Abstract
It is well established that prokaryotes and eukaryotes alike utilize phosphotransfer to regulate cellular functions. One method by which this occurs is via eukaryote-like serine/threonine kinase (ESTK)- and phosphatase (ESTP)-regulated pathways. The role of these enzymes in Staphylococcus aureus has not yet been examined. This resilient organism is a common cause of hospital-acquired and community-associated infections, infecting immunocompromised and immunocompetent hosts alike. In this study, we have characterized a major functional ESTK (STK) and ESTP (STP) in S. aureus and found them to be critical modulators of cell wall structure and susceptibility to cell wall-acting β-lactam antibiotics. By utilizing gene knockout strategies, we created S. aureus N315 mutants lacking STP and/or STK. The strain lacking both STP and STK displayed notable cell division defects, including multiple and incomplete septa, bulging, and irregular cell size, as observed by transmission electron microscopy. Mutants lacking STP alone displayed thickened cell walls and increased resistance to the peptidoglycan-targeting glycylglycine endopeptidase lysostaphin, compared to the wild type. Additionally, mutant strains lacking STK or both STK and STP displayed increased sensitivity to cell wall-acting cephalosporin and carbapenem antibiotics. Together, these results indicate that S. aureus STK- and STP-mediated reversible phosphorylation reactions play a critical role in proper cell wall architecture, and thus the modulation of antimicrobial resistance, in S. aureus.
Staphylococcus aureus constitutes a major public health threat, as it is the most common hospital-associated pathogen in the world and its prevalence in community-acquired infections is on the rise (18). This gram-positive coccus is armed with a wide variety of virulence factors that contribute to diseases ranging from mild food poisoning, skin lesions, and boils to severe and often fatal endocarditis, osteomyelitis, pneumonia, and toxic shock syndrome (28). Staphylococci are known for their evolving mechanisms of antimicrobial resistance, which have resulted in the spread of methicillin-resistant and even vancomycin-resistant S. aureus, severely limiting treatment options for those infected (41). The signaling cascades which enable the staphylococcus to evolve such resistance mechanisms and cause infection remain a major field of study.
A recent comparative analysis of several prokaryotic genomes suggests that one-component regulatory systems (in contrast to the conventional paradigm of two-component regulatory systems) are, in fact, the most abundant signaling systems in prokaryotes (43). These one-component systems include eukaryote-like serine/threonine kinases (ESTKs) and phosphatases (ESTPs), which have emerged as critical signaling molecules in prokaryotes over the past decade (5). Since the first characterization of an ESTK in soil bacteria (Myxococcus xanthus [34]), similar ESTKs have been reported in several gram-negative and -positive bacteria, including Pseudomonas aeruginosa (32, 33), Streptococcus agalactiae (39), S. pneumoniae (14, 35), S. pyogenes (22), Bacillus subtilis (30), and S. mutans (21), as well as Mycobacterium tuberculosis (reviewed in reference 3). These enzymes have been implicated in various steps of bacterial pathogenesis. More specifically, ESTKs are critical for colonization of the host and establishment of infection, as they have been shown to affect the abilities of bacteria to replicate (23, 42), adhere to host cells (22), form biofilms (21), and cause disease (14, 39, 47).
Currently, no information is available on the role of ESTK-mediated signaling in S. aureus. In the present investigation, based on a genome-wide BlastP search, we have identified the presence of a major putative ESTK (designated STK) and its cotranscribing ESTP (designated STP) in methicillin-resistant S. aureus strain N315. We have further characterized the biochemical properties of STK and STP and found them to be critical modulators of cell wall structure and hence resistance to specific cell wall-acting β-lactam antibiotics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Wild-type S. aureus N315 and derived mutant strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) (BD Diagnostic Systems). The media were supplemented with 10 μg/ml chloramphenicol, 12 μg/ml tetracycline, 5 μg/ml erythromycin, 250 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and/or 1.5 μg/ml anhydrotetracycline (Clontech) where indicated. Escherichia coli strains DH5α, XL-1-Blue, and BL21(DE3)pLysS were all grown in Luria-Bertani broth or agar (BD Diagnostic Systems) supplemented with 50 μg/ml carbenicillin or 100 μg/ml ampicillin where indicated. All bacterial strains were grown at 37°C unless otherwise stated.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pET14B | N-terminally His-tagged expression vector | Novagen |
| pET14B-STP | pET14B containing entire SA1062 gene | This study |
| pET14B-STK | pET14B containing entire SA1063 gene | This study |
| pMAD | E. coli-S. aureus shuttle vector for gene inactivation | 2 |
| pDC123 | Source of chloramphenicol acetyltransferase resistance gene (chl) | 8 |
| pMADΔSTKchl | pMAD containing up- and downstream regions of SA1063 flanking chl | This study |
| pKOR1 | E. coli-S. aureus shuttle vector with ccdB selection and lambda recombination capabilities | 4 |
| pKOR1ΔSTP | pKOR1 containing up- and downstream regions of SA1062 | This study |
| pKOR1ΔSTP/STK | pKOR1 containing regions upstream of SA1062 and downstream of SA1063 | This study |
| pCN36 | Source of tet(M) gene for tetracycline resistance | 9 |
| pCN40 | E. coli-S. aureus shuttle vector | 9 |
| pCN40tet | pCN40 with ermC gene replaced with tet(M) from pCN36 | This study |
| pCN40tet-STP | pCN40tet containing entire SA1062 gene and putative promoter upstream | This study |
| pCN40tet-STK | pCN40tet containing entire SA1063 gene and putative promoter upstream of SA1062 | This study |
| pCN40tet-STPSTK | pCN40tet containing overlapping SA1062 and SA1063 genes and putative promoter upstream of SA1062 | This study |
| E. coli strains | ||
| XL-1-Blue | endA1 gyrA96(Nalr) thi-1 recA1 relA1 lac glnV44 [F′::Tn10 proAB+lacIq Δ(lacZ)M15] hsdR17(rK− mK+) | Stratagene |
| DH5α | Subcloning efficiency F− φ80dlacZΔM15 λ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| BL21(DE3)pLysS | F−ompT hsdSB(rB− mB−) gal dcm(DE3)pLysS (Camr) | Invitrogen |
| S. aureus strains | ||
| RN4220 | Restriction-deficient derivative of NCTC 8325-4 | 24 |
| N315 | Methicillin-resistant S. aureus parent strain | 26 |
| N315ΔSTP | N315 lacking SA1062 | This study |
| N315ΔSTK | N315 lacking SA1063 | This study |
| N315ΔSTP/STK | N315 lacking both SA1062 and SA1063 | This study |
| N315pCN40tet | N315 containing pCN40tet (vector-only control) | This study |
| N315ΔSTPΩSTP | Complemented N315ΔSTP strain containing pCN40tet-STP | This study |
| N315ΔSTKΩSTK | Complemented N315ΔSTK strain containing pCN40tet-STK | This study |
| N315ΔSTP/STKΩSTP/STK | Complemented N315ΔSTP/STK strain containing pCN40tet-STPSTK | This study |
Cotranscription assay.
