Abstract
Host-pathogen interactions are of great importance in understanding the pathogenesis of infectious microorganisms. We developed in vitro models to study the host-pathogen interactions of porcine respiratory tract pathogens using two immortalized epithelial cell lines, namely, the newborn pig trachea (NPTr) and St. Jude porcine lung (SJPL) cell lines. We first studied the interactions of Actinobacillus pleuropneumoniae, an important swine pathogen, using these models. Under conditions where cytotoxicity was absent or low, we showed that A. pleuropneumoniae adheres to both cell lines, stimulating the induction of NF-κB. The NPTr cells consequently secrete interleukin 8, while the SJPL cells do not, since they are deprived of the NF-κB p65 subunit. Cell death ultimately occurs by necrosis, not apoptosis. The transcriptomic profile of A. pleuropneumoniae was determined after contact with the porcine lung epithelial cells by using DNA microarrays. Genes such as tadB and rcpA, members of a putative adhesin locus, and a gene whose product has high homology to the Hsf autotransporter adhesin of Haemophilus influenzae were upregulated, as were the genes pgaBC, involved in biofilm biosynthesis, while capsular polysaccharide-associated genes were downregulated. The in vitro models also proved to be efficient with other swine pathogens, such as Actinobacillus suis, Haemophilus parasuis, and Pasteurella multocida. Our results demonstrate that interactions of A. pleuropneumoniae with host epithelial cells seem to involve complex cross talk which results in regulation of various bacterial genes, including some coding for putative adhesins. Furthermore, our data demonstrate the potential of these in vitro models in studying the host-pathogen interactions of other porcine respiratory tract pathogens.
Porcine respiratory diseases have heavily impacted the economy of the pig rearing industry worldwide. Actinobacillus pleuropneumoniae, an exemplar of these porcine respiratory pathogens, causes porcine pleuropneumonia, a very contagious and often fatal disease characterized by necrotic and hemorrhagic lung lesions, coughing, and severe respiratory distress. Fifteen serotypes of this gram-negative facultative anaerobic bacteria are presently known based on surface polysaccharides (56). The virulence of this pathogen is accomplished with the help of many factors, including exotoxins, endotoxin, capsule polysaccharides, adhesins, and outer membrane proteins, such as iron acquisition systems.
The four pore-forming exotoxins of A. pleuropneumoniae, called Apx, are cytolytic and/or hemolytic (20, 51). In fact, the virulence of the different serotypes coincides greatly with the presence of the Apx toxins, particularly ApxI and ApxII. Serotypes 1, 5, 9, and 11 are known to be especially virulent, and all express both ApxI and ApxII (19).
As demonstrated by Jacques et al., lipopolysaccharides (LPS) are the major molecules responsible for adhesion, principally the core oligosaccharide region (45, 48). However, other putative adhesins have also been described, such as type IV fimbriae expressed under specific conditions in different serotypes (63), a 60-kDa collagen-binding protein (14), a 55-kDa outer membrane protein (61), and an autotransporter protein (4).
A close relative of A. pleuropneumoniae, Aggregatibacter (Actinobacillus) actinomycetemcomitans, was found to be invasive of the human KB cell line and primary gingival cells (17). The invasiveness of this strain has been shown to be related to the colonial morphology, since a switch from a rough to a smooth morphology leads to the loss of its invasive capacity. Haemophilus parasuis has also been shown to be invasive of porcine brain microvascular endothelial cells (43, 60). Moreover, A. actinomycetemcomitans has been demonstrated to induce cell death by apoptosis of numerous cell types, while the cytolethal distending toxin of Haemophilus ducreyi has been shown to induce apoptosis of Jurkat T cells (23, 37). Adherence, invasion, toxin secretion, and other mechanisms involved in the pathogenesis of Pasteurellaceae lead to changes in cellular processes, including the induction of nuclear factors and cytokine production. In fact, A. pleuropneumoniae stimulates the production of proinflammatory cytokines such as interleukin 1β (IL-1β), IL-8, and tumor necrosis factor alpha (TNF-α), which are detected in alveolar fluid and tissue lesions (56). Likewise, a study by our group demonstrated that the production of IL-6, TNF-α, IL-1β, MCP-1, and IL-8 by porcine alveolar macrophages is induced by purified serotype 1 A. pleuropneumoniae LPS and by heat-killed bacteria (48). IL-8, a neutrophil chemoattractant, is of particular interest, since neutrophil accumulation at the infection site is characteristic of porcine pleuropneumonia (56).
Changes in bacterial gene expression also occur during infection. Studies have been conducted to investigate the gene expression of A. pleuropneumoniae under conditions mimicking that of the host. A study by our group used microarray technology to detect changes in gene expression of A. pleuropneumoniae serotype 1 grown under iron-restricted conditions (13). In this study, many genes involved in iron acquisition were shown to be upregulated, while genes involved mainly in energy metabolism were downregulated. In vivo studies based on selective capture of transcribed sequences, in vivo expression technology, or signature-tagged mutagenesis (3, 4, 22, 31, 53) have allowed the detection of adhesin and toxin genes and of genes involved in metabolism, stress, regulation, and transport.
