Abstract
Previously, we showed that the Salmonella enterica serovar Typhimurium SR-11 tricarboxylic acid (TCA) cycle must operate as a complete cycle for full virulence after oral infection of BALB/c mice (M. Tchawa Yimga, M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen, Infect. Immun. 74:1130-1140, 2006). In the same study, we showed that for full virulence, malate must be converted to both oxaloacetate and pyruvate. Moreover, it was recently demonstrated that blocking conversion of succinyl-coenzyme A to succinate attenuates serovar Typhimurium SR-11 but does not make it avirulent; however, blocking conversion of succinate to fumarate renders it completely avirulent and protective against subsequent oral infection with the virulent serovar Typhimurium SR-11 wild-type strain (R. Mercado-Lubo, E. J. Gauger, M. P. Leatham, T. Conway, and P. S. Cohen, Infect. Immun. 76:1128-1134, 2008). Furthermore, the ability to convert succinate to fumarate appeared to be required only after serovar Typhimurium SR-11 became systemic. In the present study, evidence is presented that serovar Typhimurium SR-11 mutants that cannot convert fumarate to malate or that cannot convert malate to both oxaloacetate and pyruvate are also avirulent and protective in BALB/c mice. These results suggest that in BALB/c mice, the malate that is removed from the TCA cycle in serovar Typhimurium SR-11 for conversion to pyruvate must be replenished by succinate or one of its precursors, e.g., arginine or ornithine, which might be available in mouse phagocytes.
In sensitive mice, Salmonella enterica serovar Typhimurium causes a systemic, often fatal disease, similar to human typhoid fever (42). After ingestion, serovar Typhimurium survives passage through the acidic environment of the stomach and reaches the terminal ileum, where within 30 min it invades M cells in the Peyer's patches (21). Within 60 min, the M cells are destroyed, and serovar Typhimurium gains access to both adjacent enterocytes and underlying lymphoid cells in the mesenteric lymph follicles of the Peyer's patches (21, 23). Serovar Typhimurium then increases logarithmically in Peyer's patches for 2 days (20) and simultaneously disseminates systemically to the liver and spleen, where it grows in macrophages (39, 42). Mice usually become ruffled and lethargic 3 to 5 days after oral infection and normally die within 7 to 12 days.
Recently, we reported that the tricarboxylic acid (TCA) cycle of serovar Typhimurium strain SR-11 (hereafter called SR-11) must operate as a complete cycle for SR-11 to be fully virulent during BALB/c mouse infections (47). In addition to requiring cyclic operation of the TCA cycle, it appears that malate must be withdrawn from the TCA cycle to generate pyruvate (Fig. 1), i.e., an SR-11 ΔsfcA ΔmaeB mutant, unable to make the “malic” enzymes for conversion of malate to pyruvate (3, 25), was attenuated (47). Removing malate from the TCA cycle to make pyruvate requires that the TCA cycle be replenished so that intermediates will not be depleted. Replenishment of malate does not require the glyoxylate bypass, since an SR-11 ΔaceA mutant, which is unable to make isocitrate lyase (26) and is therefore unable to make malate or succinate from isocitrate, was fully virulent (47). Moreover, oxaloacetate was shown not to be replenished by the conversion of phosphoenolpyruvate (PEP) to oxoloacetate (Fig. 1), i.e., an SR-11 Δppc mutant, unable to make PEP carboxylase (40), was shown to be fully virulent (47).
FIG. 1.
Embden-Meyerhoff pathway, gluconeogenic pathway, and TCA cycle. The arrows indicate the physiological directions of the reactions. The genes encoding the enzymes for each reaction are listed beside the reaction.
Although a complete TCA cycle appeared to be required for full SR-11 virulence, deleting different TCA cycle genes resulted in different levels of attenuation (47). We found that an SR-11 ΔsucAB mutant, unable to convert α-ketoglutarate to succinyl-coenzyme A (CoA) via the 2-oxoglutarate dehydrogenase complex (10, 36), was avirulent; an SR-11 ΔsucCD mutant, unable to generate succinate from succinyl-CoA via succinyl-CoA synthetase (4, 10), was moderately attenuated; an SR-11 ΔsdhCDA mutant, unable to generate fumarate from succinate via succinate dehydrogenase (10, 37), was slightly attenuated; and an SR-11 Δmdh mutant, unable to convert malate to oxaloacetate via malate dehydrogenase (10, 52), was highly attenuated. On the other hand, mutants that could not run the reductive branch of the TCA cycle, i.e., an SR-11 ΔaspA mutant (aspartase), unable to convert aspartate to fumarate (10, 45), and an SR-11 ΔfrdABCD mutant (fumarate reductase), unable to convert fumarate to succinate (7, 10, 34), were fully virulent (47).
It was not surprising that the SR-11 ΔsucAB mutant was avirulent because the strain is unable to make succinyl-CoA, which, in addition to being converted to succinate in the TCA cycle, is required for biosynthesis of diaminopimelate, a precursor for the synthesis of lysine, methionine, and peptidoglycan (5, 6, 16, 17, 29). However, it was surprising that the SR-11 ΔsdhCDA mutant was far less attenuated than the SR-11 ΔsucCD mutant. This discrepancy was partially explained by our finding that an SR-11 ΔfrdABCD ΔsdhCDA double mutant was avirulent (30), which meant that fumarate reductase, which normally runs in the opposite direction to succinate dehydrogenase for branched TCA cycle operation, could substitute for succinate dehydrogenase in the SR-11 ΔsdhCDA mutant during infection to run a full TCA cycle with only a slight reduction in virulence. It is well known that fumarate reductase and succinate dehydrogenase are physiologically reversible isoenzymes that are induced under anaerobic and aerobic conditions, respectively (7, 19).
While the data suggested that the conversion of succinate to fumarate is key to SR-11 virulence (30), we wanted to know whether it was required in Peyer's patches or when SR-11 became systemic. We found that wild-type SR-11 and the SR-11 ΔfrdABCD ΔsdhCDA mutant grew at the same rate and to the same extent in Peyer's patches (30), suggesting that the conversion of succinate to fumarate was required after SR-11 became systemic, i.e., presumably in phagocytes. In support of this view, after oral infection, the SR-11 ΔfrdABCD ΔsdhCDA mutant did not grow well in the liver and spleen relative to the growth of wild-type SR-11 (30). Moreover, the SR-11 ΔfrdABCD ΔsdhCDA mutant was highly attenuated when mice were injected with 104 CFU via the intraperitoneal route (30).