Total RNA was isolated from mid-log phase (optical density at 600 nm = 0.6) cultures of strain N315 grown in TSB using Qiagen's RNeasy Mini Kit and subjected to on-column DNA digestion using Qiagen's RNase-free DNase set. A 2.5-μg sample of this RNA was used to create cDNA using Superscript RTII (Invitrogen) in accordance with the manufacturer's instructions. An identical reaction was performed without reverse transcriptase (RT) as a negative control. cDNA with or without RT and genomic DNA (gDNA) were used as templates in PCRs using primer sets specific for the overlapping (#108/#109) and outermost regions of stp and stk (#44/#107 and #110/#46), as shown in Fig. 1A.
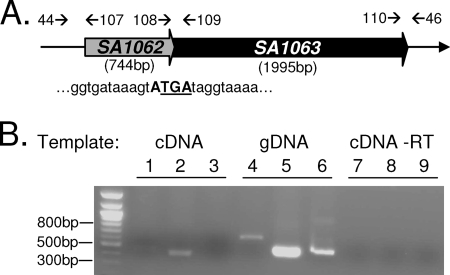
FIG. 1.
stp and stk are cotranscribed. (A) Schematic of overlapping pattern of SA1062 (stp) and SA1063 (stk) and primer annealing sites. The four-nucleotide overlap is in bold. The stop codon for SA1062 is underlined. Primer binding sites are indicated by arrows. (B) Results of PCRs using cDNA, gDNA, or cDNA reaction mixtures lacking RT as templates. Lanes 1, 4, and 7 were performed using primer set 44/107; lanes 2, 5, and 8 utilized 108/109; and lanes 3, 6, and 9 utilized 110/46, annealing on flanking or overlapping regions of stp/stk as indicated in panel A.
Production of rSTP and rSTK.
Primer sets STP-F/STP-R and STK-F/STK-R were used to create 744-bp and 1,995-bp fragments, respectively, encoding SA1062 (stp) and SA1063 (stk) from S. aureus N315 gDNA. These fragments were inserted separately into the multiple cloning site of His-tagged expression vector pET14B (Novagen) to create pET14B-STP and pET14B-STK. Expression of recombinant His-tagged STK and STP was induced in E. coli BL21 for 4 h by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Recombinant STK (rSTK) and rSTP were purified from the respective cell lysates by denaturing and nondenaturing methods, respectively, using Ni-nitrilotriacetic acid column chromatography as recommended in the manufacturer's instructions (Qiagen). rSTK was further purified by fast protein liquid chromatography using a Superdex 200 HR 10/30 column. Eluted proteins were dialyzed overnight against 25 mM Tris/HCl buffer, pH 8.
In vitro kinase assays.
Briefly, 0.5 μg rSTK and/or 1 μg myelin basic protein (MBP) were incubated separately (control reactions) and in the indicated combinations with 1 μCi [γ-32P]ATP in 40 μl of kinase buffer (50 mM Tris/HCl [pH 7.5], 1 mM dithiothreitol, 10 mM MgCl2 or MnCl2) in the absence or presence of 1 μg rSTP for up to 30 min at room temperature, as described previously (22). Phosphorylated and dephosphorylated proteins were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and identified by autoradiography.
Thin-layer chromatography.
Sites of STK-mediated autophosphorylation and phosphorylation of MBP were measured essentially as described previously (22). Briefly, phosphorylated protein bands resolved by SDS-PAGE were excised and subjected to 6 M HCl hydrolysis at 110°C for 6 h, followed by two-dimensional thin-layer chromatography using isobutyric acid-NH4OH for the 12-h first-dimension run and 2-propanol-HCl-H2O for the 8-h second-dimension run. Phosphoamino acid standards were run along with the hydrolyzed proteins. Radioactive spots of the hydrolysates were visualized by autoradiography, while phosphoamino acid standards were visualized by ninhydrin staining. Specific phosphorylated amino acids were identified by aligning the autoradiograph with the stained standards.
Construction of N315ΔSTK.
Primer sets AF/AR and BF/BR were used to amplify approximately 600 bp upstream and 750 bp downstream of SA1063 (stk). These primers were designed to keep the cotranscribing SA1062 sequence intact, including the stop codon-containing four overlapping nucleotides at its 3′ end. Similarly, primer set catF/catR was used to amplify the 743-bp chloramphenicol acetyltransferase (cat) gene and its promoter from pDC123 (8). These fragments were digested with the appropriate enzymes (Table 2) and sequentially inserted into the multiple cloning site of the temperature-sensitive vector pMAD (2) to create pMADΔSTKchl such that the cat gene was flanked by upstream and downstream regions of SA1063. This plasmid was passed through S. aureus strain RN4220 and electroporated into S. aureus strain N315 to create S. aureus strain N315ΔSA1063 as described previously (2). The resultant mutant was selected on TSA containing chloramphenicol and confirmed by PCR, gDNA sequencing, and Western blotting.
TABLE 2.