Epithelial cells play an important role as the interface between the host mucosal surfaces and the surrounding environment and are the initial site of colonization for most bacterial pathogens. Two porcine respiratory tract epithelial cell lines have been established and described in the literature, namely, the newborn pig trachea (NPTr) (15) and St. Jude porcine lung (SJPL) (52) cell lines. The NPTr cell line was established from a 2-day-old piglet from a pathogen-free herd, while the SJPL cell line was spontaneously established from the lung of a normal 4-week-old female Yorkshire pig (15, 52).
The use of these cell lines has the possibility of generating a great amount of information on the infection mechanism of A. pleuropneumoniae, as well as that of other swine bacterial or viral respiratory tract pathogens. Consequently, we developed in vitro models using these cell lines and investigated host-pathogen interactions, including adherence, invasion, and bacterial transcriptomic profile, as well as cell death, nuclear factor expression, and cytokine production by the epithelial cells. This is the first report of models in which immortalized cell lines are used to study interactions of A. pleuropneumoniae with respiratory tract epithelial cells of porcine origin.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All strains used in this study are listed in Table 1. All A. pleuropneumoniae strains and the Pasteurella multocida capsular type A and D strains were grown in brain heart infusion (BHI) broth and/or agar (Gibco, Burlington, VT) supplemented with 15 μg/ml NAD at 37°C in 5% CO2. The Actinobacillus suis strain was grown under the same conditions, with the addition of 25 μg/ml nalidixic acid and 5 μg/ml chloramphenicol. Both H. parasuis strains were grown on pleuropneumonia-like organism medium broth and on chocolate agar at 37°C without CO2.
TABLE 1.
Bacterial strains used in the present study
| Strain | Serotype | Source or reference |
|---|---|---|
| A. pleuropneumoniae S4074 | 1 | K. R. Mittala |
| A. pleuropneumoniae L20 | 5b | K. R. Mittal |
| A. pleuropneumoniae WF83 | 7 | K. R. Mittal |
| A. pleuropneumoniae FMV91-6514 | 1 (rough field strain) | 32 |
| A. pleuropneumoniae 05-4817 | 5a (field strain) | K. R. Mittal |
| A. pleuropneumoniae 05-6501 | 5b (field strain) | K. R. Mittal |
| A. pleuropneumoniae 05-3695 | 7 (field strain) | K. R. Mittal |
| H. parasuis Nagasaki | 5 | M. Gottschalka |
| H. parasuis 29755 | 5 | E. Thackerb |
| A. suis H91-0380 | O2/K2 | J. MacInnesc |
| P. multocida 88-761 | A | K. R. Mittal |
| P. multocida 1703 | D | K. R. Mittal |
Faculté de Médecine Vétérinaire, Université de Montréal, St.-Hyacinthe, Canada.
Faculty of Veterinary Medicine, Iowa State University.
Department of Pathobiology, Ontario Veterinary College, University of Guelph, Guelph, Canada.
Cell culture.
The NPTr cell line (Instituto Zooprofilattico Sperimental, Brescia, Italy) (15) was grown at 37°C in 5% CO2 in minimum essential medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% sodium pyruvate (Gibco). The SJPL cell line (St. Jude Children's Hospital, Memphis, TN) (52) was grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Gibco), supplemented with 10% fetal bovine serum, 1% sodium pyruvate, and 1.5% minimal essential medium nonessential amino acids solution (Gibco). Both cell lines were tested by PCR using porcine-specific primers, and amplicons were sequenced to ensure their origin (41).
Cytotoxicity detection assay.
The cellular cytotoxicity was measured in the different assays using the lactate dehydrogenase (LDH)-measuring CytoTox 96 nonradioactive cytotoxicity assay (Promega, Madison, WI) as prescribed by the manufacturer. Noninfected cells were used as a negative control, while total lysis of cells by a treatment with 2% Triton X-100 represented the 100%-cytotoxicity positive control. Optical densities at 490 nm were measured with a Power Wave X340 (Biotek Instruments Inc, Winooski, VT) microplate reader and used to calculate the percentage of cytotoxicity.
Apoptosis detection assays.
Apoptosis assays were performed using the cell death detection enzyme-linked immunosorbent assay (ELISA) (Roche, Laval, Québec, Canada) and the caspase-3 Western detection kit (Cell Signaling Technology Inc., Beverly, MA) as prescribed by the manufacturers. A bacterial suspension at an optical density at 600 nm (OD600) of 0.6 was added to a confluent monolayer of cells at a multiplicity of infection (MOI) of 10:1 and incubated for 3 h at 37°C in 5% CO2. Uninfected cells were used as negative controls, and cells treated with 20 μg/ml camptothecin (Sigma) for 4 h at 37°C in 5% CO2 were used as positive controls. Following the infection, the supernatant and adherent cells were recovered. Plates for the cell death detection ELISA were read at 405 nm in a Power Wave X340 (Biotek Instruments Inc.) microplate reader. For the caspase-3 Western blots, the samples were loaded on a 12% (wt/vol) polyacrylamide gel, migrated at 100 V, and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The membrane was blocked for 1 h at room temperature in 2% skim milk and then incubated overnight (O/N) at 4°C with polyclonal rabbit antibodies against cleaved caspase-3 and caspase-3. Membranes were washed three times in Tris-buffered saline and incubated with mouse anti-immunoglobulin G antibodies conjugated with horseradish peroxidase for 1 h at ambient temperature and revealed with 3,3′,5,5′-tetramethylbenzidine (Sigma).
