Abstract
Treatment failure is a major cause of concern for the Helicobacter pylori-related gastroduodenal diseases like gastritis, peptic ulcer, and gastric cancer. Curcumin, diferuloylmethane from turmeric, has recently been shown to arrest H. pylori growth. The antibacterial activity of curcumin against 65 clinical isolates of H. pylori in vitro and during protection against H. pylori infection in vivo was examined. The MIC of curcumin ranges from 5 μg/ml to 50 μg/ml, showing its effectiveness in inhibiting H. pylori growth in vitro irrespective of the genetic makeup of the strains. The nucleotide sequences of the aroE genes, encoding shikimate dehydrogenase, against which curcumin seems to act as a noncompetitive inhibitor, from H. pylori strains presenting differential curcumin MICs showed that curcumin-mediated growth inhibition of Indian H. pylori strains may not be always dependent on the shikimate pathway. The antimicrobial effect of curcumin in H. pylori-infected C57BL/6 mice and its efficacy in reducing the gastric damage due to infection were examined histologically. Curcumin showed immense therapeutic potential against H. pylori infection as it was highly effective in eradication of H. pylori from infected mice as well as in restoration of H. pylori-induced gastric damage. This study provides novel insights into the therapeutic effect of curcumin against H. pylori infection, suggesting its potential as an alternative therapy, and opens the way for further studies on identification of novel antimicrobial targets of curcumin.
Helicobacter pylori is a microaerophilic bacterium with the extraordinary ability to establish infections in human stomachs that can last for years or decades. It is carried by more than one-half of all people worldwide, and its prevalence exceeds 90% in some developing countries like India. It has attracted great attention as a major cause of peptic ulcer disease. In fact, H. pylori is the first bacterium to be classified as a group I carcinogen by the International Agency for Research on Cancer (6, 31). Based on the genetic characteristics and disease outcome, there are significant geographic differences among H. pylori strains. Indian H. pylori strains are genetically distinct from those from east Asia and the West (4, 16). Several putative virulence-associated factors, including alleles in the cag pathogenicity island (PAI) of H. pylori, contribute to its pathogenesis and augment the risk for gastric adenocarcinoma (14). As virulence markers of H. pylori are not always associated with diseases, eradication of H. pylori from infected individuals remains the best choice for an effective treatment of H. pylori-associated diseases. Several combination therapies have been formulated to eradicate this pathogen and cure or prevent associated diseases. Triple therapy, consisting of the combined usage of two antibiotics and a proton pump inhibitor, gives a high eradication rate, producing a significant improvement in the status of the disease (30). However, eradication by the triple therapy is not always successful, and the acquisition by H. pylori of resistance to antibiotics, including metronidazole and clarithromycin, could represent a serious problem that may reduce treatment efficacy (10). Many studies have indicated that the prevalence of resistance varies geographically, ranging from 10 to 90% for metronidazole and from 0 to 15% for clarithromycin (7, 18, 30). In view of the incomplete cure achieved with conventional therapy because of increasingly resistant strains, undesirable side effects (17), noncompliance among the patients (3), the cost of the antibiotic regimens (32), and a few other factors contributing to ineffectiveness, there is an urgent need to develop new treatment strategies for H. pylori infection.
Previous studies have shown that the shikimate pathway is essential for the synthesis of important metabolites such as aromatic amino acids, folic acid, and ubiquinone (20). The enzymes involved in this pathway, including shikimate dehydrogenase (SDH), have recently gained great attention as novel drug targets for developing antimicrobial agents that are nontoxic (5). In support of this idea Han et al. (11) showed curcumin to be a noncompetitive inhibitor of SDH. This enzyme, encoded by the aroE gene of H. pylori, may provide useful information for treating H. pylori-associated infection.
In India since ancient times, spices and condiments have been considered indispensable in the culinary arts, as they are used to flavor foods. Also, these spices were recognized for their physiological and medicinal properties. Curcumin (diferuloylmethane), first chemically characterized in 1910, is generally regarded as the most active constituent of the perennial herb Curcuma longa (commonly known as turmeric) (Fig. 1). Many studies have attributed a wide spectrum of activities to this compound and may provide a suitable basis for new H. pylori therapies (2, 12, 26-28). A potential role for curcumin in treating Alzheimer's disease and inflammatory bowel disease has been reported (13, 23). One study showed in vitro antimicrobial activity of curcumin against H. pylori (15), but a recent study from Italy reported that curcumin-based therapy was not effective for eradication of H. pylori infection (8).