Primers used in this study
| Use and name | Sequence (5′ → 3′)a | Restriction site |
|---|---|---|
| Cotranscription assay | ||
| #44 | GCCATGTTAACTTAATTCCTGTCAACC | |
| #107 | CGCAACCAATTTTCAGCTTGATG | |
| #108 | ACAGATAAACGTGTGAGTCCAGA | |
| #109 | CTTCTTCATCAACATCGATCATACTTAC | |
| #110 | GAATCGGTAGATGTACCATACACTG | |
| #46 | TCGACCTGTCTTCAAAATGGCA | |
| Recombinant proteins | ||
| STP-F | CTACGGCCATATGCTAGAGGCACAATTTTTTACTGATACTGGAC | NdeI |
| STP-R | CCGCGGATCCTCATACTTTATCACCTTCAATAGCCG | BamHI |
| STK-F | CTACGGCCATATGATAGGTAAAATAATAAATGAACG | NdeI |
| STK-R | CCGCGGATCCTTAAACATCATCATAGCTGACTTCTTTTTC | BamHI |
| Deletion strains | ||
| AF | GCGGATCCCATAAAGCAGGAGAAGTTGCAAG | BamHI |
| AR | CTAGCTAGCTCATACTTTATCACCTTCAATAGC | NheI |
| BF | CTAGCTAGCAATTGAAGTAAATGTACCGAGG | NheI |
| BR | TGCCATGGCACTTACCGACACTGATTGACCAC | NcoI |
| catF | CGGCTAGCCGTTGACTTTTAAAAAAGGATTGATTC | NheI |
| catR | CGGCTAGCCTTATAAAAGCCAGTCATTAGGC | NheI |
| #33 | GGGGACCACTTTGTACAAGAAAGCTGGGTGAAACAGATTTTTGAATGGTTATATCAA | |
| #34 | GTTTCACCGCGGGTTTCTACTTGTCGTTCCTTTGC | SacII |
| #58 | GTTTCACCGCGGAGTATGATAGGTAAAATAATAAATG | SacII |
| #59 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCTGGCTTTTGGAATTGCTGATGATGAG | |
| #35 | GTTTCACCGCGGATAATTGAAGTAAATGTACCGAG | SacII |
| #36 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATATCGATTTAATTCAAGAA | |
| Complementation | ||
| #73 | CGCGGATCCCTTGTGGTCAATTAAGAGCA | BamHI |
| #65 | CCGGAATTCTCATACTTTATCACCTTCAATAG | EcoRI |
| #66 | CCGGAATTCTTAAATATCATCATAGCTGACTTC | EcoRI |
| #87 | CTACGGCCATATGTTGTCTTTACCTCGTTTCTACTTGTCG | NdeI |
| #88 | CTACGGCCATATGATGATAGGTAAAATAATAAATGAACG | NdeI |
Restriction sites are in bold, and att recombination sites are in italics.
Construction of N315ΔSTP and N315ΔSTP/STK.
Both mutant strains N315ΔSTP and N315ΔSTP/STK were created by using plasmid pKOR1 (4). Primer sets #33/#34, #58/#59, and #35/#36 were used to amplify approximately 1 kb upstream of SA1062, 1 kb downstream of SA1062, and 1 kb downstream of SA1063, respectively. The resulting gene products were subjected to SacII (New England BioLabs) digestion and ligated (Epicentre Biotechnologies Fast-Link DNA ligation kit) such that two fragments containing ligated up- and downstream regions of SA1062 and SA1062-SA1063 were created. The ligation products were used as templates in two PCRs using primer sets #33/#59 and #33/#36. Respective PCR products were then inserted separately into temperature-sensitive plasmid pKOR1 by using BP Clonase (Invitrogen Gateway Clonase System). These plasmids were then transformed into E. coli DH5α, verified by DNA sequencing, passed through S. aureus strain RN4220, and ultimately electroporated separately into strain N315 for allelic replacement as described previously (4). Final selection of colonies was made on TSA containing anhydrotetracycline. A single chloramphenicol-sensitive transformant was identified for each strain, and the appropriate deletions were confirmed by PCR, gDNA sequencing, and Western blotting.
Complementation of mutants with SA1062 and/or SA1063.
To enable selection in erythromycin-resistant S. aureus strain N315, the erythromycin resistance cassette (ermC) from pCN40 was replaced with the tetracycline resistance cassette [tet(M)] from pCN36 (9) to create pCN40tet. The sequence and location of the endogenous promoter which facilitates stp and stk transcription in S. aureus are not known. pCN40 contains the endogenous S. aureus β-lactamase promoter module immediately upstream of its multiple cloning site (9). In addition to this, a 53-bp DNA fragment (nucleotides 1201589 to 1201642, located directly upstream of cotranscribing SA1062-SA1063 [stp-stk]) was included in frame and upstream of stp and/or stk to include the endogenous ribosomal binding site in all complementation constructs (20). To create the appropriate constructs, primer set #73/#65 was used to amplify SA1062, including the upstream region, to create pCN40tet-STP. In the same manner, primer set #73/#66 was used to create pCN40tetSTP/STK, again including the upstream region. Similarly, primer sets #73/#87 and #88/#66 were used to amplify SA1063 and its upstream region. These fragments were ligated together after digestion with NdeI and inserted into the complementation vector to create pCN40tet-STK. All complementation plasmids were passed through S. aureus strain RN4220 before being inserted into their respective mutants and verified by sequencing and Western blotting.
Western blotting.
Proteins in total cell lysates of N315, N315ΔSTP, N315ΔSTK, and N315ΔSTP/STK and their corresponding complemented strains were resolved by 12% SDS-PAGE under reducing conditions and transferred to polyvinylidene difluoride membranes (Bio-Rad). Antisera against rSTP and rSTK were custom made by Lampire Biologicals using a 50-day express-line rabbit protocol. These antisera were used to reveal the presence or absence of STK and STP in the appropriate lysates. As a control, N315 total lysates were also probed with prebleed serum obtained prior to immunization of the rabbits with rSTP or rSTK.
Microscopy.
Stationary-phase cultures of S. aureus N315 (control) and each mutant strain grown in TSB were fixed overnight at 4°C in 0.1 M cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde and 4% paraformaldehyde. Fixed cells were washed and embedded in 2% low-temperature-gelling agarose, cut into blocks, and postfixed in 1% osmium tetroxide. Blocks were then rinsed in water and stained in 1% uranyl acetate before being subjected to multiwash dehydration using increasing (50 to 100%) concentrations of ethanol. This was followed by propylene oxide, 1:1 propylene oxide-Eponate 12 resin, 1:2 propylene oxide-resin, and 100% resin washes. Blocks were then sectioned, stained in 2% uranyl acetate and Reynolds lead citrate, and viewed with an FEI Tecnai G2 Spirit transmission electron microscope (TEM). All procedures subsequent to fixing were performed at the Campus Microscopy and Imaging Facility at The Ohio State University.
Lysostaphin susceptibility assays.
Lysostaphin susceptibility assays were performed similarly to those described previously (16). Overnight cultures of S. aureus N315 (control), N315ΔSTP, N315ΔSTK, N315ΔSTP/STK, and the corresponding complemented strains were pelleted, washed once in lysostaphin buffer (20 mM Tris/HCl [pH 7], 150 mM NaCl, 1 mM EDTA), and resuspended in fresh buffer to the original culture volume. Lysostaphin (Sigma) was added to each culture at a final concentration of 0.5 μg/ml. The optical density at 600 nm of each culture was read (BMG POLARstar Galaxy) at the indicated time points and plotted as a percentage of the initial reading, which was set at 100% and 0 min. As a negative control and to measure autolysis, lysostaphin buffer alone (without lysostaphin) was used in identical assays. P values were determined by using Student's unpaired t test, and significance was defined as P < 0.05.
