, S., S. Sharma, M. Sharma, R. Aggarwal, and M. Bose. 2004. Induction of nitric oxide release from the human alveolar epithelial cell line A549: an in vitro correlate of innate immune response to Mycobacterium tuberculosis. Immunology 112:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirsjo, A., A. C. Gidlof, A. Olsson, H. Torma, M. Ares, H. Kleinert, U. Forstermann, and G. K. Hansson. 2000. Retinoic acid inhibits nitric oxide synthase-2 expression through the retinoic acid receptor-α. Biochem. Biophys. Res. Commun. 270:846-851. [DOI] [PubMed] [Google Scholar]
- 27.Sung, B., S. Park, B. P. Yu, and H. Y. Chung. 2006. Amelioration of age-related inflammation and oxidative stress by PPARgamma activator: suppression of NF-kappaB by 2,4-thizolidinedione. Exp. Gerontol. 41:590-599. [DOI] [PubMed] [Google Scholar]
- 28.Tentori, L., G. Graziani, S. A. Porcelli, M. Sugita, M. B. Brenner, R. Madaio, E. Bonmassar, A. Giuliani, and A. Aquino. 1998. Rifampin increases cytokine-induced expression of the CD1b molecule in human peripheral blood monocytes. Antimicrob. Agents Chemother. 42:550-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchimura, K., M. Nakamuta, M. Enjoji, I. Takashi, R. Sugimoto, T. Muta, H. Iwamoto, and J. Nawata. 2001. Activation of retinoic X receptor and peroxisome proliferator-activated receptor-γ inhibits nitric oxide and tumor necrosis factor-α production in rat Kupffer cells. Hepatology 30:91-99. [DOI] [PubMed] [Google Scholar]
- 30.Wang, A. C., X. Dai, B. Luu, and D. J. Conrad. 2001. Peroxisome proliferator-activated receptor-gamma regulates airway epithelial cell activation. Am. J. Respir. Cell Mol. Biol. 24:688-693. [DOI] [PubMed] [Google Scholar]
- 31.Werber, S., I. Shalit, I. Fabian, G. Steuer, T. Weiss, and H. Blau. 2005. Moxifloxacin inhibits cytokine-induced MAP kinase and NF-κB activation as well as nitric oxide synthesis in a human respiratory epithelial cell line. Antimicrob. Chemother. 55:293-300. [DOI] [PubMed] [Google Scholar]
- 32.Yerramasetti, R., S. Gollapudi, and S. Gupta. 2002. Rifampicin inhibits CD95-mediated apoptosis of Jurkat T cells via glucocorticoid receptors by modifying the expression of molecules regulating apoptosis. J. Clin. Immunol. 22:37-45. [DOI] [PubMed] [Google Scholar]
- 33.Yuhas, Y., E. Berent, H. Ovadia, I. Azoulay, and S. Ashkenazi. 2006. Rifampin augments cytokine-induced nitric oxide production in human alveolar epithelial cells. Antimicrob. Agents Chemother. 50:396-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuhas, Y., I. Azoulay-Alfaguter, E. Berent, and S. Ashkenazi. 2007. Rifampin inhibits prostaglandin E2 production and arachidonic acid release in human alveolar epithelial cells. Antimicrob. Agents Chemother. 51:4225-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, M., R. Wu, W. Dong, A. Jacob, and P. Wang. 2008. Endotoxin downregulates peroxisome proliferator-activated receptor-gamma via the increase in TNF-alpha release. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294:R84-R92. [DOI] [PubMed] [Google Scholar]
- 36.Ziglam, H. M., D. R. Baldwin, I. Daniels, J. M. Andrews, and R. G. Finch. 2002. Rifampin concentrations in bronchial mucosa, epithelial lining fluid, alveolar macrophages and serum following a single 600 mg oral dose in patients undergoing fibre-optic bronchoscopy. J. Antimicrob. Chemother. 50:1011-1015. [DOI] [PubMed] [Google Scholar]
- 37.Ziglam, H. M., I. Daniels, and R. G. Finch. 2004. Immunomodulating activity of rifampicin. J. Chemother. 16:357-361. [DOI] [PubMed] [Google Scholar]