FIG. 1.
Structure of curcumin.
Against this background, the present study has been conducted (i) to evaluate the efficacy of curcumin as an antimicrobial agent against H. pylori strains isolated from patients in Kolkata, India; (ii) to perform a sequence analysis of the aroE genes, encoding SDH, the fourth enzyme involved in the shikimate pathway and a promising target for antimicrobial agents, from strains for which curcumin MICs were different; and (iii) to understand curcumin's effectiveness in eradicating H. pylori infection in an animal model.
MATERIALS AND METHODS
H. pylori strains and culture.
Sixty-five archived strains of H. pylori, which were isolated from antral mucosal biopsy specimens of patients at the Post Graduate Institute of Medical Education and Research, Kolkata, India, with chronic gastritis or duodenal ulcers, and 1 reference American Type Culture Collection strain (ATCC 43504) were used for this study. The strains were identified on the basis of colony appearance, Gram staining, and positive reactions in biochemical tests (catalase, urease, and oxidase). H. pylori strains were revived and cultured on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, MI) supplemented with 5% horse serum (Invitrogen, NY), 0.4% IsoVitaleX (Becton Dickinson, MD), trimethoprim (5 μg/ml), vancomycin (8 μg/ml), and polymyxin B (10 μg/ml). The plates were incubated at 37°C in a microaerophilic atmosphere (5% O2, 10% CO2, 85% N2) (double gas incubator; Heraeus, Langenselbold, Germany) for 3 to 6 days. Stock cultures were maintained until use at −70°C. A urease test was conducted with mouse gastric tissue as mentioned earlier (4).
Mouse-adapted H. pylori strains.
Two mouse-adapted H. pylori strains (one isolated from Australia [SS1] and another one from India [AM1]) were used for the animal experiment. They were revived and cultured for mouse infection (14).
MICs.
Frozen stock cultures were streaked on BHI agar with 5% horse serum and incubated for 3 days under microaerophilic conditions as mentioned earlier. Isolates were restreaked on fresh BHI agar and incubated for 24 h. Exponentially growing H. pylori cells were suspended in sterile phosphate-buffered saline (PBS) and adjusted to an optical density of 0.1 at 600 nm. Ten microliters of the adjusted inoculum was delivered to BHI agar containing various concentrations of curcumin (Sigma Chemical Co., St. Louis, MO). Curcumin was dissolved in dimethyl sulfoxide (Sigma Chemical), and a stock concentration of 10 mg/ml was stored at −20°C. Final test concentrations consisted of 50, 40, 30, 25, 20, 15, 10, and 5 μg/ml for each sample. Growth control plates containing only BHI agar were included in each experiment. BHI agar incorporating the solvent dimethyl sulfoxide was included as a growth control to ensure that the viability of the organism was not affected by the solvent used to dissolve curcumin. A total of 65 clinical isolates and 1 reference strain (ATCC 43504) were used in the susceptibility testing. All plates were incubated under microaerophilic conditions at 37°C for 5 days. The MIC was defined as the lowest concentration of the compound at which there was no visible growth. For quality control and comparative analyses, the antibiotic amoxicillin (Sigma Chemical Co., St. Louis, MO) and clarithromycin (Abbott Laboratories, Abbott Park, IL) were also tested with each batch of curcumin.
H. pylori infection in C57BL/6 mice.
C57BL/6 mice bred in-house were used in all experiments. Experiments were designed to minimize animal suffering and to use the minimum number associated with valid statistical evaluation, according to the guidelines of the animal ethics committee of the institute. Animals were anesthetized by ketamine (12 mg/kg of body weight), followed by cervical dislocation for killing. Animals of both control and experimental groups were kept separately in standard conditions and were fasted for 6 h with free access to water before each inoculation. Groups of mice (12 mice per group) were inoculated with H. pylori cultures harvested in PBS twice in a period of 3 days, with about 108 CFU/mouse/inoculation (14). Mouse groups inoculated with SS1, AM1, or PBS (control group) were kept separately, with free access to water and food. Two weeks after the final inoculation, a group of mice were orally fed with curcumin (25 mg/kg) once daily for 7 days consecutively, while untreated infected ones received sterile water. All mouse groups were sacrificed 3 weeks postinfection, and the gastric tissues were assessed for H. pylori colonization and histology.