Antibiotic susceptibility assays.
The MICs, in micrograms per milliliter, of various antimicrobial agents for each S. aureus isolate were initially determined by using the MicroScan WalkAway system (Siemens Healthcare Diagnostics). MicroScan is an automated, commercially available system for rapid identification and susceptibility testing of gram-negative and -positive bacteria. Identification to the species level was performed by using the Gram-Positive ID/AST Combo PC29 panel. The MIC determination and quality control protocols used were in accordance with standards established by the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (10). MicroScan was performed at the Clinical Microbiology Facility at The Ohio State University Medical Center. Where indicated, MICs for the wild-type, mutant, and corresponding complemented mutant strains were determined by using Etest strips (AB Biodisk) on triplicate samples in accordance with the manufacturer's instructions.
RESULTS
Presence of cotranscribing ESTP and ESTK in the S. aureus genome.
Genome analysis (BlastP; NCBI) of methicillin-resistant and vancomycin-sensitive S. aureus strain N315 revealed the presence of SA1062 and SA1063, which encode a putative 744-bp, 247-amino-acid ESTP (possessing protein phosphatase 2C-specific motifs I to XI [6]) and a 1,995-bp, 664-amino-acid ESTK (possessing a complete set of STK-specific Hanks motifs I to XI [19]), respectively. These genes are predicted to be cotranscribed based on four overlapping nucleotides (ATGA) at the junction of their 3′ and 5′ ends (Fig. 1A). This overlapping pattern and the sequences of both genes are 100% conserved in all of the methicillin-sensitive and -resistant staphylococcal genomes we examined (Mu50, MW2, N315, RF122, COL, MRSA252, MSSA476, USA300, and NCTC8325) and are similar to those of many other gram-positive pathogens (see the supplemental material to reference 22). Further sequence analysis also supported the putative cotranscribing nature of these genes by revealing the presence a typical 13-nucleotide staphylococcal stem and 6-nucleotide loop transcriptional terminator (7, 13) downstream of SA1063 but not of SA1062.
To investigate the cotranscribing nature of these two genes, we performed RT-PCR assays using cDNA or gDNA templates and stp-stk overlapping-region (#108/#109)-specific and flanking-region (#44/#107 and #110/#46)-specific primer sets (annealing sites in Fig. 1A). Flanking region-specific primers only produced amplicons when gDNA (Fig. 1B, lanes 4 and 6), but not cDNA (Fig. 1B, lanes 1 and 3), was used as a template, while overlapping-region-specific primers produced amplicons when both cDNA and gDNA were used as a template (Fig. 1B, lanes 2 and 5). This, together with the absence of PCR products in reactions lacking RT and cDNA as a template (Fig. 1B, lanes 7 to 9) confirmed that SA1062 and SA1063 are encoded on a common RNA transcript.
STK and STP are functional proteins.
To determine biochemical and biological functions of S. aureus STP and STK, we cloned SA1062 and SA1063, produced purified recombinant proteins (rSTP and rSTK), and examined their roles in in vitro phosphorylation reactions.
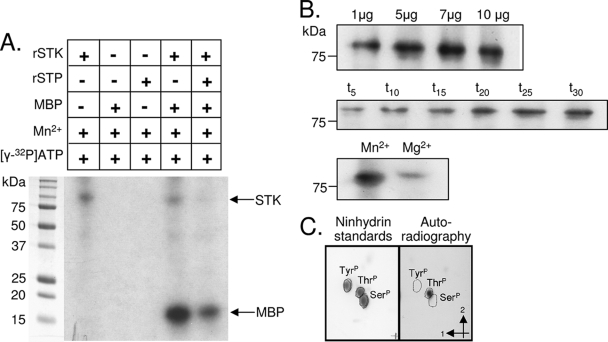
rSTK autophosphorylated and catalyzed phosphotransfer to a nonspecific model substrate, MBP (Fig. 2A). Addition of rSTP to the kinase reaction mixture resulted in dephosphorylation of both autophosphorylated STK and STK-phosphorylated MBP, indicating that both rSTK and rSTP are functionally active. STK autophosphorylation occurred in time- and dose-dependent manners (Fig. 2B) and occurred most efficiently when Mg2+ was replaced with Mn2+ as the sole cation, suggesting that S. aureus STK-mediated phosphorylation favors manganese as a cofactor. To further determine phosphorylation specificity, we performed two-dimensional thin-layer chromatography with acid hydrolysates of autophosphorylated rSTK and phosphorylated MBP. Alignment of radioactive phosphorylated amino acid spots that migrated from the hydrolysates of autophosphorylated STK (Fig. 2C) and phosphorylated MBP (data not shown) with the ninhydrin-stained phosphothreonine standard (Fig. 2C) revealed that S. aureus STK specifically targets threonine residues in its substrate(s).
FIG. 2.
Autoradiographs of in vitro kinase reactions. (A) Purified rSTK was incubated in kinase buffer with [γ-32P]ATP and Mn2+ alone and in the presence of MBP and rSTP as indicated. Proteins were resolved by SDS-PAGE and visualized by autoradiography. Shown are representative autoradiographs. (B) Increasing amounts of purified rSTK were added to separate kinase reaction mixtures to observe the dose-dependent effect. One microgram of rSTK was used for the time course and cation dependency experiments, where minutes and cations are shown above the autoradiographs. (C) Thin-layer chromatography results showing phosphoamino acid standards stained with ninhydrin (left) and autophosphorylated rSTK hydrolysis products revealed by autoradiography (right). The first and second dimensions are indicated by arrows.
Creation and confirmation of N315ΔSTP, N315ΔSTK, and N315ΔSTP/STK.