The finding that the SR-11 double mutant lacking succinate dehydrogenase and fumarate reductase was avirulent raised a new question. Why did blocking the conversion of succinate to fumarate in SR-11 result in avirulence whereas blocking the preceding step in the TCA cycle, conversion of succinyl-CoA to succinate (Fig. 1), resulted in only moderate attenuation (47)? Since malate appears to be used for the production of pyruvate during infection and therefore must be replenished (47) and since the glyoxylate bypass appeared not to be required for virulence (47), we suggested the possibility that there might be enough succinate, perhaps in the mitochondria of mouse phagocytes (e.g., neutrophils, macrophages, monocytes, and dendritic cells), to be used to replenish malate in the SR-11 TCA cycle. Alternatively, since arginine and ornithine play major roles in the nitric oxide synthesis that is thought to limit Salmonella growth in macrophages (8, 51) and since serovar Typhimurium has all the genes required to transport and degrade either arginine or ornithine to succinate (27, 38), we suggested that there might be enough arginine or ornithine present in phagocytes for use by SR-11 to replenish succinate and then malate in its TCA cycle. If so, and if there was insufficient fumarate or malate present in mouse phagocytes to replenish malate in the SR-11 TCA cycle, succinate conversion to fumarate would be key to the production of pyruvate and oxaloacetate and hence to SR-11 virulence. This scenario would explain why blocking conversion of succinyl-CoA to succinate results in attenuation whereas blocking conversion of succinate to fumarate has a far more dramatic effect. If the hypothesis that mouse phagocytes can replenish succinate, but not fumarate or malate, in SR-11 is true, we would expect that SR-11 mutants that are unable to convert fumarate to malate (18) or are unable to generate either pyruvate or oxaloacetate from malate (Fig. 1) would both be avirulent. In the present study, we examined these possibilities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. LB broth, Lennox (Difco Laboratories, Detroit, Michigan); LB agar, Lennox (Difco); and MacConkey agar (Difco) were prepared according to the package instructions. Liquid M9 minimal salts medium (32) was supplemented with either reagent grade glucose (0.4% [wt/wt]) or sodium succinate (0.4% [wt/wt]) as the sole source of carbon and energy. When necessary, the pH was adjusted to 7.2. SOC medium was prepared as described by Datsenko and Wanner (12). Agar plates were supplemented with nalidixic acid (50 μg/ml) and/or chloramphenicol (30 μg/ml), as appropriate.
TABLE 1.
Strains of serovar Typhimurium and plasmids used in this study
| Strain or plasmid | Genotypea or relevant characteristics | Defect(s) | Source or reference |
|---|---|---|---|
| SR-11 wild type | gyr-1816 | None | 11 |
| SR-11 ΔaceA | gyr-1816 ΔaceA::cat | Isocitrate lyase | 47 |
| SR-11 ΔsucCD | gyr-1816 ΔsucCD::cat | Succinyl-CoA synthetase | 47 |
| SR-11 ΔsucCD ΔaceA | gyr-1816 ΔsucCD ΔaceA::cat | Succinyl-CoA synthetase, isocitrate lyase | This study |
| SR-11 ΔfumAC | gyr-1816 ΔfumAC::cat | Fumarase A, fumarase C | This study |
| SR-11 ΔdcuB ΔfumB | gyr-1816 ΔdcuB ΔfumB::cat | DcuB dicarboxylate antiporter, fumarase B | This study |
| SR-11 ΔfumAC ΔdcuB ΔfumB | gyr-1816 ΔfumAC::cat ΔdcuB ΔfumB | DcuB dicarboxylate antiporter, fumarase A, fumarase C, fumaraseB | This study |
| “Restored” SR-11 ΔdcuB ΔfumB | gyr-1816 ΔdcuB ΔfumB | DcuB dicarboxylate antiporter, fumarase B | This study |
| SR-11 ΔsfcA ΔmaeB | gyr-1816 ΔmaeB ΔsfcA::kan | Both malate oxidoreductases | 47 |
| SR-11 Δmdh | gyr-1816 Δmdh::cat | Malate dehydrogenase | 47 |
| SR-11 ΔsfcA ΔmaeB Δmdh | gyr-1816 ΔsfcA ΔmaeB Δmdh::cat | Both malate oxidoreductases, malate dehydrogenase | This study |
| “Restored” SR-11 ΔsfcA ΔmaeB | gyr-1816 ΔsfcA ΔmaeB | Both malate oxidoreductases | This study |
| Plasmids | |||
| pKD3 | Template plasmid; contains chloramphenicol resistance cassette flanked by FLP recombinase target sites; bla cat | 12 | |
| pKD4 | Template plasmid; contains kanamycin resistance cassette flanked by FLP recombinase target sites; bla kan | 12 | |
| pKD46 | Temperature-sensitive plasmid; contains arabinose-inducible phage λ red recombinase gene for linear DNA exchange; bla | 12 | |
| pCP20 | Temperature-sensitive plasmid; contains FLP recombinase gene for removal of antibiotic resistance cassetes; bla cat | 12 |
Construction and characterization of SR-11 strains.
All mutants were constructed by allelic-exchange mutagenesis using a chloramphenicol cassette as described by Datsenko and Wanner (12). The primers used to construct the mutants are listed in Table 2. The constructs were verified by PCR and sequencing. Primers were designed by referring to the complete genome of S. enterica serovar Typhimurium LT2 (27). The 1,817-bp deletion in the SR-11 ΔdcuB ΔfumB mutant begins in the dcuB gene 171 bp upstream of the fumB start codon and includes an additional 1,646-bp deletion of the entire fumB open reading frame. The primers used for confirming the size of the ΔdcuB ΔfumB deletion by sequencing were as follows: forward, 5′-CTG CAG GGG ATG CAG 22 TTT AT-3′; reverse, 5′-GTC TTT GGC TGG ATC TTT GC-3′. The 3,216-bp deletion in the SR-11 ΔfumAC mutant begins 421 bp downstream of the fumA start codon and ends after deletion of the entire fumC open reading frame. The primers used for confirming the size of the ΔfumAC deletion by sequencing were as follows: forward, 5′-ATT CTT TTC GAT GGC GTC AC-3′; reverse, 5′-CTG GAA CGG GAA TAT CAG GA-3′. The “restored” SR-11 ΔdcuB ΔfumB mutant was constructed by replacing the ΔfumAC::cat deletion in the SR-11 ΔfumAC::cat ΔdcuB ΔfumB mutant with the SR-11 fumAC wild-type sequence using the method described by Datsenko and Wanner (12). The “restored” SR-11 ΔsfcA ΔmaeB mutant was constructed by replacing the Δmdh::cat deletion in the SR-11 ΔsfcA ΔmaeB Δmdh::cat triple mutant with SR-11 mdh wild-type sequence by the same method. The primers for these constructions are listed in Table 2. The primers used for confirming the restoration of the wild-type genes in these strains by sequencing are the same as those for restoring the wild-type sequences and are listed in Table 2. The “restored” SR-11 ΔdcuB ΔfumB mutant and the “restored” SR-11 ΔsfcA ΔmaeB mutant were selected after plating transformants on M9 minimal agar plates containing sodium succinate (0.4% [wt/wt]) as the sole carbon and energy source. As expected, the “restored” SR-11 ΔdcuB ΔfumB mutant and the “restored” SR-11 ΔsfcA ΔmaeB mutant were chloramphenicol sensitive and, unlike their parents, grew in M9 minimal medium containing succinate (0.4% [wt/wt]) as the sole carbon and energy source. For sequencing, PCR products were purified with a Qiagen Qiaquick PCR purification kit (Qiagen, MD) following the manufacturer's instructions. PCR products were submitted to the Rhode Island Genomics and Sequencing Center at the University of Rhode Island. After completion of the cycle sequencing, samples were purified with Agencourt's CleanSEQ SPRI reagent and separated on an Applied Biosystems 3130xl genetic analyzer (50-cm capillary array with POP7 polymer).