DNA methods.
Chromosomal DNA from mouse gastric tissues was extracted by standard methods (24). The presence or absence of specific bacterial genes and the specificity for the mouse genome were scored by PCR using specific primers (Table 1) and DNA from the respective tissues. PCR was carried out in 20-μl reaction volumes using 10 pmol of each primer, 0.25 mM of each deoxynucleoside triphosphate, 1 U of Taq polymerase (Takara Shuzo, Japan), and 40 ng of DNA. PCR was carried out for 40 cycles of denaturation at 94°C for 30 s, primer-template annealing at 57°C for 30 s, and DNA synthesis at 72°C for 1 min. The PCR products were analyzed by electrophoresis in 2% agarose gels and visualized by ethidium bromide staining. PCR product sizes were estimated with 100-bp standards (New England Biolabs, MA).
TABLE 1.
Details of PCR primers used for analysis of vacA middle region and SDH and mouse GAPDH genes in DNA isolated from mouse gastric tissues
| Locus name | Primer | Sequence | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|
| vacA m1/m2 | VAG-F | 5′-CAATCTGTCCAATCAAGCGAG-3′ | 567/642 | 14 |
| VAG-R | 5′-GCGTCAAAATAATTCCAAGG-3′ | |||
| aroE | aroE-F | 5′-CCAAAACGATTGGGCTGAAATTG-3′ | 963 | 11 |
| aroE-R | 5′-AAAACGCCCTTTTCTACTAG-3′ | |||
| GAPDH (mouse) | GAPDH-F | 5′-GCAGTGGCAAAGTGGAGATT-3′ | 249 | This study |
| GAPDH-R | 5′-TCTCCATGGTGGTGAAGACA-3′ |
To detect the changes in sequences associated with different MICs, the aroE gene was amplified by PCR using primers described in Table 1. PCR fragments were purified for sequencing with the QIAquick PCR purification kit (Qiagen GmBH, Germany). Sequencing reactions were carried out with a BigDye Terminator cycle sequencing kit (PerkinElmer-Applied Biosystems, Foster City, CA) and an automated sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems). DNA sequence editing and analysis were performed with the DNASTAR program. The aroE gene of fully sequenced strain 26695 was used for comparison.
Histology.
The body and the pyloric parts of stomachs from control mice and mice infected for 3 weeks and treated with curcumin were sectioned for histological studies. The tissue samples were fixed in 10% formalin and embedded in paraffin. The sections (5 μm) were cut with a microtome, stained with hematoxylin and eosin (29), and observed under an Olympus microscope. Images were captured with Camedia software (E-20P 5.0 megapixel) at original magnifications of ×10 and ×20 and processed in Adobe Photoshop version 7.0.
Nucleotide sequence accession numbers.
The sequences obtained here were deposited in GenBank under accession numbers EU939304 to EU939307.
RESULTS
Differential inhibition of H. pylori growth in vitro by curcumin.
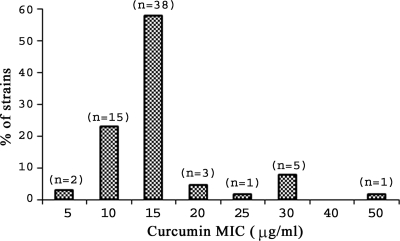
Among 65 Helicobacter pylori strains tested against curcumin, 47 strains were isolated from patients with duodenal ulcers, whereas 12 and 6 cases were isolated from patients with antral gastritis and nonulcer dyspepsia, respectively. Among these strains, 64 strains, including ATCC 43504, showed a positive amplicon for cagA and the other 2 provided an empty site amplicon, indicating absence of the total cag PAI. MICs were determined by the agar dilution method. Curcumin inhibited the growth of all 65 clinical isolates as well as the reference strain ATCC 43504. The MIC of curcumin ranged from 5 μg/ml to 50 μg/ml, and the majority of the strains (81%) showed a MIC of either 10 μg/ml (23%) or 15 μg/ml (58%) (Fig. 2). A few strains showed higher MICs (7.7% and 1.5% of the strains yielded MICs of 30 μg/ml and 50 μg/ml, respectively). These results clearly confirm that curcumin acts as a potent growth inhibitor for Indian H. pylori strains irrespective of the disease status. However, the roles of polymorphism in target genes and strain-specific differences in the MICs of curcumin need further investigation. The MICs of amoxicillin and clarithromycin ranged from 0.03 to 2 μg/ml and 0.02 to 0.12 μg/ml, respectively. The MICs of amoxicillin, curcumin, and clarithromycin against reference strain ATCC 43504 were 0.03, 15, and 0.02 μg/ml, respectively.