Microscopy.
Cells were seeded to semiconfluence in four-well LabTekII chamber microscopy slides (Nunc, Naperville, IL) and incubated O/N. One ml of a 2.5 × 106-CFU/ml suspension of A. pleuropneumoniae S4074 was added, and slides were then incubated for 2 h at 37°C in 5% CO2. Following four washes with Dubelcco's phosphate-buffered saline (DPBS) (Gibco), cells were fixed for 10 min in methanol and stained for 30 min with Giemsa stain (Sigma). Four washes with DPBS were performed, and the slides were left to dry. Noninfected cells were also stained as controls. Observation was done at a ×1,000 magnification on a Leica DMR microscope.
Adherence assay.
To quantify the adherence of the different strains on both cell lines, 2.5 × 105 epithelial cells were seeded into wells of 24-well tissue culture plates (Sarstedt, Numbrecht, Germany) and incubated O/N. Bacteria from an O/N culture grown at an OD600 of 0.6 were resuspended in the adequate cell culture medium to a concentration of 2.5 × 106 CFU/ml. One ml of this suspension was added to each well at an MOI of 10:1, and the plates were incubated from 1 to 3 h. Nonadherent bacteria were removed by washing four times with DPBS. Cell with adherent bacteria were released from the wells by adding 100 μl of 1× trypsin-EDTA (Gibco) and resuspended in 900 μl DPBS buffer. Serial dilutions were performed, with plating on agar plates to determine the number of bacteria that adhered to the epithelial cells.
Statistical analysis.
Data were log transformed prior to analysis. A two-way analysis of variance, with the cell line and the bacterial strain as factors, was used at each time point separately. The level of statistical significance was set at 0.05 throughout. Analyses were carried out using the SAS software program, version 9.1 (SAS, Cary, N C). Contrasts were used to examine differences between pairs of means.
Invasion assay.
Epithelial cells (2.5 × 105) were seeded into wells of 24-well tissue culture plates and incubated O/N. One ml of a 2.5 × 107-CFU/ml bacterial suspension was added to the wells (MOI of 100:1). Plates were incubated for 1 to 3 h. Nonadherent bacteria were removed by washing four times with DPBS buffer. One ml of DMEM containing 100 μg/ml of gentamicin was added to each well, followed by a 1-h incubation period at 37°C in 5% CO2. Killed bacteria were removed by washing twice with DPBS buffer. Cells were then lysed with 100 μl of sterile distilled H2O, and samples were plated on agar plates and incubated O/N.
Protein profiling of SJPL and NPTr cells in contact with A. pleuropneumoniae.
Protein profiling of the cells was performed to detect differentially expressed proteins. Two T175 flasks were seeded with a confluent monolayer of cells, and 25 ml of DMEM culture medium with or without 1 × 107 CFU/ml of bacteria grown at an OD600 of 0.6 was added to the flasks. Flasks were incubated for 3 h at 37°C in 5% CO2 and then washed three times with DPBS, and 500 μl of a lysis solution (20 mM morpholinepropanesulfonic acid, 0.5% triton X-100, and protease inhibitors) was added. Cells were removed from the flasks and transferred to microcentrifuge tubes on ice. Sonication at ∼180 J was performed using an ultrasonic processor (Cole-Parmer, Vernon Hills, IL) in order to lyse the cells. The samples were then ultracentrifugated at 50,000 rpm for 30 min in a Sorvall RC M100 ultracentrifuge. The supernatant was preserved and analyzed for protein concentration using the Bradford assay (Bio-Rad). Samples were diluted to 2 mg/ml and frozen at −80°C. The samples were then analyzed using the Kinexus antibody microarray, which tracks changes in protein expression of 608 different cell signaling proteins in duplicate, including phosphorylation sites and kinases (Kinex Bioinformatics Inc., Vancouver, British Columbia, Canada). Fifty μg of proteins from both the untreated (control) and treated cells were labeled with the same proprietary fluorescent dye. Each sample was separately applied to opposite sides of the antibody microarray, which contains a dam to prevent mixing of the samples. Following incubation of samples with the Kinex chip, the unbound proteins were washed away and the chips were scanned with a Perkin-Elmer ScanArray Express reader. Image analysis of the TIF files that were produced was performed using the ImaGene 7.0 software program from BioDiscovery (El Segundo, CA). Qualitative and semiquantitative analyses of the expression and phosphorylation states of the cell signaling proteins were performed.
EMSA for detection of NF-κB and AP-1.