TABLE 2.
Sequences of primers used to construct serovar Typhimurium strains
| Gene | Primer (5′ → 3′)
|
|
|---|---|---|
| Forward | Reverse | |
| ΔaceA | GTA TTG AGG CGA TTT ATC TTT CAG GCT GGC AGG TGG CGG CAG ATG GTG TAG GCT GGA GCT GCT TCG | ATG CAT GCG CCA GGT CGA ACA TGT TGA ACC ACA TGC TGT GGA TGC CAT ATG AAT ATC CTC CTT AGT |
| Δmdh | CGT ATC CAG CAT AGC GTC CAG CGA ATG TTG CTC GAA AGC GCT CAG GTG TAG GCT GGA GCT GCT TCG | CAT CGC TCC AGT GAC TCC CGG TGT GGC CGT TGA TTT GAG CCA CAT CAT ATG AAT ATC CTC CTT AGT |
| Wild-type mdh | CGA TAA GAC GTG AGG AGT | CAG CCG ATC CGG ATT ACG |
| ΔfumB | CAG GTT ATT TCG CGC AGT TAG CGC ACT GTT TGC TGA CAA TCT GCT GGA AGT GTA GGC TGG AGC TGC TTC G | TTC CGG CAC AAC CCG TAT TGG CCG CTT CGT CAT TAA CCA CAG CTT CAT TCC ATA TGA ATA TCC TCC TTA GT |
| ΔfumAC | TAA CCT GCG CTA TTC ACA GAA CGC GGC GCT GGA TAT GTA CAA AGA GGT CGT GTA GGC TGG AGC TGC TTC G | TCA TAC TGC CAA CCA TCA ACT CCG GAC GTA CCC AGG CGT CGA ACT CCT CCA TAT GAA TAT CCT CCT TAG T |
| Wild-type fumAC | ATT CTT TTC GAT GGC GTC AC | CTG GAA CGG GAA TAT CAG GA |
Growth on glucose and sodium succinate in M9 minimal medium.
For testing the growth of strains in M9 minimal medium containing glucose as the sole carbon and energy source, overnight cultures grown in LB broth, Lennox, were washed twice in M9 minimal medium (no carbon source), 100 μl of the washed cultures was transferred to 10 ml of M9 minimal medium containing reagent grade glucose (0.4% [wt/wt]) as the sole carbon and energy source, and the cultures were incubated at 37°C with shaking overnight in 125-ml tissue culture bottles. The same protocol used for growth on glucose was used for testing the growth of the strains on sodium succinate (0.4% [wt/wt]) as the sole carbon and energy source. In all growth experiments, growth was monitored spectrophotometrically (A600) using a Pharmacia Biotech Ultrospec 2000 UV/Visible spectrophotometer.
Virulence assays.
For virulence assays, the wild-type and mutant SR-11 strains were grown overnight in LB broth, Lennox. Each strain grew to an A600 of about 3.3. Wild-type SR-11 grew with a generation time of about 33 min in LB broth, Lennox, and each mutant grew with a generation time of about 39 min. Virulence assays were carried out as described previously (1, 50). Briefly, 4-week-old 13- to 15-g female BALB/c mice (Charles River Laboratories, Wilmington, MA) were housed no more than four per cage, with pine shavings as bedding. Prior to infection, the mice were starved for food and water for 4 h. Following starvation, 50 μl of 10% sodium bicarbonate was administered orally to each mouse in order to neutralize gastric acidity, and 30 min later, 108 CFU of a specific strain in 20 μl of phosphate-buffered saline (pH 7.4) containing 0.1% gelatin (BSG) was administered orally to each of four mice. After infection, food and water were returned, and the mice were inspected four times daily for obvious signs of illness (ruffled fur, eyes crusted and closed, loss of appetite, or crouching and shivering) and death. The data presented here are composites of at least two identical but independent experiments.
Definitions of fully virulent, attenuated, and avirulent.
When mice infected with a specific mutant remained healthy, i.e., showed no signs of illness throughout the 30-day duration of the experiment, the mutant was considered to be avirulent. The term fully virulent was used when mice infected with a particular mutant showed the same signs of illness as mice infected with the wild-type SR-11 strain and the survival curve was not statistically different from that of mice infected with the wild-type SR-11 strain. The term attenuated was used when the mice infected with a particular mutant showed signs of illness, death was delayed, and the survival curve was statistically different from that of mice infected with the wild-type SR-11 strain. The extent to which death was delayed is denoted by either the term slightly attenuated, attenuated, or highly attenuated.
Protection assays.
Thirty days after oral infection with an avirulent SR-11 mutant, which is sufficient time for mice to mount an intestinal mucosal immunological response and a systemic immunological response, BALB/c mice were challenged orally with 108 CFU of wild-type SR-11, as described above. In each experiment, an age-matched set of sham-vaccinated BALB/c mice were also challenged with 108 CFU of wild-type SR-11. The mice were then observed four times daily for obvious signs of illness (ruffled fur, eyes crusted and closed, loss of appetite, or crouching and shivering) and death.
Statistics.
Mouse survival curves were compared for differences using the Kaplan-Meier method (MedCalc Software, Belgium). Survival curves were considered to be different if the P value was less than 0.05.
RESULTS
An SR-11 ΔsucCD ΔaceA mutant is no more attenuated than an SR-11 ΔsucCD mutant.