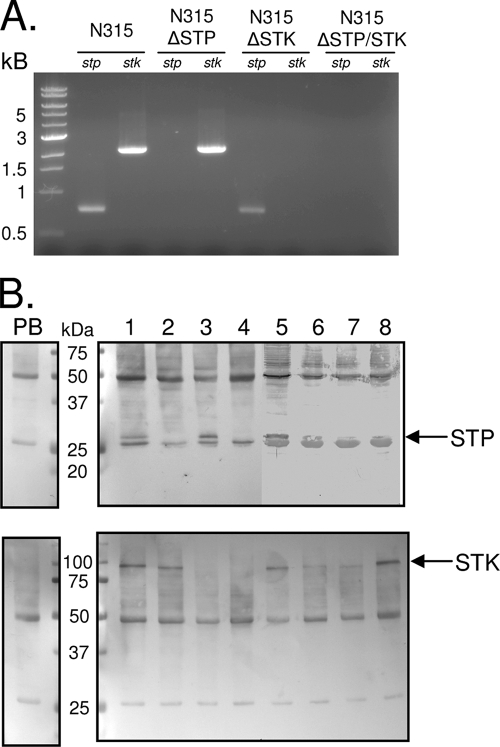
To study the contributions of STK-mediated kinase and STP-mediated phosphatase activities in staphylococcal biology, we created three mutant strains lacking stp (N315ΔSTP), stk (N315ΔSTK), or both stp and stk (N315ΔSTP/STK). We also validated their functional properties by using corresponding complemented strains (N315pCN40tet, N315ΔSTPΩSTP, N315ΔSTKΩSTK, and N315ΔSTP/STKΩSTP/STK). As shown in Fig. 3A, PCR analysis using primers specific for stp (STP-F/STP-R) and stk (STK-F/STK-R) confirmed the absence of the appropriate genes in each deletion strain. Additionally, Western blot analysis of total bacterial lysates from each strain probed with anti-rSTP (Fig. 3B, top) or anti-rSTK serum (Fig. 3B, bottom) revealed distinct antibody-reacting bands corresponding to STP (∼32 kDa) and STK (∼77 kDa) in the wild-type strain, all complemented strains, and the appropriate mutants. To express the missing gene products in the mutant strains, pCN40 vectors containing an S. aureus β-lactamase promoter (PblaZ) and the appropriate wild-type gene were used. Thus, the relatively lower expression level of STP in the corresponding complemented strain compared to the wild type is likely due to the use of PblaZ instead of its native promoter, which has not been identified. As described below, the amount of STK and/or STP expressed in the complemented strains seemed to be sufficient to regain the functionality of the wild-type strain. Importantly, the presence of an STP-reacting band in N315ΔSTK (Fig. 3B, lane 3) and an STK-reacting band in N315ΔSTP (Fig. 3B, lane 2) indicated that the deletion strategies used left the stp transcript stable in the absence of stk and had no polar effects on the stk transcript in the absence of stp.
FIG. 3.
Confirmation of mutant strains. (A) PCR analysis of gDNA isolated from the indicated strains with primers specific for stp and stk. (B) Western blot assays of total cell lysates probed separately with (top) anti-rSTP and (bottom) anti-rSTK production sera (right) and corresponding prebleed (PB, left) sera. Lanes: 1, N315; 2, N315ΔSTP; 3, N315ΔSTK; 4, N315ΔSTP/STK; 5, N315pCN40tet; 6, N315ΔSTPΩSTP; 7, N315ΔSTKΩSTK; 8, N315ΔSTP/STKΩSTP/STK. Arrows indicate the locations of STK and STP within the lysates.
To determine the function of STK and STP in S. aureus growth, the isogenic mutant strains were grown on TSA blood agar plates and their colony morphology and hemolytic patterns were compared to those of the parent N315 strain. We did not observe any obvious differences among the four strains in these assays. Additionally, the growth curves of all mutants and their complemented strains were similar to that of the wild-type strain (N315) in TSB (data not shown), prompting us to investigate other effects of the deletions of stp and/or stk in S. aureus.
Role of STP/STK in S. aureus cell wall morphology.
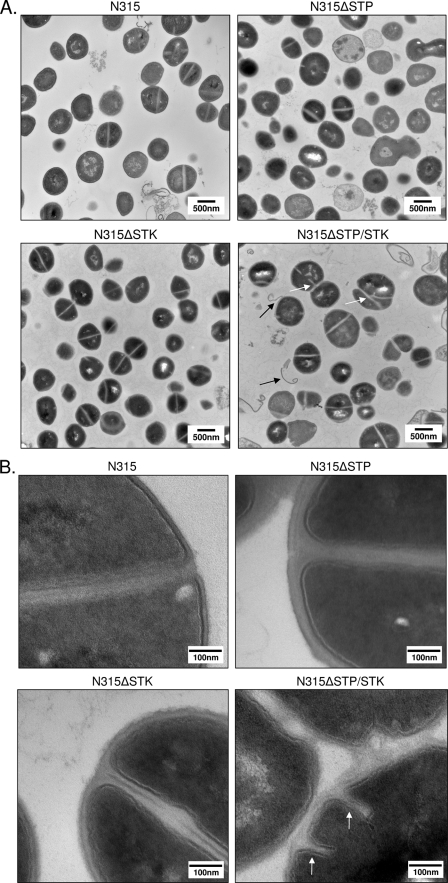
To more closely examine any morphological changes between mutants, we subjected stationary-phase cultures of the three deletion strains (N315ΔSTP, N315ΔSTK, and N315ΔSTP/STK) and the wild-type strain (N315) to TEM. Results revealed clear defects in cell division and cell septum formation when both the stp and stk genes were deleted (Fig. 4A and B, white arrows). N315ΔSTP/STK cells also displayed a noticeable increase in overall cell size (∼25% compared to N315). Although these changes were not apparent to a significant extent in N315ΔSTP or N315ΔSTK, the cell wall thickness of N315ΔSTP mutants was nearly double that of the other three strains (Fig. 4B). Further examination of cell wall and membrane regions of N315ΔSTK and N315ΔSTP/STK cells at higher resolution (Fig. 4B) revealed characteristically weak, electron-dense, wavy, and often interrupted membranes with a fragile appearance, as opposed to the thickly stained, distinctly visible, and uninterrupted cell walls seen in N315 and N315ΔSTP cells. Additional notable observations in the N315ΔSTP/STK TEM images included the apparent “peeling” of cell wall or membrane-like material away from the bacterium (Fig. 4A, black arrows). Notably, deletion of both stk and stp resulted in a more conspicuous phenotype than deletion of stk or stp alone, suggesting coordinated roles for STK and STP in the determination of cell wall structure.
FIG. 4.
Analysis of cell morphology by TEM. Shown are representative fields of TSB-grown stationary-phase wild-type (control) and mutant S. aureus strains, as indicated. White arrows indicate abnormal septation; black arrows indicate evidence of apparent wall or membrane shedding. Magnifications: A, ×30,500; B, ×120,000.
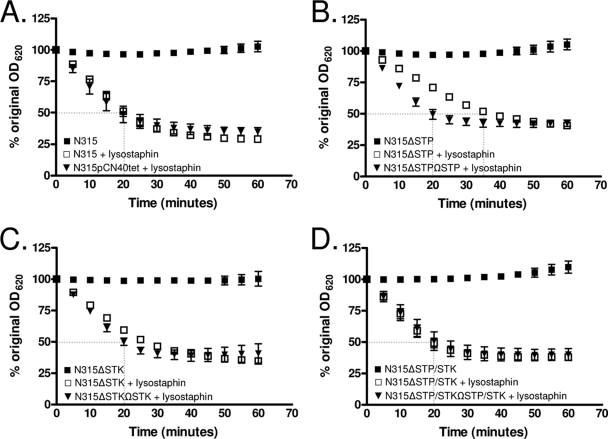
N315ΔSTP mutants display alterations in lysostaphin susceptibility.