Cells were infected at an MOI of 1:10 for 30 min, 1 h, or 3 h with A. pleuropneumoniae grown at an OD600 of 0.6. Uninfected cells were used as a control. Cell stimulation was terminated by the addition of cold phosphate-buffered saline. Six μg of nuclear proteins, extracted as described by Blanchette et al. (9), were incubated for 20 min at room temperature in 1 μl binding buffer (100 mM HEPES [pH 7.9], 40% [vol/vol] glycerol, 10% [wt/vol] Ficoll, 250 mM KCl, 10 mM dithiothreitol, 5 mM EDTA, 250 mM NaCl, and 10 mg/ml bovine serum albumin), and 200 ng/μl poly(dI-dC)-0.02% bromophenol blue with 1 μl of the labeled oligonucleotide containing a consensus sequence of NF-κB/c-Rel homodimeric and heterodimeric complexes (5′AGTTGAGGGGACTTTCCCAGGC-3′; Santa Cruz Biotechnology, Santa Cruz, CA) or of AP-1 complexes (5′CGCTTGATGACTCAGCCGGAA-3′; Santa Cruz Biotechnology) which were previously labeled using T4 polynucleotide kinase and [γ-32P]dATP (GE Healthcare, Piscataway, NJ). After incubation, DNA-protein complexes were resolved by electrophoresis in a 5% (wt/vol) nondenaturing polyacrylamide gel. Subsequently gels were dried and autoradiographed. The nonspecific probes (SP-1 [5′ATTCGATCGGGGCGGGGCGAG-3′] used to confirm the specificity of the DNA/nuclear protein reactions were synthesized in our laboratory. Cold competitor assays were conducted by adding a 100-fold molar excess of homologous unlabeled oligonucleotide for NF-κB or AP-1 and noncompetitor SP-1. For supershift assays, 2 μg of nuclear proteins were incubated with binding buffer, poly(dI-dC), 0.02% bromophenol blue, labeled ologonucleotide, and 4 μg specific antibody (anti-p50 and anti-p65N, both from Santa Cruz Biotechnology) at room temperature for 1 h and complexes resolved on a standard nondenaturing 5% (wt/vol) polyacrylamide gel. For the IRAK inhibition assays, the cells were preincubated for 1 h with 50 μM IRAK 1/4 inhibitor (Calbiochem, Darmstadt, Germany) before addition of the bacteria for an incubation of 3 h. A nuclear protein electrophoretic mobility shift assay (EMSA) was then performed as mentioned above.
Stimulation of cytokine production.
Induction assays were performed with both cell lines as described by Ramjeet et al. (48). Briefly, 1 ml culture medium containing 1 × 109 CFU A. pleuropneumoniae S4074, heat killed at 60°C for 45 min, was added to wells of 24-well tissue culture plates containing a monolayer of epithelial cells. The plates were incubated from 30 min to 48 h at 37°C in 5% CO2. The supernatant was then collected and analyzed by ELISA to detect the amounts of IL-1β, TNF-α, IL-6, and IL-8 produced by the stimulated epithelial cells as described by Ramjeet et al. (48). The stimulation tests were also performed using 35 to 3,500 endotoxin units/ml of A. pleuropneumoniae serotype 1 S4074 LPS. These LPS concentrations were shown to induce a response in porcine alveolar macrophages (48). As a control, NF-κB inhibition assays were performed where cells were preincubated for 1 h at 37°C in 5% CO2 with 25 μg/ml caffeic acid phenethyl ester (Sigma) before addition of the bacteria for an additional incubation of 12 h. The supernatant was collected and tested by ELISA for the IL-8 concentration.
RNA extractions for microarray experiments.
Monolayers of SJPL cells in T175 flasks were infected for 3 h with 100 μl of an A. pleuropneumoniae culture with an OD600 of 0.6 (MOI of 10:1). Planktonic bacteria were harvested from the culture supernatant, while adherent bacteria were harvested with the epithelial cells following two washes in phosphate-buffered saline buffer. Ice-cold RNA degradation stop solution (95% ethanol, 5% buffer-saturated phenol) was added to all samples at a 1:10 (vol/vol) ratio, and samples were then frozen at −80°C after a 5-min centrifugation at 4,000 × g. The isolation of bacterial RNA was carried out using the Qiagen RNeasy minikit with an in-column DNase treatment, as prescribed by the manufacturer. RNA was further treated with Turbo DNase (Ambion, Tx) as prescribed by the manufacturer.
Transcriptomic microarray experiments.
The AppChip1 design was part of the A. pleuropneumoniae 5b L20 genome sequencing project led by the team of John Nash (NRC-IBS, Ottawa, Canada). The microarrays used in this study contain PCR amplicons representing all the open reading frames that were identified in the A. pleuropneumoniae 5b L20 genome (13, 18). RNA was reverse transcribed to cDNA using Invitrogen Superscript II reverse transcriptase. cDNA was indirectly labeled with monofunctional Cy3 or Cy5 N-hydroxysuccinimide ester (Amersham Biosciences, Piscataway, NJ). Samples from planktonic growth or adhesion versus growth in DMEM medium were combined and cohybridized on the microarray. Four hybridizations were conducted for each condition, including a pair of microarrays for which the Cy3 and Cy5 dyes were swapped. Microarray analysis was carried out using the TM4 software suite (The Institute for Genomic Research) and the significant-analysis-of-microarrays algorithm, using a false-discovery-rate value of 0% (50). For the planktonic and adhesion experiments, the Cy5 signal was compared to the Cy3 signal in order to obtain a list of significantly differentially expressed genes. In order to obtain the list of differentially expressed genes between planktonic growth and adhesion, log2 ratios were compared in TM4, also using the significant-analysis-of-microarrays algorithm. Functional classification was performed using The Institute for Genomic Research's Comprehensive Microbial Resource (47).