FIG. 2.
Distribution of curcumin MICs among the Helicobacter pylori strains isolated from Kolkata, India.
Analysis of SDH from Indian H. pylori strains for which the curcumin MICs were different.
The shikimate pathway is essential for the synthesis of important metabolites in bacteria. Therefore, the enzymes involved in the pathway have received much attention as potential drug targets for developing nontoxic antimicrobial agents, herbicides, and antiparasite drugs. SDH catalyzes the fourth reaction in the shikimate pathway and is responsible for the NADPH-dependent reduction of 3-dehydroshikimate to shikimate. A recent study by Han et al. (11) identified a new aroE gene encoding SDH from H. pylori, and curcumin works as a potent inhibitor of SDH. This may be the mechanism for how curcumin acts on H. pylori to inhibit the growth of this organism. So, we focused on analyzing the aroE gene from H. pylori strains showing different MICs. The aroE gene of strain L1675, showing a curcumin MIC of 50 μg/ml, was sequenced, and the sequence was compared with the aroE gene sequence of 26695, showing a MIC of 20 μg/ml, and also with those of strains PG139, PG156, and PG233, showing MICs of 5 μg/ml, 30 μg/ml, and 5 μg/ml, respectively (Fig. 3). Sequence analysis showed that the aroE genes of L1675 and 26695 are identical and that the encoded proteins are also identical. But analysis of the aroE genes from the other three strains showed around 6 to 7% differences at the nucleotide and amino acid levels, which may reflect the polymorphic nature of the gene. It is interesting to note that, despite the variation observed at the nucleotide and amino acid levels for strains with different curcumin MICs, the well-conserved amino acids, which seem to be important for substrate binding, remain unchanged regardless of the MICs except at one position in a single strain. This has led us to assume that, besides the SDH gene, other genetic components may be involved in the ability of curcumin to act as a potential nontoxigenic antimicrobial agent, at least with Indian strains.
FIG. 3.
Amino acid sequences of SDHs encoded by aroE genes from five H. pylori strains were aligned. Sequences from L1675, PG139, PG233, and PG156 are presented in this study. The 26695 sequence was taken from a public database. GenBank accession numbers for sequences presented here are as follows: L1675, EU939307; PG139, EU939304; PG233, EU939305; and PG156, EU939306. Identical nucleotides are indicated by dots. Sequence alignment was performed using the CLUSTALW program within DDBJ.
Curcumin eradicates H. pylori from H. pylori-infected mouse stomach.
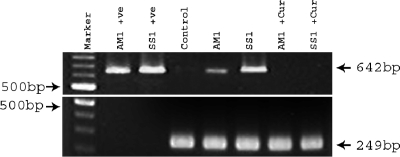
Since an in vitro assay showed that curcumin had antibacterial effects against H. pylori, we tested the effect of curcumin on H. pylori colonization in mice. A group of mice were infected with AM1 and SS1 separately. Three weeks after the infection, they were sacrificed and gastric tissues were checked by a urease test, which showed positive results indicating H. pylori colonization. Establishment of H. pylori infection was also confirmed by PCR assay of the H. pylori-specific gene vacA using DNA isolated from H. pylori-infected stomach tissues. Colonization was further reconfirmed by quantitative culture in blood agar plates. Two weeks after infection, a group of mice were treated with curcumin for 7 days. The effect of curcumin was assessed by a urease test with the respective mouse gastric tissues. It was observed from quantitative culture that curcumin, irrespective of strains, eradicated H. pylori from infected mouse stomach. To further confirm curcumin's anti-H. pylori potential, the H. pylori-specific gene vacA was amplified by PCR using DNA isolated from stomach tissues of H. pylori-infected and treated mice, while the mouse-specific GAPDH gene served as a control. Figure 4 shows that curcumin treatment completely eliminated H. pylori from mouse stomach. Thus, these in vivo data are in accordance with the in vitro results, confirming curcumin's anti-H. pylori potential.
FIG. 4.