Based on the results obtained by TEM, we suspected that deletion of stp and/or stk may directly affect cell wall structure. We therefore determined the susceptibility of each mutant and its corresponding complemented strain to the peptidoglycan-targeting glycylglycine endopeptidase lysostaphin by measuring the decline in cell density (16). Upon the addition of lysostaphin, the wild-type (N315) and double-mutant (N315ΔSTP/STK) strains and all complemented strains (triangles) reached 50% lysis by approximately 20 min (Fig. 5), while N315ΔSTP took a significantly longer incubation time (35 min, P < 0.002 compared to the wild type; Fig. 5B). Notably, N315ΔSTK reached 50% lysis by approximately 25 min (Fig. 5C), lysing more slowly than N315 but much less significantly than N315ΔSTP (P < 0.05 at 20 and 25 min compared to the wild-type strain). The slopes and standard deviations of the N315, N315ΔSTP, N315ΔSTK, and N315ΔSTP/STK lysostaphin digestion curves determined by a linear regression analysis of the data points between 5 and 25 min (the time period in which most of the lysis occurred) were −2.35 ± 0.11, −1.48 ± 0.031, −1.90 ± 0.06, and −2.17 ± 0.15, respectively. Control reactions carried out in the absence of lysostaphin showed no significant changes in kinetics, indicating that the effects observed at the given incubation times were due to lysostaphin and not autolysis. Based on these results, we concluded that N315ΔSTP bacteria are more resistant to the effects of lysostaphin than is the parent strain N315 as a direct result of the deletion of stp.
FIG. 5.
Lysostaphin sensitivity assays. Results indicate the susceptibilities of (A) N315; (B) N315ΔSTP, (C) N315ΔSTK, and (D) N315ΔSTP/STK; and their appropriate complemented strains (triangles) to lysostaphin. Autolysis (no lysostaphin) control results are shown as closed squares. Data shown are the averages of three independent experiments. Error bars indicate standard deviations. Data are presented as percentages of the original culture turbidity. OD620, optical density at 620 nm.
N315ΔSTK and N315ΔSTP/STK are more susceptible to cell wall-acting antibiotics.
Conspicuous changes in cell division and wall morphology (Fig. 4) and altered lysostaphin susceptibilities (Fig. 5) in STP- and/or STK-specific mutants suggested that S. aureus N315 STP and STK may play an important role in cell division and/or peptidoglycan structure. Changes in the cell wall of S. aureus have been shown to affect staphylococcal susceptibility to certain antibiotics, particularly those which target cell wall structure or synthesis (1, 31). Hence, we determined the susceptibilities of N315 and its isogenic mutant strains to 22 antibiotics with various known targets. N315ΔSTP did not show any change in susceptibility compared to the wild-type strain (Table 3). However, the N315ΔSTK and, to a greater degree, N315ΔSTP/STK mutant strains displayed increased susceptibility to multiple antibiotics tested (Table 3, bold values). Most appreciably, N315ΔSTP/STK bacteria displayed susceptibility increases ranging from 2-fold in the case of cefazolin to more than 50-fold in the case of ertapenem, compared to N315. The affected antibiotics for both N315ΔSTK and N315ΔSTP/STK included cephalosporins (cefazolin, ceftriaxone, cefotaxime) and carbapenems (ertapenem, imipenem, meropenem), all of which act by interfering with bacterial cell wall synthesis (12, 36, 37). To verify these effects, all of the strains were subjected to antibiotic sensitivity testing with Etest strips containing gradients of representative antibiotics from each of these groups, specifically, those in which the MICs changed for both N315ΔSTK and N315ΔSTP/STK (cefotaxime, ceftriaxone, and ertapenem). The MICs for the wild-type and deletion strains were identical to those obtained by MicroScan (data not shown), while those for N315ΔSTKΩSTK and N315ΔSTP/STKΩSTP-STK returned to those for the wild-type strain (Table 3), validating the MicroScan data. The increased selective susceptibility of N315ΔSTK and N315ΔSTP/STK to cell wall-acting antibiotics confirmed a role for STK in staphylococcal cell wall biosynthesis and structure.
TABLE 3.
MICs of antibiotics tested
| Antibiotic(s) | MIC(s) (μg/ml) for strain:
|
|||||
|---|---|---|---|---|---|---|
| N315 | N315ΔSTP | N315ΔSTK | N315ΔSTKΩSTK | N315ΔSTP/STK | N315ΔSTP/ STKΩSTP/STK | |
| Most affected | ||||||
| Cefazolin | >16 | >16 | >16 | ≤8 | ||
| Cefotaxime | >256 | >256 | 43 | >256a | 12 | >256 |
| Ceftriaxone | >32 | 32 | ≤8 | >32 | ≤8 | >32 |
| Ertapenem | >32 | >32 | 18 | >32 | 0.6 | >32 |
| Imipenem | >8 | 8 | 8 | ≤4 | ||
| Unaffected | ||||||
| Amoxicillin-clavulanate | >4/2 | >4/2 | >4/2 | >4/2 | ||
| Ampicillin-sulbactam | ≤8/4 | ≤8/4 | ≤8/4 | ≤8/4 | ||
| Ampicillin | >8 | >8 | >8 | >8 | ||
| Clindamycin | >4 | >4 | >4 | >4 | ||
| Erythromycin | >4 | >4 | >4 | >4 | ||
| Gentamicin | ≤4 | ≤4 | ≤4 | ≤4 | ||
| Levofloxacin | ≤2 | ≤2 | ≤2 | ≤2 | ||
| Linezolid | 2 | 2 | 2 | 2 | ||
| Moxifloxacin | ≤2 | ≤2 | ≤2 | ≤2 | ||
| Nitrofurantoin | ≤32 | ≤32 | ≤32 | ≤32 | ||
| Oxacillin | >2 | >2 | >2 | >2 | ||
| Penicillin | >8 | >8 | >8 | >8 | ||
| Rifampin | ≤1 | ≤1 | ≤1 | ≤1 | ||
| Synercid | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ||
| Tetracycline | ≤4 | ≤4 | ≤4 | ≤4 | ||
| Trimethoprim-sulfamethoxazole | ≤0.5/9.5 | ≤0.5/9.5 | ≤0.5/9.5 | ≤0.5/9.5 | ||
The complementation MICs shown are averages of three independent experiments with Etest strips. The antibiotic gradients on the Etest strips were 0.016 to 256 μg/ml (cefotaxime and ceftriaxone) and 0.002 to 32 μg/ml (ertapenem).