Microarray data accession number.
Data files have been submitted to the Gene Expression Omnibus (GEO accession number GSE12009).
RESULTS
Effect of bacterial infection on viability of epithelial cells.
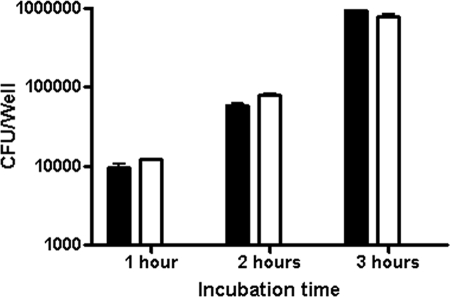
A. pleuropneumoniae expresses potent exotoxins. To ensure cell viability in our experiments, cell death assays were performed at different MOIs (10:1, 100:1, and 1,000:1) and with different incubation periods (0.5 h, 1 h, 2 h, 3 h, and 4 h) with A. pleuropneumoniae strain S4074 representing serotype 1. LDH cytotoxicity assays to detect necrosis were first performed. Important cytotoxicity was observed after an incubaction of 4 h (up to 80% at an MOI of 10:1) or with an MOI of 100:1 (up to 40% at 3 h) (data not shown). An MOI of 10:1 and incubation times not surpassing 3 h were chosen for subsequent tests as a result of low cytotoxicity under these conditions (Fig. 1). Since cell death by apoptosis cannot be detected by the LDH test, apoptosis assays were also preformed. An ELISA assay detecting DNA degradation and a Western blot assay detecting caspase-3 activation were carried out and demonstrated that neither cell line undergoes apoptosis after 3 h of bacterial infection at an MOI of 10:1 (data not shown).
FIG. 1.
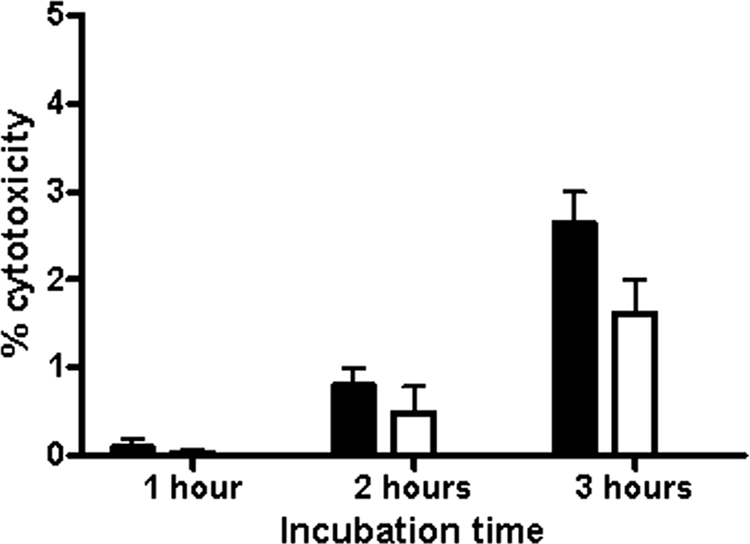
SJPL (filled bars) and NPTr (empty bars) cells were assessed for cytotoxicity following an infection with A. pleuropneumoniae strain S4074 at an MOI of 10:1.
Adherence of A. pleuropneumoniae.
Standardization of adherence models was performed using both cell lines and the A. pleuropneumoniae reference strain S4074. Microscopy assays visually demonstrated the adhesion of the bacteria to the cells (Fig. 2). The increase of adherence over time was well demonstrated in the adherence assay, with an increase of about 1 log every hour (Fig. 3).
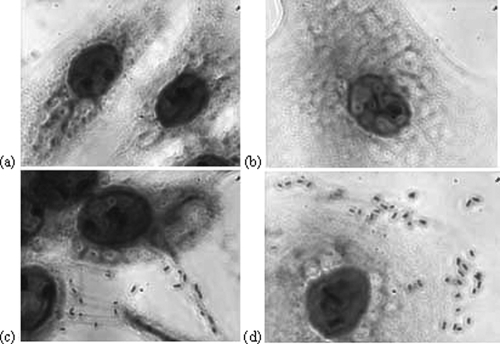
FIG. 2.
NPTr (a and c) or SJPL (b and d) cells stained with Giemsa stain in the presence (c and d) or absence (a and b) of A. pleuropneumoniae S4074, seen through a Leica DMR microscope at an original magnification of ×1,000.
FIG. 3.
Adherence of A. pleuropneumoniae S4074 to SJPL (filled bars) or NPTr (empty bars) cells from 1 to 3 h.
Protein profiling of SJPL and NPTr cells incubated with A. pleuropneumoniae.