Effect of curcumin on H. pylori viability in H. pylori-infected mice. Lanes 1 (second from the left) and 2 represent the amplification of the vacA middle region using DNA from the mouse-colonizing strains AM1 and SS1 with primers VAG-F and VAG-R (Table 1). Lane 3 represents the amplification of bacterium-specific vacA and the mouse-specific GAPDH gene using DNA isolated from the gastric tissue of control mice. Lanes 4 and 5 represent the amplification of vacA and GAPDH using DNA isolated from the gastric tissue of H. pylori-infected mice after 3 weeks of infection. Lanes 6 and 7 indicate the amplification of vacA and GAPDH using DNA isolated from the gastric tissue of curcumin-treated H. pylori-infected mice. +ve, positive control.
Histological analysis of mouse gastric tissues during H. pylori infection and the effect of curcumin.
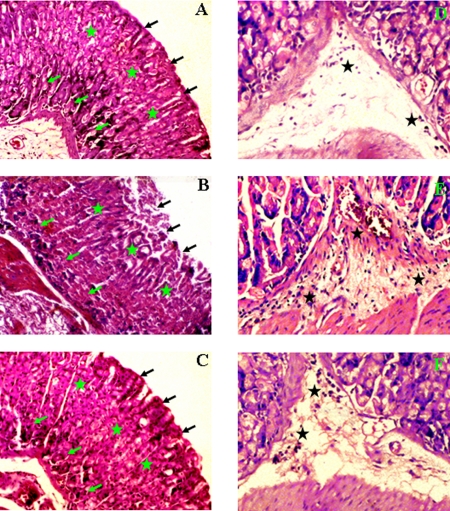
Groups of C57BL/6 mice were intragastrically inoculated with SS1 strain while the control group received sterile PBS. Two weeks after the final inoculation, a group of H. pylori-infected mice were orally fed with 25 mg/kg curcumin for 7 days consecutively and sacrificed. Microscopic observations revealed considerable damage in the mouse gastric tissues infected with H. pylori SS1 strain for 3 weeks (Fig. 5B) compared to the control (Fig. 5A). The gastric mucosa of control tissue had an intact epithelial layer and glandular cells with continuous gastric pits (Fig. 5A). In contrast, denudation of the surface epithelial layer was clearly observed in the SS1-infected mouse gastric tissues; this layer was restored to almost normal after curcumin treatment. Moreover, inflammation in the gastric pit cells, as observed in the infected tissues (Fig. 5B), was checked markedly by curcumin treatment (Fig. 5C). Disruption in the submucosal and muscularis mucosal layers of the mouse gastric tissues that occurred due to H. pylori infection (Fig. 5B) was also almost completely abolished by curcumin treatment (Fig. 5C). Glandular atrophy that occurred because of H. pylori infection (Fig. 5B) was considerably abated by curcumin therapy (Fig. 5C). In control tissues the presence of inflammatory cells was negligible (Fig. 5D). However, infiltration of inflammatory cells was observed in the submucosal region of H. pylori-infected mouse gastric tissues (Fig. 5E) and was prevented to a significant degree in curcumin-treated mice (Fig. 5F). Altogether, these results indicate that curcumin is highly effective in healing the overall damage caused by H. pylori infection.
FIG. 5.
Histology of control, H. pylori-infected, and curcumin-treated mouse gastric tissues. Histological sections were stained with hematoxylin and eosin, and photographs were taken at ×10 and ×20 magnifications, respectively. (A to C) Histological appearance of gastric tissues from (A) control mice, (B) mice infected with SS1 for 3 weeks, and (C) mice infected with SS1 and treated with curcumin at 10× magnification. (D to F) Higher-magnification (20×) views of (D) control, (E) SS1-infected, and (F) SS1-infected, curcumin-treated mouse gastric tissues. The gastric mucosal epithelium (black arrows), gastric pits (green stars), gastric glands (green arrows), and inflammatory cell infiltration (black stars) are shown.
DISCUSSION
Eradication of H. pylori with a proton pump inhibitor-based triple therapy is presently used to treat H. pylori infection (21). Though it has a success rate of 80 to 90%, problems like treatment failure and contraindications for some patients are common. Furthermore, rapidly emerging drug resistance in H. pylori strains during treatment with various antibiotics is a major obstacle for successful eradication therapies (1). Because of the prevalence of antibiotic-resistant H. pylori strains, there is an increasing search for safe and effective nonantibiotic compounds that inhibit H. pylori growth. In the Indian traditional medical system, a number of plants and plant products are known to possess potent medicinal properties, suggesting their usefulness in treatment. Recent studies have shown that curcumin possesses anti-H. pylori potential (11, 15). This prompted us to explore its antimicrobial potential against Indian H. pylori strains that are geographically distinct from east Asian and Western strains. Moreover, a majority of the Indian population harbors H. pylori and quite a number of them suffer from H. pylori-associated gastrointestinal diseases.