DISCUSSION
In this study, we report for the first time functional roles for an ESTK and an ESTP in S. aureus. ESTKs and ESTPs are characterized in a variety of bacteria with different genetic backgrounds and pathogenic potentials; thus, deletion of one or more of these enzymes results in different phenotypes based on the bacteria being studied. For example, deletion of ESTKs results in extremely long chain formation in Streptococcus agalactiae (39), aggregation in group A Streptococcus (22), and no significant changes in Enterococcus faecalis grown in nutritionally enriched medium (25). In the present investigation, deletion of both stk and stp in S. aureus resulted in dysregulation of cell wall components and division machinery, as revealed by TEM and susceptibility testing to cell wall-acting antibiotics.
Like many other gram-positive ESTKs (22, 40), the extracellular domain of S. aureus STK contains a domain containing three PASTA (penicillin-binding protein- and serine/threonine kinase-associated) repeats. PASTA domains contain one to five PASTA repeats (25, 48), bind peptidoglycan (40), and are believed to recruit penicillin binding proteins (PBPs) to sites of cell growth (48). Thus, it is possible that S. aureus STK also uses PASTA repeats as an external sensor to recognize the level of unlinked peptidoglycan and mediate appropriate signaling cascades intracellularly via its cytoplasmic N-terminal kinase domain. Future experimental verification is required to determine whether this signaling cascade indeed facilitates the transcription of one or more cell wall synthesis genes or involves direct phosphorylation of PBPs, as has been shown in mycobacteria (11).
Although dephosphorylation is an essential step in nearly all signaling networks, information regarding the regulatory role(s) of ESTPs is limited. This may be due in part to their requirement for bacterial survival, as ESTP-specific mutations have proven lethal in S. pyogenes (22). Mutants and variants of S. agalactiae (39) and S. pneumoniae (35) have been reported, although detailed characteristics of these mutants are not available. In the present study, we were able to create both stp and stp/stk deletion mutants, indicating that in S. aureus N315, STP is not essential for survival. We speculatively attribute this to the unique presence of other putative and nonconserved ESTPs in the N315 genome (SA0140, SA1662, and SA2225). Notably, out of eight other S. aureus genomes we examined, homologs of SA2225, but not SA0140 and SA1662, were found in only five (Mu50, MW2, RF122, MRSA252, and MSSA476, missing from USA300, NCTC8325, and COL) (http://genomics.ornl.gov/mist; 44). The presence of an additional noncotranscribing putative STK (SA0077) (44) may play a role in compensating for the loss of stk in the appropriate deletion strains.
Staphylococcal cell wall peptidoglycan is composed of strands of alternating N-acetylglucosamine and N-acetylmuramic acid sugars joined together by amino acid side peptides which are cross-linked by pentaglycine chains (46). These pentaglycine chains serve as a substrate for lysostaphin-mediated cleavage. Changes in this structure directly affect lysostaphin binding and cleavage activities in S. aureus (17). The most notable effects of the stp deletion alone were thicker cells walls compared to those of wild-type S. aureus (Fig. 4B) and the perhaps consequential increased resistance to lysostaphin (Fig. 5B). This suggests an alteration in peptidoglycan composition and/or structure, specifically, pentapeptide bridge composition and/or cross-linking frequency. Deletion of stk had a minor effect on lysostaphin susceptibility, while deletion of both stp and stk did not affect lysostaphin susceptibility (Fig. 5B). This could be due to compensatory mechanisms (such as the aforementioned noncotranscribing STK and STPs) being activated in the absence of this primary STP-STK pair, as it is not uncommon for STKs to share substrates (23). Additionally, the absence of the extracellular, environment-sensing PASTA domain upon deletion of stk or stp/stk may result in mutants with defects in the ability to sense unlinked peptidoglycan (40) and regulate pentaglycine chain formation by way of PBP recruitment (48). The synchronized role of STK and STP may therefore be maintaining equilibrium in cross-linking. Increased resistance to lysostaphin digestion in the absence of stp could thus be due to an absence of substrate or to uncontrolled recruitment of PBPs and resultant increases in cross-linking. While our ongoing experiments analyzing cell wall structures in these mutants may reveal alterations, increased lysostaphin resistance in the absence of stp is consistent with the increased cell wall thickness observed (Fig. 4B).
Deletion of stk alone resulted in increased susceptibility to cell wall-acting β-lactam antibiotics (Table 3), as has been reported for E. faecalis ESTK PrkC deletion strains (25), and increased cell wall fragility (Fig. 4). Importantly, deletion of both stp and stk significantly enhanced these phenotypes, resulting in further selective antibiotic susceptibility (Table 3) and pronounced defects in cell division (Fig. 4). These additive effects, combined with the ability of rSTP to reverse STK-mediated phosphorylation (Fig. 2A) and the cotranscribing nature of stp and stk (Fig. 1B), suggest that STK and STP are indeed cognate proteins. Thus, we believe that although STP and STK, on their own, may contribute to the regulation of certain cell wall synthesis steps, it is probably their coordinated reversible phosphorylation which is most critical.
It is important to note that only MICs of β-lactam antibiotics which specifically target cell wall synthesis were affected by the deletion of stk or stk and stp. PBPs, which have transpeptidase, transglycosylase, and/or carboxypeptidase activities to facilitate peptidoglycan synthesis (29), are often the primary target of β-lactam antibiotics. Meropenems, and specifically ertapenem, bind a variety of PBPs to inhibit cell wall synthesis, particularly PBP2 and -3 in E. coli (36). Cephalosporins also directly interact with PBPs. Ceftriaxone has been shown to bind tightly to S. aureus PBP1 (12), and a more potent cefotaxime binds S. aureus PBP1 and -2 with a higher affinity than other PBPs (15, 37). Based on the ceftriaxone- and cefotaxime-sensitive phenotypes observed in N315ΔSTK and N315ΔSTP/STK in this study, it is reasonable to believe that STK and coordinated STP/STK signaling may, in some way, modulate the functionality of PBP1 and/or -2. PBPs have also been implicated in a variety of functions, including cell division and septum formation, by directly interacting with cell division proteins, including FtsZ (29, 38, 45), and modulating peptidoglycan synthesis in S. aureus specifically (27). Thus, the alterations in cell wall structure and division and increased antibiotic susceptibility of S. aureus in the absence of STP and/or STK in the present study are likely attributable to the altered recruitment and regulation of S. aureus PBPs and other necessary cell wall synthesis and division proteins.
