Abstract
Platensimycin, which is isolated from Streptomyces platensis MA7327, and platencin, which is isolated from S. platensis MA7339, are two recently discovered natural products that serve as important antibiotic leads. Here we report on the identification of S. platensis MA7327 as a dual producer of both platensimycin and platencin. A PCR-based approach was used to locate and clone the locus involved in platensimycin and platencin production, including ptmR1, which encodes a putative GntR-like transcriptional regulator. Deletion of this gene from the producing organism allowed us to isolate strains that overproduce platensimycin and platencin with yields of 323 ± 29 mg/liter and 255 ± 30 mg/liter, respectively. These results illustrate the effectiveness of genetic manipulation for the rational engineering of improvements in titers.
The discovery of platensimycin (16, 20) and platencin (8, 19) as members of an entirely new class of antibacterial antibiotics with a mode of action not exploited by current drugs represents an important step in the fight against antibiotic resistance (Fig. 1a). Both compounds are potent and selective inhibitors of bacterial fatty acid synthesis. Platensimycin specifically targets the elongation β-ketoacyl-acyl carrier protein (ACP) synthase I/II, FabF/B (20), while platencin has a dual mode of action and targets both FabF/B and the initiation β-ketoacyl-ACP synthase III, FabH (19). Both natural products are effective against a broad spectrum of gram-positive pathogens, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci, and show no cross-resistance with other classes of commercially available antibiotics (19). Although platensimycin has proven effective in clearing methicillin-resistant S. aureus infection from a mouse model, the high doses and the suboptimal delivery system required highlight the need for further refinement of its structure prior to the conduct of clinical trials. Multiple total syntheses of both compounds, as well as numerous analogs, underscore the excitement generated by these compounds as leads for novel anti-infectives (10-14, 18).
FIG. 1.
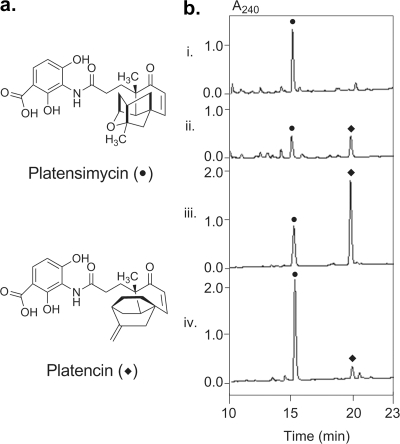
Production of platensimycin (•) and platencin (⧫) by S. platensis MA7327, SB12001, and SB12002. (a) Structures of platensimycin and platencin; (b) HPLC traces of the crude extracts from 0.5 ml of MA7327 (i), 30 μl of SB12001 (iii), and 30 μl of SB12002 (iv) fermented under the reported platensimycin production conditions (14) and from 3 ml of MA7327 (ii) fermented under the reported platencin production conditions (8). The amount of production culture analyzed by HPLC was adjusted for different strains to ensure that the final peak areas fell within the linear range for quantitative determination of the platensimycin and platencin yields.
Platensimycin was isolated from Streptomyces platensis MA7327 at a yield of 2 to 4 mg/liter (16), and platencin was isolated from Streptomyces platensis MA7339 at a yield of 1 mg/liter (8). Subsequent fermentation optimization has led to a recent report that S. platensis MA7327 produced platensimycin at yields up to 56 mg/liter (6). No such improvement of the platencin titer has been reported to date. Strains capable of producing higher yields of platensimycin or platencin, or both, will facilitate the development of these promising leads into clinical agents. Toward this end, we have determined an appropriate set of protocols for the genetic manipulation of platensimycin-producing strain S. platensis MA7327 and exploited the regulatory mechanism of platensimycin and platencin biosynthesis to engineer S. platensis strains that are capable of overproducing both platensimycin and platencin at titers of 323 ± 29 mg/liter and 255 ± 30 mg/liter, respectively.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli DH5α (15) was used for routine cloning, E. coli XL1-Blue MRF and Gigapack III XL (Stratagene, La Jolla, CA) were used for library construction, E. coli BW25113/pIJ790 was used for PCR targeting (3), and E. coli S17-1 (9) was used for intergenic conjugation. Vectors SuperCos1 (Stratagene) and pGEM-3zf and pGEM-5zf (Promega, Madison, WI) were obtained from commercial sources. Platensimycin and platencin producer S. platensis MA7327 was kindly provided by Merck Research Laboratories (Rahway, NJ). E. coli strains carrying plasmids were grown in Luria-Bertani (LB) medium and were selected with appropriate antibiotics (15). Standard media and protocols were used for Streptomyces growth and sporulation (9). Medium components and all other chemicals were from standard commercial sources.