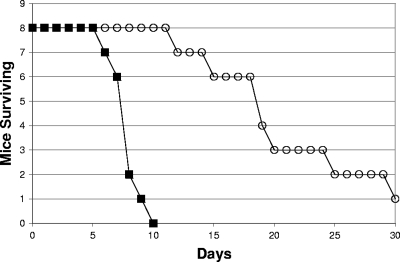
A major question relating to the requirement for a complete TCA cycle for SR-11 virulence was raised by our previous data (30, 47). Why did blocking the conversion of succinate to fumarate (ΔfrdABCD ΔsdhCDA) in SR-11 result in avirulence whereas blocking the conversion of succinyl-CoA to succinate (ΔsucCD), which precedes succinate conversion to fumarate in the TCA cycle (Fig. 1), resulted in only moderate attenuation (47)? Since neither fatty acid degradation nor the glyoxylate bypass was required for full SR-11 virulence (47) and a full TCA cycle appeared to be required for SR-11 growth only after it became systemic (30), we suggested the possibility that succinate or one of its precursors, i.e., arginine or ornithine, present in mouse phagocytes normally replenishes TCA cycle intermediates. In this case, some malate would be made for conversion to pyruvate and oxaloacetate in an SR-11 ΔsucCD mutant in vivo, resulting in attenuation, whereas there would be no malate generated in the SR-11 ΔfrdABCD ΔsdhCDA mutant, resulting in avirulence. However, there was the alternative possibility that the glyoxylate bypass was induced in vivo in an SR-11 ΔsucCD mutant, thereby generating both succinate and malate for conversion to pyruvate and oxaloacetate (Fig. 1). To test this possibility, we constructed an SR-11 ΔsucCD ΔaceA double mutant, unable to convert succinyl-CoA to succinate in the TCA cycle and unable to generate succinate and malate from isocitrate via the glyoxylate bypass but, unlike the SR-11 ΔfrdABCD ΔsdhCDA mutant, still able to grow using succinate as the sole source of carbon and energy (not shown), and tested it for virulence in BALB/c mice (108 CFU/mouse by the oral route, about 103 times the wild-type 50% lethal dose [LD50] [11]). As illustrated in Fig. 2, the SR-11 ΔsucCD ΔaceA double mutant was attenuated (P < 0.0001) but not avirulent and was no more attenuated than the SR-11 ΔsucCD mutant (see Fig. 4B in reference 47). It therefore appears that the glyoxylate bypass plays no role in the attenuated virulence of the SR-11 ΔsucCD mutant, making it more likely that replenishment of succinate in the SR-11 TCA cycle by succinate or a precursor of succinate present in mouse phagocytes is required for full SR-11 virulence in BALB/c mice.
FIG. 2.
Survival of BALB/c mice infected orally with 108 CFU of either wild-type SR-11 (▪) or the SR-11 ΔsucCD ΔaceA mutant (○).
FIG. 4.
Survival of BALB/c mice infected orally with 108 CFU of either wild-type SR-11 (▪) or the SR-11 ΔsfcA ΔmaeB Δmdh mutant (○) (A) or 108 CFU of either wild-type SR-11 (▪) or the “restored” SR-11 ΔsfcA ΔmaeB mutant (○) (B).
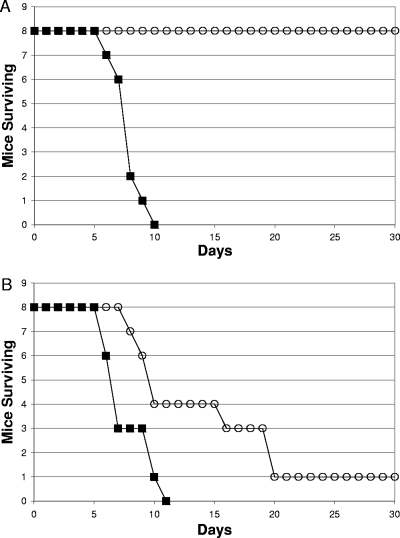
An SR-11 ΔdcuB ΔfumB mutant is fully virulent (108 CFU/mouse by the oral route).
If the hypothesis that the conversion of succinate to fumarate is key to SR-11 virulence because it is the major route to production of malate for conversion to both pyruvate and oxaloacetate (47) is true, then an SR-11 mutant unable to convert fumarate to malate should be avirulent. There are three fumarases in Escherichia coli, fumarase A, fumarase B, and fumarase C, encoded by fumA, fumB, and fumC, respectively (2, 31, 54). Fumarase A is the major active E. coli fumarase under microaerophilic conditions (49). E. coli fumarase B has a higher affinity for malate than for fumarate and appears to operate under anaerobic conditions to generate fumarate from malate (49, 55). Fumarase C is highly active under aerobic growth conditions (49). Homologs of fumarase A, fumarase B, and fumarase C that are, respectively, 95%, 92%, and 91% identical to those present in E. coli MG1655 are present in serovar Typhimurium (3, 27).
In E. coli and serovar Typhimurium, the fumB gene is immediately downstream of the dcuB gene (3, 27). The dcuB gene (for dicarboxylate uptake) encodes a C4 dicarboxylate antiporter for malate influx and succinate efflux (44). The fumB and dcuB genes are induced under anaerobic conditions when fumarate acts as a terminal electron acceptor (2, 44, 49). In the example cited above, succinate would be exchanged for malate, which would then be converted to fumarate by fumarase B, and fumarate would be converted to succinate by fumarate reductase. This scenario seems unlikely to be of value to SR-11 in vivo in view of the fact that fumarate reductase is not required for full SR-11 virulence (47). However, since DcuB also transports fumarate anaerobically (44), it was also possible that DcuB and fumarase B play roles in vivo in the conversion of fumarate to malate, in which case an SR-11 ΔdcuB ΔfumB mutant would be attenuated. To determine whether the dcuB and fumB genes play roles in SR-11 virulence, BALB/c mice were infected orally with 108 CFU (i.e., about 103 times the wild-type LD50 [11]) of either wild-type SR-11 or an SR-11 ΔdcuB ΔfumB mutant. The SR-11 ΔdcuB ΔfumB mutant appeared to be fully virulent (Fig. 3A), although statistically it was slightly less virulent than its parent (P = 0.03). These results suggest that neither C4 dicarboxylate transport by DcuB nor fumarase B plays a major role in the conversion of fumarate to malate by SR-11 during infection of BALB/c mice.