In the present study, we have primarily shown that curcumin potentially inhibited the growth of all the 65 H. pylori strains in vitro that were isolated from infected patients suffering from gastrointestinal disorders. It is noteworthy that a majority of these strains were metronidazole resistant, with MICs ranging from 16 μg/ml to >64 μg/ml (7). So, our results suggest that curcumin acts through mechanisms distinctly different from the mode of action of these antibiotics for inhibition of H. pylori growth. Han et al. (11) demonstrated that the growth-inhibitory activity of curcumin against H. pylori is due to inhibition of the shikimate pathway, necessary for synthesis of aromatic amino acids in bacteria. Thus, the aroE genes, encoding SDH, from H. pylori strains showing different MICs were analyzed for nucleotide and amino acid sequence variations. Striking differences in the nucleotide sequences of the genes from these strains corroborated the fact that curcumin targets the shikimate pathway and indicate that polymorphism in aroE may lead to differential inhibition of H. pylori growth. However, strains having identical sequences also had different curcumin MICs. This may be due to the strain-to-strain variations in the rate of curcumin uptake/efflux, or it may be that the antimicrobial effect of curcumin on H. pylori follows distinct mechanisms that may not be solely dependent on inhibition of the shikimate pathway. A recent study by Rai et al. suggests that curcumin may inhibit bacterial cell proliferation by inhibiting the assembly dynamics of FtsZ (a bacterial protofilament), which polymerizes to form a Z ring at the midcell that orchestrates bacterial cell division (22).
The mouse model of H. pylori infection has been widely used in investigations of host responses to H. pylori infection as well as in eradication studies. In the present study, we have shown that curcumin treatment completely eradicated H. pylori from infected mouse stomach. This eradication by curcumin was irrespective of the bacterial genotype, which is independent of the presence of the cag PAI. These data are of immense importance for the development of alternative therapy against H. pylori infection since studies on high doses of curcumin in animals and humans have confirmed a lack of any toxic side effects (9, 25). Histological analysis clearly showed that curcumin is highly effective in repairing damaged tissue. All these observations not only indicate the therapeutic potential of curcumin against H. pylori infections but also highlight the anti-inflammatory effect of curcumin. Various clinical trials suggest a therapeutic potential for curcumin in diseases such as familial adenomatous polyposis, inflammatory bowel disease, ulcerative colitis, colon cancer, pancreatic cancer, hypercholesteremia, atherosclerosis, pancreatitis, psoriasis, and arthritis (9). It has been claimed that curcumin and related compounds have anti-human immunodeficiency virus type 1 and 2 activity in a recent patent application (19).
In conclusion, our study highlights the potential antibacterial activity of curcumin against H. pylori in vitro, as curcumin was highly effective in inhibiting H. pylori growth irrespective of the genetic makeup of the strains, although its MIC is relatively high; this may be due to the poor bioavailability of curcumin (22). Our studies also showed that curcumin-mediated inhibition of H. pylori growth involved mechanisms that may not always be dependent on the shikimate pathway, which opens the way for further studies directed toward determination of novel antimicrobial targets of curcumin. Curcumin showed immense therapeutic potential against H. pylori infections and H. pylori-associated gastroduodenal diseases, as it was equally effective in eradicating H. pylori from infected mouse stomach. Moreover, the gastric damage induced by H. pylori infection was almost completely restored by curcumin, thus highlighting its potential as an alternative therapy against H. pylori infection. Overall, this study provides novel insights in the therapeutic potential of curcumin against H. pylori infections, although further studies are required to extrapolate its effect on humans.
Acknowledgments
The work was supported in part by the Indian Council of Medical Research, Government of India, and Program of Founding Research Center for Emerging and Reemerging Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan. P.K. is a recipient of Senior Research Fellowship from University Grants Commission.
Footnotes
Published ahead of print on 9 February 2009.