DNA isolation, manipulation, and sequencing.
Plasmid preparation from E. coli and gel extraction were carried out with commercial kits (Qiagen, Valencia, CA). Total S. platensis DNA was isolated by standard procedures (9), and all restriction endonuclease digestions and ligations were performed by standard procedures (15). For Southern analysis, digoxigenin labeling of DNA probes, hybridization, and detection were performed as described in the protocols provided by the manufacturer (Roche Diagnostics, Indianapolis, IN). Automated DNA sequencing and oligonucleotide primer synthesis were performed at the Biotechnology Center, University of Wisconsin—Madison. Long primers for PCR targeting were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
PCR of 3-amino-4-hydroxybenzoic acid (AHBA) synthase.
The PCR primers were designed as shown in Fig. 2. Each PCR mixture (12.5 μl) consisted of 6.25 μl 2× GC buffer II, 400 μM each deoxynucleoside triphosphate, 400 nM each primer, 5% dimethyl sulfoxide, 500 ng S. platensis MA7327 genomic DNA, and 1 U LA Taq polymerase (Takara Bio, Inc., Shiga, Japan). The PCRs were performed on a Mastercycler ep gradient thermocycler (Eppendorf AG, Hamburg, Germany), with the following program: prerun denaturation at 95°C for 5 min, followed by 30 cycles of amplification (1 min of denaturation at 95°C, 30 s annealing with a temperature gradient from 48°C to 55°C, and a 1-min extension at 72°C) and, finally, a postrun extension of 6 min at 72°C.
FIG. 2.
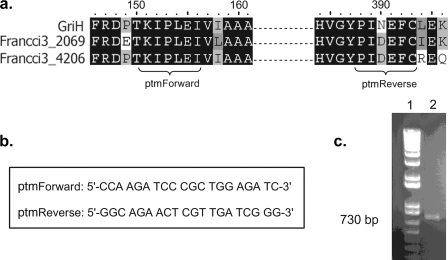
Primer design for amplification of the AHBA synthase gene from S. platensis MA7327 genomic DNA. (a) Primary sequence alignment of GriH, the AHBA synthase from Streptomyces griseus (17), and two highly similar open reading frames from the genome sequence of Frankia sp. strain CCI3 (strain Francci3_2069, GenBank accession no. YP_48177; strain Francci3_4026, GenBank accession no. YP_483283). (b) Primers ptmForward and ptmReverse were designed to amplify an ∼730-bp internal fragment. (c) PCR amplification of an internal fragment of the AHBA synthase gene by use of the annealing temperature to control the specificities of the primers resulted in a single product with the predicted size of 730 bp (lane 2) as determined by comparison to 1 kb Plus DNA Ladder (lane 1) (Invitrogen, Carlsbad, CA).
S. platensis MA7327 genomic library construction and screening.
A genomic library of S. platensis MA7327 was constructed in the SuperCos1 cosmid following the instructions provided by the manufacturer (Stratagene). A Gigapack III XL packaging extract (Stratagene) was used for library construction in E. coli XL1-Blue MRF. The genomic library (∼8,000 colonies) was screened by colony hybridization with the digoxigenin-labeled AHBA synthase PCR product as the probe. Sublibraries of the cosmids harboring the AHBA synthase sequence were constructed by cloning the EcoRI or BamHI digestion fragments into the cloning vector pGEM-3zf. Preliminary sequence data came from the end-in sequencing of these inserts with the M13 primers.
Construction of ΔptmR1 mutant by gene replacement.