Protein profiling of SJPL and NPTr cells incubated with A. pleuropneumoniae was performed as a rapid screening method using the Kinexus antibody microarray. Six hundred eight cell signaling proteins, including 250 phospho sites, 240 protein kinases, and 110 cell signaling proteins that regulate cell proliferation, stress,and apoptosis, were represented on the microarray. Only proteins with an n-fold change of ±1 and higher on a log2 scale were deemed differentially expressed. Twenty proteins were upregulated for the SJPL cells, in contrast to 21 for the NPTr cells, while 25 proteins were downregulated for both the SJPL and NPTr cells (see Tables S1 and S2 in the supplemental material). Among the upregulated proteins, most were implicated in stress response. Mostly proteins implicated in cell growth and proliferation were observed to be downregulated. Among the proteins differentially expressed, IκB kinase α (IKKα), IKKβ, IRAK4, and three different MEKK proteins were detected and directed our focus toward the NF-κB and AP-1 pathways and ultimately to an examination of the production of cytokines by the epithelial cells.
NF-κB induction and cytokine production.
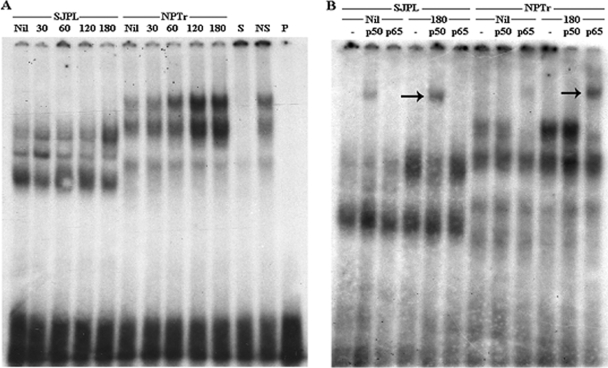
EMSAs detecting the induction of NF-κB and AP-1 were performed on both cell lines following incubation with A. pleuropneumoniae S4074, since proteins involved in these pathways were observed to be differentially expressed in our previous experiment. In comparison to the basal level of uninfected cells, a clear induction of NF-κB was noticed for the NPTr cells as soon as at 1 h postinfection which is represented by the appearance of bands of higher density in the upper part of the gel, corresponding to a band shift (Fig. 4A). Only a slight increase in density was observed for the SJPL cells at 3 h postinfection. In order to assess the specificity of the NF-κB induction, a supershift assay using specific antibodies was carried out for the detection of two subunits of NF-κB, p50 and p65. The p50 subunit was found to be induced for the SJPL cells but not the NPTr cells after 3 h of incubation with A. pleuropneumoniae S4074, and inversely, the p65 subunit was induced in the NPTr cells only (Fig. 4B). No induction of the AP-1 transcription factor was detected for either cell line in comparison to results for the uninfected cells (data not shown).
FIG. 4.
EMSA (A) or supershift assay (B) performed on nuclear proteins of SJPL and NPTr cells following an incubation (30 to 180 min) with A. pleuropneumoniae S4074 or no treatment for the control (Nil). For controls, proteins were incubated with specific oligonucleotides (S) and nonspecific oligonucleotides (NS). The probe alone was also loaded on the gel (P). For the supershift assay (B), proteins were incubated with p50 antibodies (p50), p65 antibodies (p65), or no antibodies (-). Arrows demonstrate the subunit band shifts (B).
To further investigate the bacterium-based induction of the NF-κB transcription factor, we evaluated cytokine production by both cell lines under stimulation conditions. Incubations for up to 48 h of the SJPL and NPTr cells with heat-killed A. pleuropneumoniae were then performed to quantify the production of IL-1β, IL-6, IL-8 and TNF-α, proinflammatory cytokines involved in innate immunity, by the epithelial cells. ELISAs performed on the supernatant samples showed that under these conditions, the NPTr cells but not the SJPL cells secrete IL-8. Production of IL-8 by the NPTr cells increased over time to reach 2,500 pg/ml at 48 h (Fig. 5), in comparison to 800 pg/ml with purified LPS. However, no IL-1β, IL-6, or TNF-α was detected in the samples from both cell lines (data not shown). Following inhibition of NF-κB by caffeic acid phenethyl ester in the NPTr cells, IL-8 concentrations decreased to basal levels (data not shown). This demonstrates that the production of IL-8 observed for the NPTr cells is indeed due to the induction of NF-κΒ.
FIG. 5.
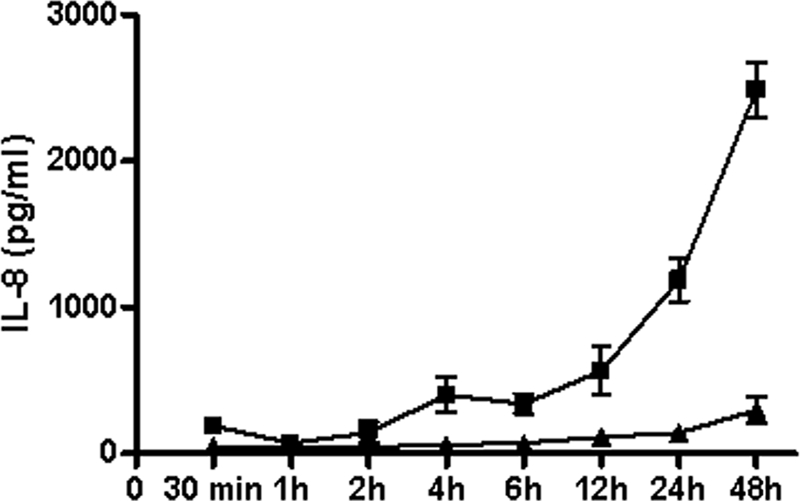
Production of IL-8 by NPTr cells following an induction with heat-killed A. pleuropneumoniae S4074 (▪) or when not stimulated (▴).