REFERENCES
- 1.Alarcón, T., D. Domingo, and M. López-Brea. 1999. Antibiotic resistance problems with Helicobacter pylori. Int. J. Antimicrob. Agents 12:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Apisariyakul, A., N. Vanittanakom, and D. Buddhasukh. 1995. Antifungal activity of turmeric oil extracted from Curcuma longa (Zingiberaceae). J. Ethnopharmacol. 49:163-169. [DOI] [PubMed] [Google Scholar]
- 3.Broutet, N., S. Tchamgoué, E. Pereira, H. Lamouliatte, R. Salamon, and F. Megraud. 2003. Risk factors for failure of Helicobacter pylori therapy—results of an individual data analysis of 2751 patients. Aliment. Pharmacol. Ther. 17:99-109. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay, S., R. Patra, T. Ramamurthy, A. Chowdhury, A. Santra, G. K. Dhali, S. K. Bhattacharya, D. E. Berg, G. B. Nair, and A. K. Mukhopadhyay. 2004. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. J. Clin. Microbiol. 42:2821-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coggins, J. R., C. Abell, L. B. Evans, M. Frederickson, D. A. Robinson, A. W. Roszak, and A. P. Lapthorn. 2003. Experiences with the shikimate-pathway enzymes as targets for rational drug design. Biochem. Soc. Trans. 31:548-552. [DOI] [PubMed] [Google Scholar]
- 6.Covacci, A., J. L. Telford, G. D. Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 7.Datta, S., S. Chattopadhyay, R. Patra, R. De, T. Ramamurthy, J. Hembram, A. Chowdhury, S. K. Bhattacharya, D. E. Berg, G. B. Nair, and A. K. Mukhopadhyay. 2005. Most Helicobacter pylori strains of Kolkata in India are resistant to metronidazole but susceptible to other drugs commonly used for eradication and ulcer therapy. Aliment. Pharmacol. Ther. 22:51-57. [DOI] [PubMed] [Google Scholar]
- 8.Di Mario, F., L. G. Cavallaro, A. Nouvenne, N. Stefani, G. M. Cavestro, V. Iori, M. Maino, G. Comparato, L. Fanigliulo, E. Morana, A. Pilotto, L. Martelli, M. Martelli, G. Leandro, and A. Franzè. 2007. A curcumin-based 1-week triple therapy for eradication of Helicobacter pylori infection: something to learn from failure? Helicobacter 12:238-243. [DOI] [PubMed] [Google Scholar]
- 9.Goel, A., A. B. Kunnumakkara, and B. B. Aggarwal. 2008. Curcumin as “curecumin”: from kitchen to clinic. Biochem. Pharmacol. 75:787-809. [DOI] [PubMed] [Google Scholar]
- 10.Graham, D. Y. 1998. Antibiotic resistance in Helicobacter pylori: implication for therapy. Gastroenterology 115:1272-1277. [DOI] [PubMed] [Google Scholar]
- 11.Han, C., L. Wang, K. Yu, L. Chen, L. Hu, K. Chen, H. Jiang, and X. Shen. 2006. Biochemical characterization and inhibitor discovery of shikimate dehydrogenase from Helicobacter pylori. FEBS J. 273:4682-4692. [DOI] [PubMed] [Google Scholar]
- 12.Han, S. S., S. T. Chung, D. A. Robertson, D. Ranjan, and S. Bondada. 1999. Curcumin causes the growth arrest and apoptosis of B cell lymphoma by downregulation of egr-1, c-myc, bcl-XL, NF-kappa B, and p53. Clin. Immunol. 93:152-161. [DOI] [PubMed] [Google Scholar]
- 13.Holt, P. R., S. Katz, and R. Kirshoff. 2005. Curcumin therapy in inflammatory bowel disease: a pilot study. Dig. Dis. Sci. 50:2191-2193. [DOI] [PubMed] [Google Scholar]
- 14.Kundu, P., A. K. Mukhopadhyay, R. Patra, A. Banerjee, D. E. Berg, and S. Swarnakar. 2006. Cag pathogenicity island-independent up-regulation of matrix metalloproteinases-9 and -2 secretion and expression in mice by Helicobacter pylori infection. J. Biol. Chem. 281:34651-34662. [DOI] [PubMed] [Google Scholar]
- 15.Mahady, G. B., S. L. Pendland, G. Yun, and Z. Z. Lu. 2002. Turmeric (Curcuma longa) and curcumin inhibit the growth of Helicobacter pylori, a group 1 carcinogen. Anticancer Res. 22:4179-4181. [PubMed] [Google Scholar]
- 16.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Kolkata, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myllyluoma, E., L. Veijola, T. Ahlroos, S. Tynkkynen, E. Kankuri, H. Vapaatalo, H. Rautelin, and R. Korpela. 2005. Probiotic supplementation improves tolerance to Helicobacter pylori eradication therapy—a placebo-controlled, double-blind randomized pilot study. Aliment. Pharmacol. Ther. 21:63-72. [DOI] [PubMed] [Google Scholar]
- 18.Nahar, S., A. K. Mukhopadhyay, R. Khan, M. M. Ahmad, S. Datta, S. Chattopadhyay, S. C. Dhar, S. A. Sarker, L. Engstrand, D. E. Berg, G. B. Nair, and M. Rahman. 2004. Antimicrobial susceptibility of Helicobacter pylori strains isolated from Bangladesh. J. Clin. Microbiol. 42:4856-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardee, A. B., J. Li, C. Crumpacker, and L. Zhang. March 1994. Treatment of human viral infections. U.S. patent US933470, WO9404139.