The ptmR1 gene located on the insert of cosmid pBS12001 was replaced by the aac(3)IV-oriT resistance cassette from pIJ773 by λRED-mediated PCR targeting (3) with the primers ptmR1Forward (5′-AAGGGACCCCAGAAGCGAAACGGGGCGGCACTTCGTGTGATTCGGGGGATCCGTCGACC-3′) and ptmR1Reverse (5′-ATGGGCCTCGGCCTCGTCGGCCTCGCCCGCCTGAATCAGTTGAGG CTGGAGCTGCTTC-3′) to yield the modified cosmid pBS12002. This inactivation construct was introduced to S. platensis MA7327 by intergenic conjugation with E. coli S17-1 as the donor strain (1, 9). Briefly, E. coli S17-1 cells harboring pBS12001 were grown to an optical density at 600 nm of 0.6 in LB medium supplemented with 20 mM MgCl2 at 37°C. During this incubation, ∼108 S. platensis MA7327 spores were heat shocked in modified TSB++ medium (30 g/liter tryptic soy broth, 100 g/liter sucrose, 4 g/liter glycine) at 50°C for 10 min and were then incubated at 28°C. The donor E. coli cells and the recipient S. platensis cells were then mixed and plated on IWL4 medium (37 g/liter Difco ISP Medium 4, 0.5 g/liter yeast extract, 1 g/liter tryptone) supplemented with 20 mM MgCl2. The plates were incubated for 16 h at 30°C before they were overlaid with 1 ml H2O containing final concentrations of 25 μg/ml nalidixic acid (to select against E. coli) and 50 μg/ml apramycin (to select for exconjugants). The plates were incubated for ∼6 days at 30°C, and the resultant colonies were replica plated to find apramycin-resistant and kanamycin-sensitive clones that had undergone homologous recombination on each side of the locus of gene replacement.
Platensimycin and platencin production conditions.
The S. platensis strains were cultured under conditions taken from the literature (8, 16), with modifications. An aliquot of 500 μl of dense cultures grown in R2YE [103 g/liter sucrose, 0.25 g/liter K2SO4, 10.12 g/liter MgCl2 · 6H2O, 10 g/liter dextrose, 0.1 g/liter Difco Casamino Acids, 5 g/liter Difco yeast extract, and 5.73 g/liter N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] was adjusted to pH 7.0 and autoclaved, and then 6 ml of 1 M NaOH, 10 ml of 0.5% KH2PO4, 20 ml of 1 M CaCl2, 15 ml of 20% proline, and 2 ml of trace elements per liter of medium were added] (9) was used to inoculate 50 ml of ISM-3 seed medium (15 g/liter Difco yeast extract, 10 g/liter Difco malt extract, 0.244 g/liter MgSO4, 0.3 g/liter FeCl3 · 6H2O, 20 g/liter dextrose, pH 7.0) in 250-ml baffled flasks. Seed cultures were grown at 28°C and 250 rpm in incubation shakers for 48 h. Five hundred microliters of the seed culture was used to inoculate 50 ml of the following production media: platensimycin medium (PTM), which consisted of 40 g/liter Stadex 60K dextrin, 40 g/liter lactose, and 5 g/liter Difco yeast extract, pH 7.0; modified PTM (PTMM), which consisted of PTM with 20 g/liter morpholinepropanesulfonic acid (MOPS), sodium salt, pH 7.3; platencin medium (PTN), which consisted of 6 g/liter yeast extract, 15 g/liter malt extract, 6 g/liter dextrose, 20 g/liter MOPS, sodium salt, pH 7.4. The media were autoclaved in 250-ml flasks supplemented with 250 μl trace elements (9). Resin-containing flasks were supplemented with 1.5 g Amberlite XAD-16 resin after autoclaving. Production cultures were incubated for 8 days at 28°C and 250 rpm before the cells were harvested.
Platensimycin and platencin titer determinations.
Harvested resin and mycelial fragments from 50 ml of cultures were separated from the broth by centrifugation and were washed three times with H2O. The resin was then extracted with acetone (four times with 6 ml each time) to recover >99% of the platensimycin and the platencin. The acetone was removed under reduced pressure, and the crude extract was resuspended in methanol prior to analysis on a Waters 510 high-pressure liquid chromatography (HPLC) system with a photodiode array detector (Waters, Milford, MA) by using an Apollo C18 column (particle size, 5 μm; 4.6 by 250 mm; Grace Davison Discovery Sciences, Deerfield, IL) and a 20-min solvent gradient (1 ml/min) from 15% acetonitrile in H2O-0.1% formic acid to 90% acetonitrile in H2O-0.1% formic acid. The peak area at 240 nm was used to quantify platensimycin and platencin on the basis of standard calibration curves.
Platensimycin and platencin isolation and confirmation of their structures.