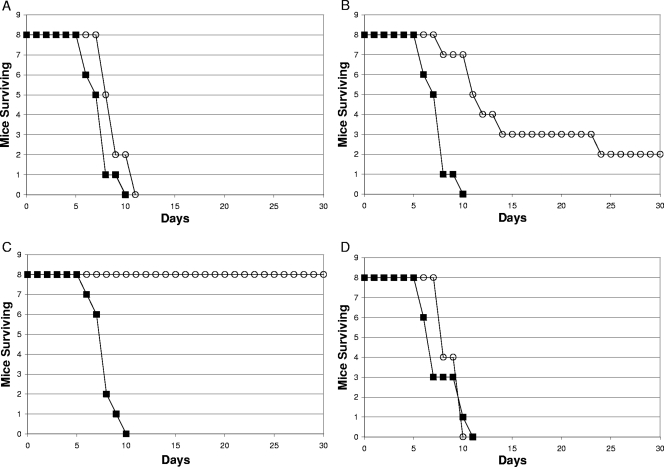
FIG. 3.
Survival of BALB/c mice infected orally with 108 CFU of either wild-type SR-11 (▪) or the SR-11 ΔdcuB ΔfumB mutant (○) (A), 108 CFU of either wild-type SR-11 (▪) or the SR-11 ΔfumAC mutant (○) (B),108 CFU of either wild-type SR-11 (▪) or the SR-11 ΔfumAC ΔdcuB ΔfumB mutant (○) (C), or 108 CFU of either wild-type SR-11 (▪) or the SR-11 “restored” ΔdcuB ΔfumB mutant (○) (D).
An SR-11 ΔfumAC mutant is attenuated and an SR-11 ΔfumAC ΔdcuB ΔfumB mutant is avirulent (108 CFU/mouse by the oral route).
Since it appears that fumarase B plays no major role in SR-11 virulence in BALB/c mice (Fig. 3A), we tested the virulence of an SR-11 ΔfumAC mutant, unable to make both fumarase A and fumarase C. BALB/c mice were infected orally with 108 CFU (i.e., about 103 times the wild-type LD50 [11]) of either wild-type SR-11 or SR-11 ΔfumAC. The SR-11 ΔfumAC mutant was attenuated (P = 0.0004) but not avirulent (Fig. 3B). In fact, mice infected with the SR-11 ΔfumAC double mutant that survived the 30-day duration of the experiment still appeared sick (ruffled fur, crusted eyes, and lethargic). These results suggest that either fumarase A or fumarase C or both play a role in SR-11 virulence.
It was possible that the SR-11 ΔfumAC mutant was not avirulent because endogenous fumarase B could substitute somewhat for fumarase A and/or fumarase C. BALB/c mice were therefore infected orally with 108 CFU (i.e., about 103 times the wild-type LD50 [11]) of either wild-type SR-11 or an SR-11 ΔfumAC ΔdcuB ΔfumB mutant. The SR-11 ΔfumAC ΔdcuB ΔfumB mutant was indeed avirulent (P < 0.0001), i.e., mice infected with wild-type SR-11 all died within 10 days postinfection (Fig. 3C), whereas the mice infected with the SR-11 ΔfumAC ΔdcuB ΔfumB mutant remained completely healthy throughout the 30-day duration of the experiment (Fig. 3C). Since SR-11 became avirulent only when all three fumarase genes were deleted, it appears that if both fumarase A and fumarase C are not available in vivo, fumarase B can substitute and convert fumarate to malate, albeit not at the rate required for full virulence (Fig. 3B).
The SR-11 ΔfumAC ΔdcuB ΔfumB mutant was constructed by inserting the ΔfumAC deletion into the SR-11 ΔdcuB ΔfumB mutant. To prove conclusively that the deletions were responsible for the avirulence of the SR-11 ΔfumAC ΔdcuB ΔfumB mutant, the wild-type fumAC genes were reinserted into the mutant to regenerate the SR-11 ΔdcuB ΔfumB mutant (see Materials and Methods). Although the SR-11 ΔfumAC ΔdcuB ΔfumB mutant grows well with glucose as the sole source of carbon and energy (not shown), it fails to grow using succinate as the sole carbon and energy source (not shown) because it cannot carry out the conversion of fumarate to malate and therefore is unable to perform gluconeogenesis; however, the “restored” SR-11 ΔdcuB ΔfumB mutant grew with succinate as the sole source of carbon and energy (not shown) and, like the original SR-11 ΔdcuB ΔfumB mutant, was fully virulent (P = 0.44) (Fig. 3D). Therefore, collectively, the data strongly suggest that preventing fumarate from being converted to malate by the three fumarases in SR-11 renders SR-11 completely avirulent in BALB/c mice.
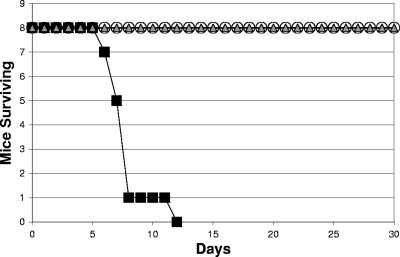
An SR-11 ΔsfcA ΔmaeB Δmdh triple mutant is avirulent (108 CFU/mouse by the oral route).
Previously, we showed that an SR-11 ΔsfcA ΔmaeB mutant, unable to convert malate to pyruvate, and an SR-11 Δmdh mutant, unable to convert malate to oxaloacetate, were both attenuated in BALB/c mice but not completely avirulent (47). If the conversion of succinate to fumarate is key to SR-11 virulence because the fumarate then provides the major source of malate for conversion to both pyruvate and oxaloacetate during infection of BALB/c mice, an SR-11 ΔsfcA ΔmaeB Δmdh mutant should be avirulent. To address this question, BALB/c mice were infected orally with 108 CFU of either wild-type SR-11 or an SR-11 ΔsfcA ΔmaeB Δmdh mutant. Mice infected with wild-type SR-11 all died within 10 days postinfection (Fig. 4A), whereas the mice infected with the SR-11 ΔsfcA ΔmaeB Δmdh mutant remained completely healthy throughout the 30-day duration of the experiment (Fig. 4A), i.e., the triple mutant was completely avirulent (P < 0.0001).
The SR-11 ΔsfcA ΔmaeB Δmdh mutant was constructed by inserting the Δmdh deletion into the SR-11 ΔsfcA ΔmaeB mutant. To prove conclusively that the inability of the SR-11 ΔsfcA ΔmaeB Δmdh mutant to convert malate to pyruvate and oxaloacetate was responsible for its avirulence, the wild-type mdh gene was reinserted into the SR-11 ΔsfcA ΔmaeB Δmdh mutant to regenerate the SR-11 ΔsfcA ΔmaeB mutant (see Materials and Methods). While the SR-11 ΔsfcA ΔmaeB Δmdh mutant grows well with glucose as the sole source of carbon and energy (not shown), it fails to grow using succinate as the sole carbon and energy source because it is unable to convert malate to either pyruvate or oxaloacetate and is therefore unable to carry out gluconeogenesis. The SR-11 ΔsfcA ΔmaeB mutant grows with succinate as the sole source of carbon and energy, since it is able to convert oxaloacetate to PEP and is therefore able to carry out gluconeogenesis (Fig. 1). As expected, the “restored” SR-11 ΔsfcA ΔmaeB mutant grew with succinate as the sole source of carbon and energy (not shown) and, like the original SR-11 ΔsfcA ΔmaeB mutant, was attenuated (P = 0.015) but not avirulent (Fig. 4B). Collectively, these data suggest that preventing malate from being converted to either pyruvate or oxaloacetate attenuates SR-11 virulence but preventing malate from being converted to both pyruvate and oxaloacetate renders it completely avirulent in BALB/c mice.
