Abstract
blaVEB-6 was found on the Proteus mirabilis chromosome in a context similar to those of blaVEB-1a and blaVEB-1b, in a truncated gene cassette flanked by 135-bp elements and duplications of the 3′-conserved segment of class 1 integrons. A linked aacA4-aadB-dfrA1-orfC cassette array includes components of Tn1331, illustrating the complex mosaicism of multiresistance regions.
Resistance to extended-spectrum β-lactams is an important clinical problem. In addition to the large TEM, SHV, and CTX families, several minor extended-spectrum β-lactamases have been identified (22), including VEB. All VEB enzymes identified to date (Table 1) are minor variants of VEB-1, which confers a high level of resistance to ceftazidime, cefotaxime, and aztreonam (26). blaVEB genes have been identified in a variety of species of Enterobacteriaceae and in nonfermenting bacilli from Asia, Europe, the Middle East, Africa, and North and South America (22) (Table 2) on both plasmids and the chromosome.
TABLE 1.
Amino acid differences among VEB variants
| VEB variant | Amino acid position for indicated type of peptidea
|
GenBank accession no.b | Reference or source | ||||
|---|---|---|---|---|---|---|---|
| Leader
|
Mature
|
||||||
| 18 | 19 | 25 | 56 | 104 | |||
| 1 | Ile | Val | Thr | Leu | Thr | AF010416 | 26 |
| 1a | Val | Val | Thr | Leu | Thr | AF324833 | 28 |
| 1b | Val | Glu | Thr | Leu | Thr | AF324834 | 28 |
| 2 | Ile | Val | Ala | Leu | Thr | AY027870 | 8 |
| 3 | Val | Val | Thr | Phe | Thr | AY536519 | 11 |
| 4 | Ile | Val | Ala | Leu | Met | EF136375 | 1 |
| 5 | Val | Glu | Thr | Leu | Met | EF420108 | |
| 6 | Val | Val | Ala | Leu | Met | EU259884 | This work |
Positions were numbered consecutively from the start codon. Amino acids differing from those in VEB-1 are in boldface.
Accession numbers in boldface type are listed at http://www.lahey.org/Studies.
TABLE 2.
Distribution and contexts of blaVEB genes
| blaVEB variant | Speciesa | Country | GenBank accession no. | Genetic contextd | Locationf | Reference(s) or sourceg |
|---|---|---|---|---|---|---|
| 1 | E. coli | Vietnam | AF205943b | qacI-aadB-aacA1/orfG-veb-aadB-arr2-cmlA-oxa10/aadA1 | P | 20 and 26 |
| K. pneumoniae | Vietnam | Not available | Not available | P | 26 | |
| P. mirabilis | Vietnam | AF220758 | qacI-aadB-aacA1/orfG-veb-aadB-?-oxa10-? | P | 18 | |
| A. xylosoxidans | France | DQ393569 | dfrA14-veb-aadB | P | 23 | |
| A. baumannii | France | CT025832c | veb-aadB-arr2-cmlA-oxa10/aadA1 | C | 7 | |
| A. baumannii | Belgium | Not available | veb-aadB-arr2-cmlA-oxa10/aadA1 | C | 19 | |
| E. coli | Canada | Not available | oxa10/aadA1-qacI-aadB-aacA1/orfG-veb-aadB-arr2-cmlA-oxa10/aadA1 | P | 27 | |
| E. coli | Canada | Not available | Not available | NA | 25 | |
| P. aeruginosa | China | AY536743 | veb-aadB-oxa10/aadA1 | NA | ||
| P. aeruginosa | China | DQ333895 | Incomplete veb gene sequence | NA | ||
| P. aeruginosa | Thailand | AF133699 | veb-aadB | C | 21 | |
| P. aeruginosa | Thailand | AF078527 | veb-aadB-arr2-cmlA-? | NA | 34 | |
| P. aeruginosa | Thailand | Not available | veb-aadB-arr2-cmlA-oxa10/aadA1 | C | 8 | |
| E. coli | Thailand | Not available | Various cassette arrays were partially characterized by PCR | P | 9 | |
| K. pneumoniae | Thailand | |||||
| E. cloacae | Thailand | |||||
| E. sakazakii | Thailand | |||||
| P. aeruginosa | Bulgaria | Not available | Not available | NA | 31 | |