The isolation of platensimycin and platencin was accomplished by using strains S. platensis SB12002 and SB12001, respectively. The following procedure describes the volumes appropriate for 1.0 liter of fermentation broth. Following production, the contents of each culture were centrifuged at 8,000 rpm for 30 min to pellet the XAD-16 resin. The supernatant was removed from each sample, and the mycelial cake (which was a layer on top of the compacted XAD-16 resin) was removed by scraping. The remaining XAD-16 resin was made into a slurry with H2O (250 ml), and the mixture was filtered through a WypAll L10 utility wipe towel (Kimberly-Clark, Appleton, WI) to permit passage of the aqueous contents and retention of the XAD-16 resin. The resin cake was subjected to three iterations of washing with H2O (250 ml). While it was still moist, the resin cake was then subjected to four iterations of washing with acetone (250 ml), and the combined acetone extracts were concentrated under reduced pressure with gentle warming (∼40°C). The residual H2O was removed by lyophilization. The resulting dried crude extracts from each strain were then subjected to a two-step chromatographic purification. Initial silica gel column chromatography (230 to 400 mesh) with 25% methanol (MeOH) in CHCl3 (for platensimycin) or 20% MeOH in CHCl3 (for platencin) generally afforded ∼85% pure material. Dry loading of the crude extracts into the columns after they were adsorbed to ∼2 g of silica per 1 g of crude extract was critical to the success of this step. Silica extract slurries were made by using a mixture of equal parts acetone, MeOH, and CHCl3. Following silica gel chromatography, semipure platensimycin and platencin samples (separately) were subjected to benchtop C18 silica gel (230- to 400-mesh) column chromatography by using a mobile phase of H2O-acetone (2:1) under ∼10 lb/in2 of pressure. Fractions containing pure platensimycin or platencin were combined, and then the acetone was removed under reduced pressure with mild warming (∼40°C). The residual H2O was removed from each sample by lyophilization, and the purity of each sample was confirmed to be in excess of 95% by reversed-phase HPLC by the protocol noted above for titer determination. The mass spectrometry and 1H nuclear magnetic resonance spectral data for purified platensimycin and platencin precisely matched those reported previously (8, 10, 16).
RESULTS AND DISCUSSION
S. platensis MA7327 is a dual producer.
Upon receiving S. platensis MA7327, we first verified its ability to produce platensimycin. We fermented the strain under the platensimycin production conditions reported previously (16) and confirmed its ability to produce platensimycin. The level of production was slightly above the reported levels (10.1 ± 3.6 mg/liter) (Fig. 1 and 3b). We subsequently confirmed the ability of this strain to produce both platensimycin and platencin in equal amounts (∼1 mg/liter) when it was grown under the previously reported platencin production conditions (Fig. 1 and Table 1) (8). This titer is on par with the levels reported for the platencin producer S. platensis MA7339 (8). While a rigorous medium optimization was not performed, we added Amberlite XAD-16 resin to the production cultures to facilitate the isolation of platensimycin and platencin; the inclusion of the resin had no adverse effects on the titer of either antibiotic (Table 1).
FIG. 3.
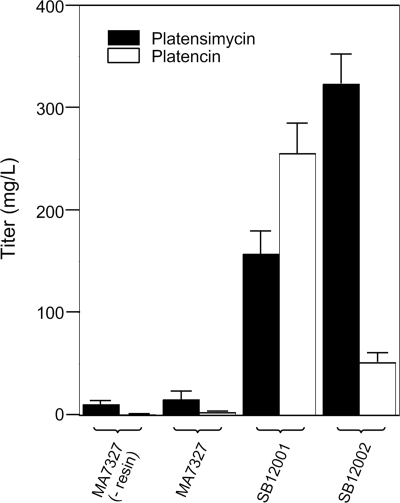
Production of platensimycin (black bars) and platencin (white bars) by wild-type and engineered S. platensis strains under the reported platensimycin production conditions (16). Error bars denote 1 standard deviation from the statistical mean, calculated from at least three independent trials.
TABLE 1.
Titers of platensimycin and platencin in various production media
| Compound and medium | Titera (mg/liter) for the following S. platensis strain:
|
|||
|---|---|---|---|---|
| MA7327b | MA7327 | SB12001 | SB12002 | |
| Platensimycin | ||||
| PTM | 10.1 ± 3.6 | 15.1 ± 8.2 | 157 ± 22 | 323 ± 29 |
| PTMM | 1.3 ± 2.1 | 1.2 ± 0.54 | 62 ± 14 | 202 ± 42 |
| PTN | 1.7 ± 1.0 | 2.1c | 43c | 122c |
| Platencin | ||||
| PTM | 0.2 ± 0.1 | 2.5 ± 0.7 | 255 ± 30 | 51 ± 9.2 |
| PTMM | 0.3 ± 0.2 | 0.8 ± 0.4 | 242 ± 21 | 56 ± 5.7 |
| PTN | 1.2 ± 0.5 | 0.8c | 122c | 29c |
Unless indicated otherwise, the values are averages of at least three independent trials and are reported with standard deviations.