The SR-11 ΔfumAC ΔdcuB ΔfumB mutant and the ΔsfcA ΔmaeB Δmdh mutant are immunogenic.
To this point, the data suggested that the SR-11 ΔfrdABCD ΔsdhCDA mutant, the SR-11 ΔfumAC ΔdcuB ΔfumB mutant, and the SR-11 ΔsfcA ΔmaeB Δmdh mutant are avirulent for the same reason, i.e., because they are all unable to make malate for conversion to both pyruvate and oxaloacetate. In addition to being avirulent, we found previously that the SR-11 ΔfrdABCD ΔsdhCDA mutant was immunogenic, i.e., mice orally vaccinated with the SR-11 ΔfrdABCD ΔsdhCDA mutant were completely protected when challenged 30 days later with the virulent wild-type SR-11 (30). It therefore seemed likely that if the three mutants are avirulent for the same reason, they should all be immunogenic. To this end, BALB/c mice originally fed either 108 CFU of the SR-11 ΔfumAC ΔdcuB ΔfumB mutant or the ΔsfcA ΔmaeB Δmdh mutant were orally challenged with 108 CFU of the wild-type SR-11 30 days later. The mice never appeared ill and remained healthy both throughout the initial 30 days of the experiment and during the ensuing 30 days after challenge with wild-type SR-11, whereas sham-infected mice orally challenged with 108 CFU of the wild-type SR-11 30 days after sham infection all died within 12 days (Fig. 5). Therefore, like mice infected with the SR-11 ΔfrdABCD ΔsdhCDA mutant, mice infected with either the SR-11 ΔfumAC ΔdcuB ΔfumB mutant or the ΔsfcA ΔmaeB Δmdh mutant were fully protected against challenge with wild-type SR-11 (P < 0.0001).
FIG. 5.
Survival of BALB/c mice initially sham infected orally and infected 30 days later with 108 CFU of wild-type SR-11 (▪), initially infected orally with 108 CFU of the SR-11 ΔfumAC ΔdcuB ΔfumB mutant and infected 30 days later with 108 CFU of wild-type SR-11 (○), or initially infected orally with 108 CFU of the SR-11 ΔsfcA ΔmaeB Δmdh mutant and infected 30 days later with 108 CFU of wild-type SR-11 (▵).
DISCUSSION
In a recent study, we showed that fatty acid degradation, the glyoxylate bypass, gluconeogenesis, and the anaplerotic replenishment of oxaloacetate in the TCA cycle from PEP did not appear to be needed for full SR-11 virulence in BALB/c mice, but cyclic operation of the TCA cycle and the formation of both pyruvate and oxaloacetate from malate were required (47). Removal of malate from the TCA cycle to produce pyruvate requires that TCA cycle intermediates be replenished to keep the TCA cycle operative. However, since it appeared that the glyoxylate bypass was not needed for full virulence (47), replenishment of TCA cycle intermediates did not seem to occur by this pathway. Thus, the data were consistent with the hypothesis that during infection of BALB/c mice, SR-11 grows in a mixed mode using limiting glycolytic sugars and either amino acids or intermediates of the TCA cycle to replenish it, thereby allowing continuous formation of pyruvate from malate.
In a subsequent report, we showed that although an SR-11 ΔfrdABCD mutant, unable to make fumarate reductase, was fully virulent in BALB/c mice and an SR-11 ΔsdhCDA mutant, unable to make succinate dehydrogenase, was only slightly attenuated, an SR-11 ΔfrdABCD ΔsdhCDA mutant was avirulent (30). These data strengthened the hypothesis that a complete TCA cycle is necessary for SR-11 virulence and suggested that fumarate reductase, which normally runs in the opposite direction to succinate dehydrogenase for branched TCA cycle operation, can substitute for succinate dehydrogenase in the SR-11 ΔsdhCDA mutant during infection to run a full TCA cycle with only a slight reduction in virulence. In addition, since an SR-11 ΔsucCD mutant that could not convert succinyl-CoA to succinate was only moderately attenuated (47), the data suggested that the conversion of succinate to fumarate is key to SR-11 virulence.
In this study, we have presented data showing that in addition to an SR-11 ΔfrdABCD ΔsdhCDA mutant being avirulent, both an SR-11 ΔfumAC ΔdcuB ΔfumB mutant and an SR-11 ΔsfcA ΔmaeB Δmdh mutant are also avirulent. The common feature shared by these mutants is that they are all unable to generate malate for the production of both pyruvate and oxaloacetate (47). However, blocking the conversion of succinyl-CoA to succinate, which precedes the conversion of succinate to fumarate in the TCA cycle (Fig. 1), should also result in the inability to generate malate, yet an SR-11 ΔsucCD mutant is only moderately attenuated (47), and as shown here, an SR-11 ΔsucCD ΔaceA mutant is no more attenuated than an SR-11 ΔsucCD mutant (Fig. 2), ruling out the possibility that the glyoxylate bypass becomes important in the SR-11 ΔsucCD mutant to generate malate. Moreover, unlike SR-11 ΔfrdABCD ΔsdhCDA, SR-11 ΔfumAC ΔdcuB ΔfumB, and SR-11 ΔsfcA ΔmaeB Δmdh, the SR-11 ΔsucCD ΔaceA mutant grows with succinate as the sole source of carbon and energy.
Collectively, these data can be explained as follows. We suggest that succinate, ornithine, or arginine present in mouse phagocytes replenishes succinate in the SR-11 TCA cycle. Succinate is then converted to fumarate, then to malate, and then to pyruvate and oxaloacetate. Since malate and subsequently pyruvate and oxaloacetate could be synthesized from replenished succinate in the SR-11 ΔsucCD mutant but could not be made by theSR-11 ΔfrdABCD ΔsdhCDA, SR-11 ΔfumAC ΔdcuB ΔfumB, or SR-11 ΔsfcA ΔmaeB Δmdh mutant (Fig. 1), the SR-11 ΔsucCD mutant could retain some virulence. More work will be necessary to test this hypothesis; however, it will be of great interest if arginine present in mouse phagocytes is found to be used by SR-11 to replenish succinate in its TCA cycle, since arginine is used for nitric oxide synthesis (8) and nitric oxide is thought to limit the growth of Salmonella in macrophages (51). Consequently, if this scenario is correct, arginine utilization by SR-11 in macrophages would not only support SR-11 virulence by replenishing TCA cycle intermediates, but might limit nitric oxide synthesis and thereby further promote SR-11 virulence.