| 1a | A. baumannii | Argentina | Not available | PCR suggests that veb is not located in a cassette array or typical Re/3′-CS structure | C | 24 |
| P. aeruginosa | Bangladesh | DQ315788 | Re | C | 17 | |
| P. aeruginosa | India | AY444815 | Re | C | 2 | |
| P. aeruginosa | Kuwait | AF324833 | PCR suggests that veb is the last cassette in an array | P | 28 | |
| P. aeruginosa | United Kingdom | Not available | Not available | NA | 36 | |
| 1b | P. aeruginosa | Kuwait | AF324834 | PCR suggests that veb is the last cassette in an array | C | 28 |
| C. freundii | Kuwait | Not available | Not available | NA | 4 | |
| P. mirabilis | Korea | Not available | Cassette array; aadB also present | C | 13 | |
| P. stuartii | Algeria | Not available | Re | P | 3 | |
| 2 | P. aeruginosa | Thailand | AY027870 | veb-aadB-arr2-cmlA-oxa10/aadA1 | C | 8 |
| P. aeruginosa | Thailand | Not available | ?-veb-aadB-arr2-cmlA-oxa10/aadA1 | C | 8 | |
| 3 | E. cloacae | China | AY536519 | No arraye | C | 11 |
| P. aeruginosa | China | Not available | Not available | NA | 12 | |
| A. baumannii | Taiwan | Not available | Not in cassette array? | 10 | ||
| 4 | P. mirabilis | Spain | EF136375 | veb gene sequence only | C | 1 |
| 5 | E. coli | United States | EF420108 | Sequence includes start of veb cassette, suggesting a cassette array | NA | |
| 6 | P. mirabilis | Australia | EU259884 | Re | C | This work |
K. pneumoniae, Klebsiella pneumoniae; A. baumannii, Acinetobacter baumannii; A. xylosoxidans, Achromobacter xylosoxidans; C. freundii, Citrobacter freundii; E. cloacae, Enterobacter cloacae; E. sakazakii, Enterobacter sakazakii.
GenBank accession no. AF205943 contains a longer sequence from the same strain as in AF010416 (Table 1).
Partial genome sequence of A. baumannii AYE. The same sequence is also available from the whole-genome sequence of AYE under GenBank accession no. CU459141.
Contexts available from the GenBank entry or a published paper are listed. ?, the remainder of the array was not determined; Re, a truncated veb cassette is found in a structure with repeat elements (Fig. 1A).
The sequence includes the first 7 bp of the blaVEB-3 cassette, but the cassette is interrupted after 937 of 1,070 bp and the attC site is missing. A short region (34 bp) matching the right end of ISCR1 immediately follows, and then the left end of IS6100, probably explaining why an amplicon carrying blaVEB-3 was not obtained using primers in the 5′-CS and 3′-CS (11).
P, blaVEB gene found on a plasmid obtained by conjugation or electroporation; C, blaVEB gene in a whole-genome sequence or found on the chromosome by hybridization (in boldface type) or presumed to be on the chromosome from the absence of transconjugants/transformants and/or plasmid DNA; NA, information not available.
Reference 8 includes several other cassette arrays, but the blaVEB variant is not specified.
blaVEB-1 was first described in a gene cassette in a class 1 integron (26), and most other examples of blaVEB genes where enough sequence data are available are also found in cassette arrays in class 1 integrons. These arrays are mostly related (Table 2), containing different combinations from a limited set of cassettes in different configurations, suggesting rearrangements mediated by both homologous and IntI-catalyzed recombination. The blaVEB cassette is followed by the aadB cassette in almost all of these arrays, and the 5′-conserved segment (5′-CS) is interrupted in several cases by IS1999, with or without IS2000 (21).