No resin was included in the production medium.
Values are the averages of two independent trials.
Locating putative gene cluster via PCR amplification of AHBA synthase.
Having confirmed the production of both platensimycin and platencin in the S. platensis MA7327 strain, we next set out to identify the locus of the biosynthetic gene cluster(s) responsible for producing these compounds. A PCR-based approach was used with primers designed to amplify a fragment of the AHBA acid synthase gene, which is thought to be involved in the biosynthesis of the 3-amino-2,4-dihydroxybenzoic acid moiety of platensimycin and platencin (4, 5). Primers specific for the conserved regions of the AHBA synthases retrieved from sequence databases were designed (Fig. 2a and b) and used to amplify a 730-bp fragment by using S. platensis MA7327 genomic DNA as the template (Fig. 2c). Sequencing of this PCR product confirmed that it is comprised of a single sequence, suggesting that a single AHBA synthase gene is present within the genome and that platensimycin and platencin share at least a portion of their biosynthetic machineries.
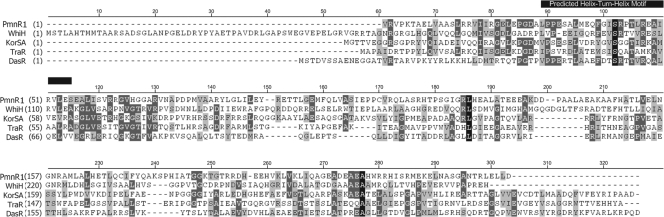
To identify neighboring genes, a genomic library from S. platensis MA7327 was constructed and screened by colony hybridization with the labeled AHBA synthase PCR product as the probe. Six cosmids containing inserts of genomic DNA that contain the AHBA synthase gene were isolated. A sublibrary of these cosmids was constructed by cloning EcoRI or BamHI fragments into the corresponding restriction site in the cloning vector pGEM-3fz. Although complete sequencing and analysis of the surrounding gene cluster are under way, preliminary sequencing revealed ptmR1 (GenBank Accession number EU805802) in the cloned locus. PtmR1 shows sequence similarity to the GntR family of transcriptional repressors (Fig. 4), leading us to postulate that it may be involved in the regulation of platensimycin and platencin production and that the inactivation of ptmR1 could lead to altered levels of production of platensimycin or platencin, or both.
FIG. 4.
Primary sequence alignment of PtmR1 with other characterized GntR family transcriptional repressors from the genus Streptomyces (7), including WhiH (GenBank accession no. SC05819) from S. coelicolor A3(2), DasR (GenBank accession no. BAB79296) from S. griseus, KorSA (GenBank accession no. CAA79637) from S. ambofaciens, and TraR (GenBank accession no. CAA56754) from S. ghanaensis. The sequence encoding a predicted helix-turn-helix DNA binding motif is noted with a black bar.
Inactivation of ptmR1 and characterization of overproducing strains.
S. platensis MA7327 proved to be amenable to previously developed methods for the genetic manipulation of Streptomyces spp., thus enabling the detailed characterization of antibiotic production in this native producer. Namely, intergenic conjugation with E. coli S17-1 as the donor strain allowed the delivery of DNA into S. platensis MA7327. The conjugation frequency mediated by homologous recombination ranges from 10−5 for φC31-mediated site-specific integration vectors such as pSET152 (1) to 10−8 for nonreplicating vectors such as pSET151 (1). S. platensis MA7327 is sensitive to apramycin, thiostrepton, erythromycin, and kanamycin, allowing the use of the genes for resistance to these agents as possible selection markers for in vivo genetic manipulation.