- 20.Parish, T., and N. G. Stoker. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148:3069-3077. [DOI] [PubMed] [Google Scholar]
- 21.Peitz, U., M. Menegatti, D. Vaira, and P. Malfertheiner. 1998. The European meeting on Helicobacter pylori: therapeutic news from Lisbon. Gut 43:S66-S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rai, D., J. K. Singh, N. Roy, and D. Panda. 2008. Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem. J. 410:147-155. [DOI] [PubMed] [Google Scholar]
- 23.Ringman, J. M., S. A. Frautschy, G. M. Cole, D. L. Masterman, and J. L. Cummings. 2005. A potential role of the curry spice curcumin in Alzheimer's disease. Curr. Alzheimer Res. 2:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Shishodia, S., M. M. Chaturvedi, and B. B. Aggarwal. 2007. Role of curcumin in cancer therapy. Curr. Probl. Cancer 31:243-305. [DOI] [PubMed] [Google Scholar]
- 26.Singh, R., R. Chandra, M. Bose, and P. M. Luthra. 2002. Antibacterial activity of Curcuma longa rhizome extract on pathogenic bacteria. Curr. Sci. 83:737-740. [Google Scholar]
- 27.Srinivas, L., V. K. Shalini, and M. Shylaja. 1992. Turmerin: a water soluble antioxidant peptide from turmeric (Curcuma longa). Arch. Biochem. Biophys. 292:617-623. [DOI] [PubMed] [Google Scholar]
- 28.Surh, Y. 1999. Molecular mechanisms of chemopreventive effects of selected dietary and medicinal phenolic substances. Mutat. Res. 428:305-327. [DOI] [PubMed] [Google Scholar]
- 29.Swarnakar, S., K. Ganguly, P. Kundu, A. Banerjee, P. Maity, and A. V. Sharma. 2005. Curcumin regulates expression and activity of matrix metalloproteinases 9 and 2 during prevention and healing of indomethacin-induced gastric ulcer. J. Biol. Chem. 280:9409-9415. [DOI] [PubMed] [Google Scholar]
- 30.Toracchio, S., L. Cellini, E. Di Campli, G. Cappello, M. G. Malatesta, A. Ferri, A. F. Ciccaglione, L. Grossi, and L. Marzio. 2000. Role of antimicrobial susceptibility testing on efficacy of triple therapy in Helicobacter pylori eradication. Aliment. Pharmacol. Ther. 14:1639-1643. [DOI] [PubMed] [Google Scholar]
- 31.Westblom, T. U., S. J. Czinn, and J. G. Nedrud (ed.). 1999. Current topics in microbiology and immunology, vol. 241. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Springer Verlag, Berlin, Germany.
- 32.Wong, W. M., Q. Gu, S. K. Lam, F. M. Y. Fung, K. C. Lai, W. H. C. Hu, Y. K. Yee, C. K. Chan, H. H. X. Xia, M. F. Yuen, and B. C. Y. Wong. 2003. Randomized controlled study of rabeprazole, levofloxacin and rifabutin triple therapy vs. quadruple therapy as second-line treatment for Helicobacter pylori infection. Aliment. Pharmacol. Ther. 17:553-560. [DOI] [PubMed] [Google Scholar]