Why is the formation of pyruvate from malate necessary for full SR-11 virulence? The ability of SR-11 to generate PEP from pyruvate has a minimal effect on virulence, i.e., we previously showed that an SR-11 ΔppsA ΔpckA double mutant, unable to make PEP either from oxaloacetate via PEP carboxykinase, encoded by the pckA gene (28), or from pyruvate via PEP synthase, encoded by the ppsA gene (35), was at most slightly attenuated (47). It therefore appears that the requirement for malate to be converted to pyruvate for full virulence is not for the generation of PEP and suggests that insufficient pyruvate is produced by glycolytic reactions to fulfill the precursor metabolite requirements for biosynthesis of one or more of the following classes of compounds: (i) alanine, valine, isoleucine, and leucine for protein synthesis; (ii) acetyl-CoA for the synthesis of fatty acids and membrane phospholipids; (iii) acetyl-CoA to keep the TCA cycle operative; and (iv) acetyl-CoA for the synthesis of acetyl-phosphate, a known regulator of virulence genes (22). In any case, the fact that not enough pyruvate is generated through glycolysis for full virulence suggests that SR-11 grows in an environment scarce in glycolytic sugars in vivo.
Upon entry into a host phagocyte, serovar Typhimurium resides and replicates within a membrane-bound compartment called a Salmonella-containing vacuole (SCV) (14, 39). Recent evidence suggests that replication of serovar Typhimurium in the SCV of a single phagocyte in either the liver or spleen is limited during acute infection, usually not exceeding two or three doublings (41, 43). Furthermore, it appears that serovar Typhimurium increases in numbers in the liver by spreading from one phagocyte to others nearby, thereby forming a focus of infection (43). However, it appears that during acute infection some serovar Typhimurium cells leave one focus of infection to form other foci of infection (43). Thus, although serovar Typhimurium grows poorly in infected phagocytes, it grows to high numbers in the liver because it spreads to form numerous foci of infection (43). Although several nonnutritional host factors have been implicated in limiting serovar Typhimurium growth in the SCV (15, 48), our data suggest that the poor growth may, at least in part, be due to a scarcity of carbon sources.
While gluconeogenesis and isocitrate lyase of the glyoxylate bypass appear not to be necessary for acute serovar Typhimurium infection in BALB/c mice (13, 47), isocitrate lyase does appear to be required for chronic serovar Typhimurium infection (13). Moreover, several other bacterial and fungal pathogens appear to require either gluconeogenesis or isocitrate lyase, or both, for virulence (9, 24, 33, 46, 53, 56). It therefore appears that different intracellular pathogens have evolved different strategies for success within phagocytes. In fact, recent evidence suggests the possibility that serovar Typhimurium, and perhaps other intracellular pathogens, orchestrates host responses to reduce the growth rate within phagocytes for successful infection (15). One of the orchestrations that intracellular pathogens choose may be to alter the phagosome or the phagolysosome membrane to limit the transport of various carbon sources. If so, since, as described above, different intracellular pathogens appear to have chosen different strategies for growth, it may be that different pathogens limit the transport of different carbon sources during infection.
On a practical note, it is clear that the SR-11 ΔfrdABCD ΔsdhCDA, SR-11 ΔfumAC ΔdcuB ΔfumB, and SR-11 ΔsfcA ΔmaeB Δmdh mutants are not only avirulent, they effectively protect BALB/c mice against subsequent infection with wild-type SR-11. In this context, it would be of great interest to determine whether these mutants are avirulent in humans and animals and can also protect them against infection by their virulent parents and whether, if the mutants are able to meet the energy requirements for foreign-antigen production, they might be effective as vehicles for genes that express virulence antigens of unrelated pathogens to induce protective immunity against those pathogens.
Acknowledgments
This research was supported by a USDA Strengthening Research Grant entitled Environmental Biotechnology at URI to P.S.C. and in part by Public Health Service grant AI 48945 to T.C. and P.S.C.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Allen, J. H., M. Utley, H. Van den Bosch, P. Nuijten, M. Witvliet, B. A. McCormick, K. A. Krogfelt, T. R. Licht, D. Brown, M. Mauel, M. P. Leatham, D. C. Laux, and P. S. Cohen. 2000. A functional cra gene is required for Salmonella enterica serovar Typhimurium virulence in BALB/c mice. Infect. Immun. 683772-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, P. J., S. C. Andrews, M. N. Sivak, and J. R. Guest. 1989. Nucleotide sequence of the FNR-regulated fumarase gene (fumB) of Escherichia coli K-12. J. Bacteriol. 1713494-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, and D. J. Rose. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Buck, D., M. E. Spencer, and J. R. Guest. 1986. Cloning and expression of the succinyl-CoA synthetase genes of Escherichia coli K12. J. Gen. Microbiol. 1321753-1762. [DOI] [PubMed] [Google Scholar]
- 5.Bukhari, A. I., and A. L. Taylor. 1971. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J. Bacteriol. 105844-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo-Castañeda, G., and M. V. Ortega. 1970. Mutants of Salmonella typhimurium lacking phosphoenolpyruvate carboxykinase and α-ketoglutarate dehydrogenase activities. J. Bacteriol. 102524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecchini, G., I. Schroder, R. P. Gunsalus, and E. Maklashina. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553140-157. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. I., J. C. Liao, and L. Kuo. 1998. Arginase modulates nitric oxide production in activated macrophages. Am. J. Phys. 274H342-H348. [DOI] [PubMed] [Google Scholar]
- 9.Collins, D. M., T. Wilson, S. Campbell, B. M. Buddle, B. J. Wards, G. Hotter, and G. W. de Lisle. 2002. Production of avirulent mutants of Mycobacterium bovis with vaccine properties by the use of illegitimate recombination and screening of stationary-phase cultures. Microbiology 1483019-3027. [DOI] [PubMed] [Google Scholar]
- 10.Cronan, J. E., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass. p. 206-216. In F. C. Neihardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella Typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 11.Curtiss, R., III, and S. M. Kelly. 1987. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 553035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, F. C., S. J. Libby, M. E. Castor, and A. M. Fung. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 732547-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlay, B. B., B. Gumbiner, and S. Falkow. 1988. Penetration of Salmonella through a polarized Madin-Darby canine kidney epithelial cell monolayer. J. Cell Biol. 107221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-del Portillo, F., C. Núñez-Hernández, B. Eisman, and J. Ramos-Vivas. 2008. Growth control in the Salmonella-containing vacuole. Curr. Opin. Microbiol. 1146-52. [DOI] [PubMed] [Google Scholar]