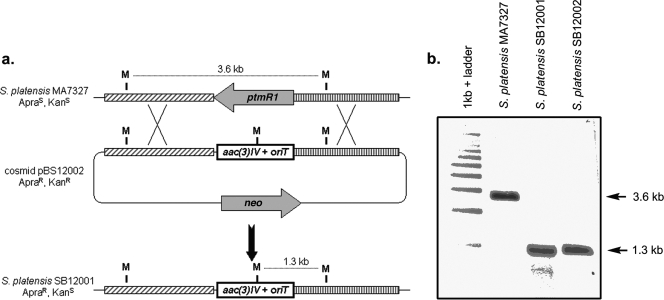
This genetic system was used to replace ptmR1 with the apramycin resistance cassette aac(3)IV by using the Redirect Technology (3). Two exconjugants, S. platensis SB12001 and SB12002, were isolated and their genotypes were confirmed by Southern analysis (Fig. 5). HPLC analysis of the crude extract produced by culturing these mutant strains under the reported platensimycin or platencin production conditions showed significant improvements in the titers of each compound (Fig. 1b and 3; Table 1). Surprisingly, the two overproducing strains were found to have distinct metabolite profiles, with S. platensis SB12002 routinely producing more platensimycin and S. platensis SB12001 routinely favoring platencin production (Fig. 1b). As the induced mutation in these strains has been shown to be identical by Southern analysis (Fig. 5) and the opposite orientation of neighboring genes precludes a polar effect of the gene replacement, this difference in titer can best be explained by a fortuitous genetic variation that existed between the two S. platensis MA7327 parent cells that received DNA during the original conjugation. Under the reported platensimycin production conditions (16), S. platensis SB12002 produced platensimycin at a yield of 323 ± 29 mg/liter and S. platensis SB12001 produced platencin at a yield of 255 ± 30 mg/liter (Fig. 3). A summary of the platensimycin and platencin titers in the various production media, including PTM, PTMM (PTM buffered with MOPS at pH 7.3), and PTN is given in Table 1. For a given strain, PTMM and PTN gave a higher ratio of the platencin yield to the platensimycin yield; however, the greatest absolute yields of each compound were seen in PTM medium.
FIG. 5.
Replacement of ptmR1 with the apramycin resistance gene, aac(3)IV, via homologous recombination. (a) Restriction maps of the S. platensis MA7327 wild-type and SB12001 mutant strains showing predicted fragment sizes upon digestion with MluI. (b) Southern analysis of MA7327 and isolated mutant SB12001 and SB12002 genomic DNAs digested with MluI. The digests were probed with a digoxigenin-labeled nucleotide fragment complementary to the DNA just 3′ of ptmR1 in the orientation depicted above to yield hybridized bands of 3.6 kb for the wild-type strain and 1.3 kb for the mutant strains, as expected. ApraS, apramycin sensitive; ApraR, apramycin resistant; KanS, kanamycin sensitive; KanR, kanamycin resistant.
It should be pointed out that these experiments do not define the biosynthetic relationship between platensimycin and platencin, nor do they address the direct mechanism by which PtmR1 regulates antibiotic production. However, these data vividly demonstrate the effectiveness of the rational genetic manipulation of a natural biosynthetic pathway as a means of enhancing the titers of important drug leads. This process stands in contrast to the lengthy, empirical strain improvement programs that have traditionally been implemented for the generation of overproducing strains (2).
In summary, we confirmed that S. platensis MA7327, the original producer of platensimycin (20), produces both platensimycin and platencin and implemented a strategy for the genetic manipulation of this organism to achieve significantly improved levels of platensimycin and platencin production. The resultant new recombinant strains, S. platensis SB12002 and S. platensis SB12001, produce platensimycin and platencin with yields of 323 ± 29 mg/liter and 255 ± 30 mg/liter, respectively. These titers are ∼100-fold greater than the original yields reported for the native producing strains (8, 16), making the production and isolation of large amounts of these compounds vastly more efficient. These findings underscore once again the effectiveness of the judicious application of metabolic pathway engineering principles to achieve improvements in titers. The overproducing strains reported here provide a solution to the concerns over platensimycin and platencin availability and should greatly facilitate the development of these promising lead compounds into clinical antibacterial agents.
Acknowledgments
We thank Sheo Singh, Merck Research Laboratories, Rahway, NJ, for providing the S. platensis MA7327 strain; the Analytic Instrumentation Center of the School of Pharmacy, University of Wisconsin—Madison, for support in obtaining mass spectrometry and nuclear magnetic resonance data; and the John Innes Center, Norwich, United Kingdom, for providing the Redirect Technology kit.
This work was supported in part by the MERC program, University of Wisconsin—Madison. M.J.S is supported in part by NIH grant T32 GM008349.