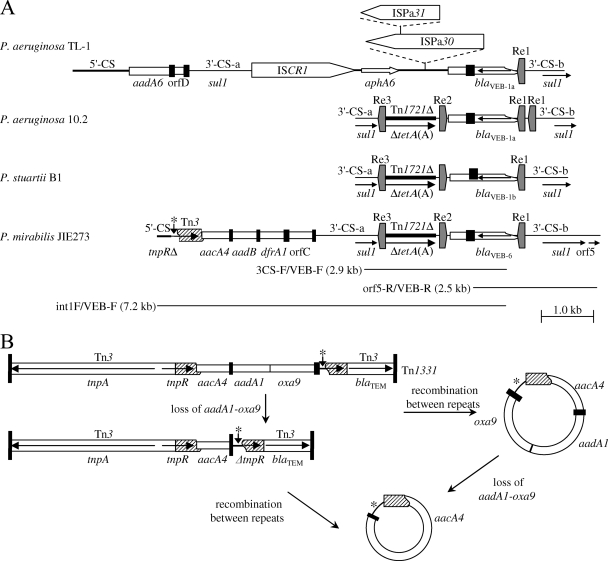
blaVEB-1a and blaVEB-1b cassettes missing the first 7 bp have been found outside arrays in regions containing one or more 135-bp repeat elements (Re1, Re2, Re3) (Fig. 1A). In both Pseudomonas aeruginosa 10.2 (GenBank accession no. AY444815) (2) and Providencia stuartii B1 (3), Re1 and Re2 (71% identical) flank a region containing a truncated blaVEB cassette, and Re2 and Re3 (98% identical to Re1) flank a region apparently largely derived from Tn1721 (GenBank accession no. X61367) that includes the tetA(A) gene truncated at both ends. This whole structure is flanked by duplications of the 3′-CS of class 1 integrons (3′-CS-a and 3′-CS-b) (Fig. 1A), and strain 10.2 has an extra copy of Re1. In P. aeruginosa TL-1 (GenBank accession no. DQ315788) (17), only Re1 is present, and blaVEB-1a is linked to ISCR1, the aphA6 gene, and the aadA6-orfD cassette array. A strong promoter in Re1 was found to drive expression of blaVEB-1a (2), and the Re and/or the 3′-CS duplications in the first two structures may provide an alternative means for the movement of blaVEB genes by homologous recombination.
FIG. 1.
(A) Genetic structures containing blaVEB genes and repeat elements. The species and isolate names are shown on the left. Insertion sequences are shown as pointed boxes labeled with the insertion sequence number, and Re1, Re2, and Re3 and their orientations are indicated. Gene cassettes are shown as open boxes, with small, filled boxes representing their attC sites. The extents of other resistance genes are shown, and the 5′-CS and 3′-CS of class 1 integrons and transposon fragments are labeled. Selected PCR products used for mapping are indicated by lines labeled with the primers used to amplify them. The region marked with an asterisk represents the first 1 to 111/114 bp of the 3′-CS. The structures for P. aeruginosa strains TL-1 and 10. 2 were drawn from sequences available under GenBank accession no. DQ315788 and AY444815, respectively, and P. stuartii B1 was drawn from the work of Aubert et al. (3). No additional information about flanking regions is available for 10.2 or B1. (B) Structure of Tn1331 and model for creation of a circular cassette containing aacA4 fused to the start of the blaTEM-1a gene. Circular molecules are not shown to scale. Tn1331 was drawn from the sequence available under GenBank accession no. AF479774.
We have previously reported a novel blaVEB variant, designated blaVEB-6, in a Proteus mirabilis clinical isolate (JIE273) that is resistant to cefotaxime and ceftazidime (37). VEB-6 is essentially identical to VEB-4 (1), the A52G variation, predicting only a conservative amino acid substitution (Ile18Val) in the leader peptide.
Repeated attempts to transfer blaVEB-6 from JIE273 to DH5αRf (a rifampin-resistant variant of Escherichia coli DH5α) by conjugation (38) and by electroporation with alkaline lysis preparations were unsuccessful. Whole-cell DNA from JIE273 was digested with I-CeuI (New England Biolabs, Ipswich, MA) (15), electrophoresed (for 36 h at a switch time of 5 to 60 s) (CHEF-DR II; Bio-Rad, Hercules, CA), and transferred to a Hybond-N+ membrane (GE Healthcare, Piscataway, NJ). Digoxigenin-labeled (DIG DNA labeling and detection kit; Roche, Penzberg, Germany) blaVEB and 16S rRNA PCR amplicons (see Table 3) hybridized to the same (ca. 291-kb) I-CeuI fragment (data not shown), suggesting that blaVEB-6 is located on the chromosome of JIE273.