Taken together, the results shown in the present study clearly indicate that S. aureus STP and STK and their coordinated phosphorylation play an important role in cell wall structure, cell division, and β-lactam antibiotic susceptibility. In the context of the dramatic rise in antimicrobial resistance, as well as the continued prevalence of staphylococci in hospital environments, the present study provides an important insight to consider, as these enzymes have the potential to serve as novel targets for alternative antibiotic therapy.
Acknowledgments
We thank Olaf Schneewind and Taeok Bae for pKOR1, Emmanuelle Charpentier for pCN36 and pCN40, Hong Jin for thin-layer chromatography, and Preeti Pancholi for antibiotic susceptibility testing by MicroScan.
A.B. is a recipient of two subsequent training grant awards from the National Institutes of Health (T32GM068412, a Systems in Integrative Biology fellowship administered through the Integrative Biomedical Graduate Program, and T32AI065411, an NRSA training grant administered by the Center for Microbial Interface Biology). This work was also supported by NIH grant AI064912 and an OSUMC Strategic Initiative grant (V.P.).
Editor: A. Camilli
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Antignac, A., K. Sieradzki, and A. Tomasz. 2007. Perturbation of cell wall synthesis suppresses autolysis in Staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. J. Bacteriol. 1897573-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8238-244. [DOI] [PubMed] [Google Scholar]
- 4.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 5558-63. [DOI] [PubMed] [Google Scholar]
- 5.Bakal, C. J., and J. E. Davies. 2000. No longer an exclusive club: eukaryotic signaling domains in bacteria. Trends Cell Biol. 1032-37. [DOI] [PubMed] [Google Scholar]
- 6.Bork, P., N. P. Brown, H. Hegyi, and J. Schultz. 1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 51421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky, L. I., V. V. Ivanov, Y. L. Kalaydzidis, A. M. Leontovich, V. K. Nikolaev, S. I. Feranchuk, and V. A. Drachev. 1995. GeneBee-NET: internet-based server for analyzing biopolymers structure. Biochemistry 60923-928. [PubMed] [Google Scholar]
- 8.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 21991-99. [DOI] [PubMed] [Google Scholar]
- 9.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 706076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Dasgupta, A., P. Datta, M. Kundu, and J. Basu. 2006. The serine threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152493-504. [DOI] [PubMed] [Google Scholar]
- 12.Davies, T. A., M. G. P. Page, W. Shang, T. Andrew, M. Kania, and K. Bush. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 512621-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Hoon, M. J. L., Y. Makita, K. Nakai, and M. Miyano. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M. C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 722434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopapadakou, N. H., B. A. Dix, and Y. R. Mauriz. 1986. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob. Agents Chemother. 29333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gründling, A., D. M. Missiakas, and O. Schneewind. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 1886286-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 1882463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 19.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 24142-52. [DOI] [PubMed] [Google Scholar]
- 20.Haugen, S. P., W. Ross, and R. L. Gourse. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 1881628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 3571351-1372. [DOI] [PubMed] [Google Scholar]
- 23.Kang, C. M., D. A. Abbott, S. T. Park, C. C. Dascher, L. C. Cantley, and R. N. Husson. 2005. The Mycobacterium tuberculosis serine/threonine kinase PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 191692-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 25.Kristich, C. J., C. L. Wells, and G. M. Dunny. 2007. A eukaryotic-type ser/thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. USA 1043508-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 27.Łeski, T. A., and A. Tomasz. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 1871815-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 29.Macheboeuf, P., C. Contreras-Martel, V. Job, D. Otto, and A. Dessen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30673-691. [DOI] [PubMed] [Google Scholar]
- 30.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46571-586. [DOI] [PubMed] [Google Scholar]
- 31.Mainardi, J. L., R. Villet, T. D. Bugg, C. Mayer, and M. Arthur. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in gram-positive bacteria. FEMS Microbiol. Rev. 32386-408. [DOI] [PubMed] [Google Scholar]
- 32.Motley, S. T., and S. Lory. 1999. Functional characterization of a serine/threonine protein kinase of Pseudomonas aeruginosa. Infect. Immun. 675386-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay, S., V. Kapatral, W. Xu, and A. M. Chakrabarty. 1999. Characterization of a Hank's type serine/threonine kinase and serine/threonine phosphoprotein phosphatase in Pseudomonas aeruginosa. J. Bacteriol. 1816615-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz-Dorado, J., S. Inouye, and M. Inouye. 1991. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67995-1006. [DOI] [PubMed] [Google Scholar]
- 35.Nováková, L., L. Sasková, P. Pallová, J. Janeèek, J. Novotná, A. Ulrych, J. Echenique, M.-C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS Lett. 2721243-1254. [DOI] [PubMed] [Google Scholar]
- 36.Odenholt, I. 2001. Ertapenem: a new carbapenem. Expert Opin. Investig. Drugs 101157-1166. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, S. M., S. L. Kang, D. M. Cappelletty, and M. J. Rybak. 1995. Bactericidal killing activities of cefepime, ceftazidime, cefotaxime, and ceftriaxone against Staphylococcus aureus and β-lactamase-producing strains of Enterobacter aerogenes and Klebsiella pneumoniae in an in vitro infection model. Antimicrob. Agents Chemother. 391764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira, S. F. F., A. O. Henriques, M. G. Pinho, H. de Lencastre, and A. Tomasz. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 1893525-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 27814429-14441. [DOI] [PubMed] [Google Scholar]
- 40.Shah, I. M., M.-H. Laaberki, D. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel, J. D., E. Rhinehart, M. Jackson, and L. Chiarello. 2006. Management of multidrug-resistant organisms in healthcare settings. Centers for Disease Control and Prevention, Atlanta, GA.
- 42.Thakur, M., and P. K. Chakraborti. 2006. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic type ser/thr kinase, PknA. J. Biol. Chem. 28140107-40113. [DOI] [PubMed] [Google Scholar]
- 43.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 1352-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulrich, L. E., and I. B. Zhulin. 2007. MiST: a microbial signal transduction database. Nucleic Acids Res. 35D386-D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varma, A., and K. D. Young. 2004. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 1866768-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32149-167. [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., C. Li, H. Yang, A. Mushegian, and S. Jin. 1998. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J. Bacteriol. 1806764-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a β-lactam-binding protein. Trends Biochem. Sci. 27438-440. [DOI] [PubMed] [Google Scholar]