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Demain, A. 1981. Industrial microbiology. Science 214:987-995. [DOI] [PubMed] [Google Scholar]
- 3.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herath, K., A. B. Attygalle, and S. B. Singh. 2008. Biosynthetic studies of platencin. Tetrahedron Lett. 49:5755-5758. [Google Scholar]
- 5.Herath, K. B., A. B. Attygalle, and S. B. Singh. 2007. Biosynthetic studies of platensimycin. J. Am. Chem. Soc. 129:15422-15423. [DOI] [PubMed] [Google Scholar]
- 6.Herath, K. B., C. Zhang, H. Jayasuriya, J. G. Ondeyka, D. L. Zink, B. Burgess, J. Wang, and S. B. Singh. 2008. Structure and semisynthesis of platensimide A produced by Streptomyces platensis. Org. Lett. 10:1699-1702. [DOI] [PubMed] [Google Scholar]
- 7.Hillerich, B., and J. Westpheling. 2006. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 188:7477-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayasuriya, H., K. B. Herath, C. Zhang, D. L. Zink, A. Basilio, O. Genilloud, M. T. Diez, F. Vicente, I. Gonzalez, O. Salazar, F. Palaez, R. Cummings, S. Ha, J. Wang, and S. B. Singh. 2007. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew. Chem. Int. Ed. Engl. 46:4684-4688. [DOI] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 10.Nicolaou, K. C., A. Li, and D. J. Edmonds. 2006. Total synthesis of platensimycin. Angew. Chem. Int. Ed. Engl. 45:7086-7090. [DOI] [PubMed] [Google Scholar]
- 11.Nicolaou, K. C., D. J. Edmonds, A. Li, and G. S. Tria. 2007. Asymmetric total syntheses of platensimycin. Angew. Chem. Int. Ed. Engl. 46:3942-3945. [DOI] [PubMed] [Google Scholar]
- 12.Nicolaou, K. C., G. S. Tria, and D. J. Edmonds. 2008. Total synthesis of platencin. Angew. Chem. Int. Ed. Engl. 47:1780-1783. [DOI] [PubMed] [Google Scholar]
- 13.Nicolaou, K. C., T. Lister, R. M. Denton, A. Montero, and D. J. Edmonds. 2007. Adamantaplatensimycin: a bioactive analogue of platensimycin. Angew. Chem. Int. Ed. Engl. 46:4712-4714. [DOI] [PubMed] [Google Scholar]
- 14.Nicolaou, K. C., Y. Tang, and J. Wang. 2007. Formal synthesis of (±)-platensimycin. Chem. Commun. 2007:1922-1923. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 16.Singh, S. B., H. Jayasuriya, J. G. Ondeyka, K. B. Herath, C. Zhang, D. L. Zink, N. N. Tsou, R. G. Ball, A. Basilio, O. Genilloud, M. T. Diez, F. Vicente, F. Pelaez, K. Young, and J. Wang. 2006. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J. Am. Chem. Soc. 128:11916-11920. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki, H., Y. Ohnishi, Y. Furusho, S. Sakuda, and S. Horinouchi. 2006. Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes. J. Biol. Chem. 281:36944-36951. [DOI] [PubMed] [Google Scholar]
- 18.Tiefenbacher, K., and J. Mulzer. 2008. Synthesis of platensimycin. Angew. Chem. Int. Ed. Engl. 47:2548-2555. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J., S. Kodali, S. H. Lee, A. Galgoci, R. Painter, K. Dorso, F. Racine, M. Motyl, L. Hernandez, and E. Tinney. 2007. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. USA 104:7612-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, J., S. M. Soisson, K. Young, W. Shoop, S. Kodali, A. Galgoci, R. Painter, G. Parthasarathy, Y. S. Tang, R. Cummings, S. Ha, K. Dorso, M. Motyl, H. Jayasuriya, J. Ondeyka, K. Herath, C. Zhang, L. Hernandez, J. Allocco, A. Basilio, J. R. Tormo, O. Genilloud, F. Vicente, F. Pelaez, L. Colwell, S. H. Lee, B. Michael, T. Felcetto, C. Gill, L. L. Silver, J. D. Hermes, K. Bartizal, J. Barrett, D. Schmatz, J. W. Becker, D. Cully, and S. B. Singh. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358-361. [DOI] [PubMed] [Google Scholar]