TABLE 3.
Primers used in this work
| Primer | Sequence (5′-3′) | Target | Reference or source |
|---|---|---|---|
| VEB-F | CGACTTCCATTTCCCGATGC | blaVEB | 19 |
| VEB-R | GGACTCTGCAACAAATACGC | ||
| hep58 | TCATGGCTTGTTATGACTGT | 5′-CS | 35 |
| hep59 | GTAGGGCTTATTATGCACGC | 3′-CS | |
| Int1F | CAGTGGACATAAGCCTGTTC | intI1 | 14 |
| 3CS-F | CTATTGGTCTCGGTGTCG | 3′-CS | 6 |
| 3CS-R | ATCGTTCAGGTAGCCCAC | 3′-CS | |
| orf5-R | ACGAAGGTCTCCGCGAATGTC | 3′-CS | This work |
| A | AGAGTTTGATCHTGGYTYAGA | 16S rRNA | 16 |
| B | ACGGYTACCTTGTTACGACTT |
PCR mapping (Table 3; Fig. 1A), including by long-range PCR (Expand Long Template PCR system; Roche, Mannheim, Germany), and sequencing revealed a genetic context most similar to blaVEB-1b in P. stuartii B1. The aacA4-aadB-dfrA1-orfC array found adjacent to the 3′-CS-a was preceded by an unusual structure: the end of the 5′-CS is followed by the first 111 to 114 bp of the 3′-CS (Fig. 1) and then by 517 to 520 bp from the transposon Tn3 (three A residues could be derived from either the 3′-CS or Tn3). The Tn3 region includes the end of tnpR and the start of blaTEM-1a and is followed by a short sequence (AAACAAAG) derived from the attI1 sites of class 1 integrons (Fig. 1A).
These components are all found in a different configuration in Tn1331 (32) and related transposons. Tn1331 (Fig. 1B) is a derivative of Tn3, with a 517- to 520-bp duplication at the end of the Tn3 tnpR gene and at the start of the blaTEM-1a gene flanking the aacA4-aadA1-blaOXA-9 cassette array and the first 111 to 114 bp of the 3′-CS (Fig. 1B). The sequence AAACAAAG derived from the attI1 site links the first 19 bp of blaTEM to the start of the aacA4 cassette, providing both a promoter and a ribosome binding site for aacA4 expression and giving an AacA4 protein with a 17-amino-acid N-terminal extension (33). The aadA1 and blaOXA-9 cassettes are separated by part of the attI1 site rather than a complete aadA1 attC site.
The configuration seen in pJIE273 may be derived from Tn1331 or a related transposon, but in Tn1331, the 111- to 114-bp 3′-CS region follows the cassette array, while in JIE273, it precedes the cassette array. This region would be brought to the start of the aacA4 cassette in a circular molecule created by homologous recombination between the duplicated regions of Tn1331 (Fig. 1B). Tn1331.2 has a duplication of the tnpR-aacA4-aadA1-blaOXA-9 region, and experimentally observed conversion to Tn1331 (30) could also occur by loss of this circular molecule. Loss of aadA1-blaOXA-9 from the circular molecule would effectively give an extended aacA4 cassette and could occur either before or after circularization. It has recently been demonstrated that aadA1-blaOXA-9 can be readily excised from Tn1331 (29), and circles containing more than one cassette were found to separate into individual cassettes before insertion into class 1 integrons (5). The extended aacA4 cassette could then be inserted in front of an aadB-dfrA1-orfC array by IntI1-mediated recombination to give the configuration seen in pJIE273.