To further investigate the mechanism of NF-κB induction in both cell lines, we performed an EMSA on cells pretreated with an IRAK1/4 inhibitor. IRAK1/4 is recruited by the MyD88 protein immediately after Toll receptor activation at the beginning of the NF-κB pathway (27). The level of NF-κΒ induction in comparison to that of noninhibited cells consequently demonstrated indirectly if the activation of Toll-like receptors by the bacterium is responsible for this induction. Our EMSA results indicate that for the SJPL cells, NF-κΒ induction occurs through the Toll receptor pathway (data not shown), but for the NPTr cells, NF-κΒ induction occurs through a different pathway.
A. pleuropneumoniae transcriptomic profiling.
To assess the transcriptional response of A. pleuropneumoniae to both planktonic life over and adherence to SJPL cells, transcript profiling experiments using DNA microarrays were performed after an incubation time of 3 h. Overall, 170 genes were significantly differentially expressed during planktonic growth (Tables 2 and 3), with this number dropping to 131 during adhesion to SJPL cells (Tables 4 and 5). While some genes showed similar patterns of expression during the two conditions, 150 were differentially expressed(see Table S3 in the supplemental material).
TABLE 2.
A. pleuropneumoniae genes which are upregulated during planktonic life over SJPL cellsa
| Locus tag | Gene | Description | Fold change |
|---|---|---|---|
| Hypothetical/unclassified/unknown | |||
| APL_1839 | udp | COG2820: uridine phosphorylase, probable outer membrane protein, possible efflux protein | 3.937 |
| APL_1833 | COG2717: predicted membrane protein, conserved hypothetical protein | 3.388 | |
| APL_1435 | Unassigned protein | 3.214 | |
| APL_1285 | Hypothetical protein | 2.894 | |
| APL_0145 | COG1611: predicted Rossmann fold nucleotide-binding protein | 2.726 | |
| APL_1976 | yedF | COG0425: predicted redox protein, regulator of disulfide bond formation | 2.669 |
| APL_0116 | Hypothetical protein | 2.523 | |
| APL_0093 | DUF1260 domain-containing protein | 2.020 | |
| APL_1374 | Unassigned protein | 1.946 | |
| APL_0465 | DUF74 domain-containing protein | 1.943 | |
| APL_1919 | Predicted enzyme related to aldose 1 epimerase | 1.939 | |
| APL_1138 | DUF526 domain-containing protein | 1.933 | |
| APL_0443 | COG5295: autotransporter adhesin | 1.918 | |
| APL_1656 | COG0561: predicted hydrolases of the HAD superfamily | 1.842 | |
| APL_1203 | DUF479 domain-containing protein, hypothetical protein | 1.811 | |
| APL_1609 | DUF533 domain-containing protein | 1.724 | |
| APL_1881 | Putative carbamoylphosphate synthase large subunit | 1.703 | |
| APL_1244 | Hypothetical protein | 1.555 | |
| Biosynthesis of cofactors | |||
| APL_1622 | cbiM | Predicted ABC cobalt transport permease protein CbiM | 2.419 |
| APL_0931 | iscS | Cysteine desulfurase, iron-sulfur cluster assembly | 2.374 |
| APL_0930 | nifU | NifU-like protein, involved in Fe-S cluster formation | 2.011 |
| APL_1555 | hemL | Glutamate-1-semialdehyde 2,1-aminomutase | 1.619 |
| Energy metabolism | |||
| APL_1832 | COG2041: sulfite oxidase and related enzymes | 13.073 | |
| APL_0892 | fdxG | Formate dehydrogenase, nitrate inducible, major subunit | 12.558 |
| APL_0895 | fdnI | Formate dehydrogenase, cytochrome b556 subunit | 10.946 |
| APL_0894 | fdxH | Formate dehydrogenase, iron-sulfur subunit | 6.0988 |
| APL_0687 | dld | d-Lactate dehydrogenase | 5.442 |
| APL_0100 | nrfA | Ammonia-forming cytochrome c552 nitrite reductase | 5.039 |
| APL_0106 | putA | Bifunctional protein PutA | 4.634 |
| APL_0101 | nrfB | Nitrate reductase, cytochrome-c-type protein | 4.295 |
| APL_1091 | aspA | Aspartate ammonia lyase | 4.254 |
| APL_1528 | frdC | Fumarate reductase subunit C | 3.713 |
| APL_1137 | pgi | Glucose-6-phosphate isomerase | 3.136 |
| APL_1959 | adhI | Alcohol dehydrogenases 1 | 2.992 |
| APL_1414 | mqo | Putative malate:quinone oxidoreductase | 2.802 |