- 16.Gilvarg, C. 1957. N-Succinyl-l-diaminopimelic acid, an intermediate in the biosynthesis of diaminopimelic acid. Biochim. Biophys. Acta 24216-217. [DOI] [PubMed] [Google Scholar]
- 17.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neihardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella Typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 18.Guest, J. R., J. S. Miles, R. E. Roberts, and S. A. Woods. 1985. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC). J. Gen. Microbiol. 1312971-2984. [DOI] [PubMed] [Google Scholar]
- 19.Hirsh, C. A., M. Rasminsky, B. D. Davis, and E. C. Lin. 1963. A fumarate reductase in Escherichia coli distinct from succinate dehydrogenase. J. Biol. Chem. 2383770-3774. [PubMed] [Google Scholar]
- 20.Hohmann, A. W., G. Schmidt, and D. Rowley. 1978. Intestinal colonization and virulence of Salmonella typhimurium. Infect. Immun. 22763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 18015-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal shortchain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 461451-1464. [DOI] [PubMed] [Google Scholar]
- 23.Lindgren, S. W., I. Stojiljkovic, and F. Heffron. 1996. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 934197-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 41283-86. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan, S. K., C. C. Chu, D. K. Willis, A. Templin, and A. J. Clark. 1990. Physical analysis of spontaneous and mutagen-induced mutants of Escherichia coli K-12 expressing DNA exonuclease VIII activity. Genetics 125261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka, M., and B. A. McFadden. 1988. Isolation, hyperexpression, and sequencing of the aceA gene encoding isocitrate lyase in Escherichia coli. J. Bacteriol. 1704528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 28.Medina, V., R. Pontarollo, D. Glaeske, H. Tabel, and H. Goldie. 1990. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J. Bacteriol. 1727151-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mengin-Lecreulx, D., C. Michaud, C. Richaud, D. Blanot, and J. van Heijenoort. 1988. Incorporation of ll-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J. Bacteriol. 1702031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercado-Lubo, R., E. J. Gauger, M. P. Leatham, T. Conway, and P. S. Cohen. 2008. A Salmonella enterica serovar Typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infect. Immun. 761128-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miles, J. S., and J. R. Guest. 1984. Complete nucleotide sequence of the fumarase gene fumA of Escherichia coli. Nucleic Acids Res. 123631-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Muñoz-Elias, E. J., and J. D. McKinney. 2005. M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vitro growth and virulence. Nat. Med. 11638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., J. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA.
- 35.Niersbach, M., F. Kreuzaler, R. H. Geerse, P. W. Postma, and H. J. Hirsch. 1992. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol. Gen. Genet. 231332-336. [DOI] [PubMed] [Google Scholar]
- 36.Park, S. J., G. Chao, and R. P. Gunsalus. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode α-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 1794138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, S. J., C. P. Tseng, and R. P. Gunsalus. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15473-482. [DOI] [PubMed] [Google Scholar]
- 38.Reitzer, L. 25 July 2005, posting date. Module 3.4.7, Catabolism of amino acids and related compounds. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 39.Richter-Dahlfors, A., A. M. J. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabe, H., T. Miwa, T. Kodaki, K. Izui, S. Hiraga, and H. Katsuki. 1984. Molecular cloning of the phosphoenolpyruvate carboxylase gene, ppc, of Escherichia coli. Gene 31279-283. [DOI] [PubMed] [Google Scholar]
- 41.Salcedo, S. P., M. Noursadeghi, J. Cohen, and D. W. Holden. 2001. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol. 3587-597. [DOI] [PubMed] [Google Scholar]
- 42.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 31335-1344. [DOI] [PubMed] [Google Scholar]
- 43.Sheppard, M., C. Webb, F. Heath, V. Mallows, R. Emilianus, D. Maskell, and P. Mastroeni. 2003. Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell Microbiol. 5593-600. [DOI] [PubMed] [Google Scholar]
- 44.Six, S., S. C. Andrews, G. Unden, and J. R. Guest. 1994. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct). J. Bacteriol. 1766470-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagi, J. S., N. Ida, M. Tokushige, H. Sakamoto, and Y. Shimura. 1985. Cloning and nucleotide sequence of the aspartase gene of Escherichia coli W. Nucleic Acids Res. 132063-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, D. J., Y. Q. He, J. X. Feng, B. R. He, B. L. Jiang, G. T. Lu, B. Chen, and J. L. Tang. 2005. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J. Bacteriol. 1876231-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tchawa Yimga, M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. The Role of gluconeogenesis and the TCA cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 741130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tierrez, A., and F. Garcia-del Portillo. 2005. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell Microbiol. 7901-909. [DOI] [PubMed] [Google Scholar]
- 49.Tseng C. P., C. C. Yu, H. H. Lin, C. Y. Chang, and J. T. Kuo. 2001. Oxygen and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J. Bacteriol. 183461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utley, M., D. P. Franklin, K. A. Krogfelt, D. C. Laux, and P. S. Cohen. 1998. A Salmonella typhimurium strain unable to utilize fatty acids and citrate is avirulent and immunogenic in mice. FEMS Microbiol. Lett. 163129-134. [DOI] [PubMed] [Google Scholar]
- 51.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ischiropoulos, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel, R. F., K. D. Entian, and D. Mecke. 1987. Cloning and sequence of the mdh structural gene of Escherichia coli coding for malate dehydrogenase. Arch. Microbiol. 14936-42. [DOI] [PubMed] [Google Scholar]
- 53.Wall, D. M., P. S. Duffy, C. DuPont, J. F. Prescott, and W. G. Meijer. 2005. Isocitrate lyase activity is required for virulence of the intracellular pathogen Rhodococcus equi. Infect. Immun. 736736-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woods, S. A., J. S. Miles, R. E. Roberts, and J. R. Guest. 1986. Structural and functional relationships between fumarase and aspartase. Nucleotide sequences of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K12. Biochem. J. 237547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woods, S. A., S. D. Schwartzbach, and J. R. Guest. 1988. Two biochemically distinct classes of fumarases in Escherichia coli. Biochim. Biophys. Acta 95414-26. [DOI] [PubMed] [Google Scholar]
- 56.Yang, W., Y. Liu, L. Chen, G. Qian, H. Liu, B. Hu, and F. Liu. 2008. Involvement of gluconeogenic pathway in virulence of Xanthomonas oryzae pv. oryzae. J. Phytopathol. 156174-180. [Google Scholar]