The only example of an aadB-dfrA1-orfC array currently in GenBank has three nucleotide differences from the corresponding part of the aacA4-aadB-dfrA1-orfC array in JIE273 and is found in P. aeruginosa TL-1 (GenBank accession no. DQ315789, where orfC is referred to as orfX), although it is not linked to blaVEB-1a (17). It is also interesting that in P. aeruginosa TL-1, the aadA6-orfD array is linked to blaVEB-1a, while a related array, aacA8-blaOXA-2a-aacA7-aadA6-orfD, was found in P. aeruginosa 10.2 but could not be linked to blaVEB-1a by PCR. These similarities suggest multiple recombination events. The association of blaVEB-6 with a class 1 integron and components of both Tn1721 and Tn1331 illustrates recombinations and rearrangements of a limited set of different components which, in addition to the actions of individual mobile elements, all contribute to the mosaicism characteristic of many complex multiresistance regions.
The patient carrying JIE273 was born in and had recently traveled to India, and blaVEB was not detected in other enterobacterial isolates from Sydney (38). P. aeruginosa strains 10.2 and TL-1 were also isolated from patients in India/Bangladesh (2, 17), and the similarities between the genetic contexts of blaVEB-6 in JIE273 and blaVEB-1a in these strains suggest that this structure, if not this bacterial strain, was acquired in that region.
Nucleotide sequence accession number.
The nucleotide sequence surrounding blaVEB-6 has been added to GenBank accession no. EU259884.
Acknowledgments
We are grateful to Lee Thomas for collecting isolate JIE273. We also thank Laurent Poirel and Thierry Naas (Hôpital de Bicêtre, Paris, France) for helpful discussions.
Z.Z. is supported by an Endeavor International Postgraduate Student Scholarship from the Australian Government Department of Education, Science and Training. S.R.P. is supported by grants from the National Health and Medical Research Council of Australia.
Footnotes
Published ahead of print on 12 January 2009.
REFERENCES
- 1.Aragón, L. M., B. Mirelis, E. Miró, C. Mata, L. Gómez, A. Rivera, P. Coll, and F. Navarro. 2008. Increase in β-lactam-resistant Proteus mirabilis strains due to CTX-M- and CMY-type as well as new VEB- and inhibitor-resistant TEM-type β-lactamases. J. Antimicrob. Chemother. 61:1029-1032. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended-spectrum β-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, D., T. Naas, M. F. Lartigue, and P. Nordmann. 2005. Novel genetic structure associated with an extended-spectrum β-lactamase blaVEB gene in a Providencia stuartii clinical isolate from Algeria. Antimicrob. Agents Chemother. 49:3590-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattoir, V., L. Poirel, V. Rotimi, C. J. Soussy, and P. Nordmann. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394-397. [DOI] [PubMed] [Google Scholar]
- 5.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 6.Espedido, B. A., S. R. Partridge, and J. R. Iredell. 2008. blaIMP-4 in different genetic contexts in Enterobacteriaceae from Australia. Antimicrob. Agents Chemother. 52:2984-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 9.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, L. Y., T. L. Chen, P. L. Lu, C. A. Tsai, W. L. Cho, F. Y. Chang, C. P. Fung, and L. K. Siu. 2008. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin. Microbiol. Infect. 14:1010-1019. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, X., Y. Ni, Y. Jiang, F. Yuan, L. Han, M. Li, H. Liu, L. Yang, and Y. Lu. 2005. Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 β-lactamase in China. J. Clin. Microbiol. 43:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, X., Z. Zhang, M. Li, D. Zhou, F. Ruan, and Y. Lu. 2006. Detection of extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J. Y., Y. J. Park, S. I. Kim, M. W. Kang, S. O. Lee, and K. Y. Lee. 2004. Nosocomial outbreak by Proteus mirabilis producing extended-spectrum β-lactamase VEB-1 in a Korean university hospital. J. Antimicrob. Chemother. 54:1144-1147. [DOI] [PubMed] [Google Scholar]
- 14.Koeleman, J. G., J. Stoof, M. W. Van Der Bijl, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2001. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 39:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri, H., S. Bellais, and P. Nordmann. 2002. Chromosome-encoded β-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob. Agents Chemother. 46:3561-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas, T., D. Aubert, T. Lambert, and P. Nordmann. 2006. Complex genetic structures with repeated elements, a sul-type class 1 integron, and the blaVEB extended-spectrum β-lactamase gene. Antimicrob. Agents Chemother. 50:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naas, T., F. Benaoudia, S. Massuard, and P. Nordmann. 2000. Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46:703-711. [DOI] [PubMed] [Google Scholar]
- 19.Naas, T., P. Bogaerts, C. Bauraing, Y. Degheldre, Y. Glupczynski, and P. Nordmann. 2006. Emergence of PER and VEB extended-spectrum β-lactamases in Acinetobacter baumannii in Belgium. J. Antimicrob. Chemother. 58:178-182. [DOI] [PubMed] [Google Scholar]
- 20.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 22.Naas, T., L. Poirel, and P. Nordmann. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):42-52. [DOI] [PubMed] [Google Scholar]
- 23.Neuwirth, C., C. Freby, A. Ogier-Desserrey, S. Perez-Martin, A. Houzel, A. Pechinot, J. M. Duez, F. Huet, and E. Siebor. 2006. VEB-1 in Achromobacter xylosoxidans from cystic fibrosis patient, France. Emerg. Infect. Dis. 12:1737-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasteran, F., M. Rapoport, A. Petroni, D. Faccone, A. Corso, M. Galas, M. Vazquez, A. Procopio, M. Tokumoto, and V. Cagnoni. 2006. Emergence of PER-2 and VEB-1a in Acinetobacter baumannii strains in the Americas. Antimicrob. Agents Chemother. 50:3222-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitout, J. D., P. Le, D. L. Church, D. B. Gregson, and K. B. Laupland. 2008. Antimicrobial susceptibility of well-characterised multiresistant CTX-M-producing Escherichia coli: failure of automated systems to detect resistance to piperacillin/tazobactam. Int. J. Antimicrob. Agents 32:333-338. [DOI] [PubMed] [Google Scholar]
- 26.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum β-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., V. O. Rotimi, E. M. Mokaddas, A. Karim, and P. Nordmann. 2001. VEB-1-like extended-spectrum β-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg. Infect. Dis. 7:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez, M. S., T. R. Parenteau, D. Centron, and M. E. Tolmasky. 2008. Functional characterization of Tn1331 gene cassettes. J. Antimicrob. Chemother. 62:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soler Bistue, A. J., D. Birshan, A. P. Tomaras, M. Dandekar, T. Tran, J. Newmark, D. Bui, N. Gupta, K. Hernandez, R. Sarno, A. Zorreguieta, L. A. Actis, and M. E. Tolmasky. 2008. Klebsiella pneumoniae multiresistance plasmid pMET1: similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS ONE 3:e1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strateva, T., V. Ouzounova-Raykova, B. Markova, A. Todorova, Y. Marteva-Proevska, and I. Mitov. 2007. Problematic clinical isolates of Pseudomonas aeruginosa from the university hospitals in Sofia, Bulgaria: current status of antimicrobial resistance and prevailing resistance mechanisms. J. Med. Microbiol. 56:956-963. [DOI] [PubMed] [Google Scholar]
- 32.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 33.Tran van Nhieu, G., and E. Collatz. 1987. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded β-lactamase. J. Bacteriol. 169:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, P. A., C. J. McIver, Y. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 36.Woodford, N., J. Zhang, M. E. Kaufmann, S. Yarde, M. Tomas Mdel, C. Faris, M. S. Vardhan, S. Dawson, S. L. Cotterill, and D. M. Livermore. 2008. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum β-lactamases in the United Kingdom. J. Antimicrob. Chemother. 62:1265-1268. [DOI] [PubMed] [Google Scholar]
- 37.Zong, Z., S. R. Partridge, and J. R. Iredell. 2008. RmtC 16S rRNA methyltransferase in Australia. Antimicrob. Agents Chemother. 52:794-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong, Z., S. R. Partridge, L. Thomas, and J. R. Iredell. 2008. Dominance of blaCTX-M within an Australian ESBL gene pool. Antimicrob. Agents Chemother. 52:4198-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]