| APL_1379 | ccp | Cytochrome c peroxidase | 2.796 |
| APL_1450 | fbp | Fructose-1,6-bisphosphatase | 2.528 |
| APL_1529 | frdA | Fumarate reductase, flavoprotein subunit | 2.420 |
| APL_0187 | pykA | Pyruvate kinase | 2.322 |
| APL_1197 | 3-Hydroxyacid dehydrogenase | 2.270 | |
| APL_0102 | nrfC | Nitrate reductase | 2.250 |
| APL_1526 | frdD | Fumarate reductase subunit D | 2.178 |
| APL_0486 | maeA | Malic enzyme (NADP dependent) | 2.074 |
| APL_1674 | dmsA | Anaerobic dimethylsulfoxide reductase chain A precursor | 1.934 |
| APL_0483 | Predicted nitroreductase | 1.912 | |
| APL_0084 | trxB | Thioredoxin reductase | 1.486 |
| Transport and binding proteins: cations and iron | |||
| APL_1265 | copA | Copper-transporting P-type ATPase | 1.518 |
| Transport and binding proteins: others | |||
| APL_0107 | putP | Na+/proline symporter | 6.218 |
| APL_1173 | pnuC | Nicotinamide mononucleotide transporter | 2.448 |
| APL_0377 | glpT | Glycerol-3-phosphate transporter | 2.331 |
| APL_1254 | COG0471: di- and tricarboxylate transporters | 2.237 | |
| APL_1319 | ptsB | PTS system sucrose-specific EIIBC component | 2.206 |
| APL_0447 | lctP | l-Lactate permease | 2.204 |
| APL_0262 | modA | Molybdate-binding periplasmic protein precursor | 2.049 |
| APL_1620 | cbiO | Predicted ABC-type cobalt transport system, ATPase component | 1.902 |
| APL_1627 | Putative di- and tricarboxylate transporters | 1.876 | |
| APL_1624 | cbiK | Putative periplasmic binding protein CbiK | 1.823 |
| APL_1902 | yrhG | COG2116: formate/nitrite family of transporters | 1.718 |
| APL_1371 | ccmB | Heme exporter protein B, cytochrome-c-type biogenesis protein | 1.596 |
| Regulatory functions | |||
| APL_108 | iclR | Putative HTH-type transcriptional regulator | 2.832 |
| APL_0395 | rseA | Putative sigma E factor negative regulatory protein | 2.260 |
| APL_0823 | glpR | Glycerol-3-phosphate regulon repressor | 2.175 |
| APL_0997 | lacZ | Beta-galactosidase | 1.654 |
| Transcription | |||
| APL_1475 | rpoD | RNA polymerase sigma 70 factor | 2.135 |
| APL_0560 | rhlB | ATP-dependent RNA helicase RhlB | 1.911 |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| APL_0646 | cpdB | 2′,3′-cyclic-nucleotide 2′-phosphodiesterase precursor | 2.087 |
| Protein fate | |||
| APL_0742 | degS | Protease DegS precursor | 1.768 |
| APL_0871 | pepE | Peptidase E | 1.686 |
| Protein synthesis | |||
| APL_0484 | rimK | Ribosomal protein S6 modification enzyme | 3.093 |
| APL_0146 | dusA | tRNA-dihydrouridine synthase A | 2.456 |
| Cellular processes | |||
| APL_0004 | sodC | Cu/Zn superoxide dismutase precursor | 3.003 |
| APL_0251 | sodA | Manganese superoxide dismutase | 2.356 |
| APL_1241 | Probable carbon starvation protein A, predicted membrane protein | 2.098 | |
| APL_1405 | oapA | Cell envelope opacity-associated protein A | 1.793 |
| Cell envelope | |||
| APL_1494 | ftpA | DNA-binding ferritin-like protein (oxidative damage protectant), fine-tangled pilus major subunit (24-kDa surface protein) | 6.378 |
| Fatty acid and phospholipid metabolism | |||
| APL_0397 | lcfA | Acyl-CoA synthetases (AMP forming)/AMP-acid ligases IIb | 1.746 |
| Mobile and extrachromosomal element functions | |||
| APL_1501 | Transposase | 2.078 | |
| APL_0990 | Transposase | 1.814 | |
| APL_0985 | Transposase | 1.680 | |
| DNA metabolism | |||
| APL_1143 | recA | RecA recombinase | 1.491 |
| Central intermediary metabolism | |||
| APL_0109 | Possible 5-formyltetrahydrofolate cycloligase | 2.607 | |
| APL_0375 | glpK | Glycerol kinase | 2.316 |
Eighty-two genes.
CoA, coenzyme A.
TABLE 3.
A. pleuropneumoniae genes which are downregulated during planktonic life over SJPL cellsa
| Locus tag | Gene | Description | Fold change |
|---|---|---|---|
| Hypothetical/unclassified/unknown | |||
| APL_1387 | Predicted metal-binding, possibly nucleic acid-binding protein | −2.431 | |
| APL_0053 | typA | Predicted membrane GTPase involved in stress response | −2.386 |
| APL_0173 | Hypothetical cytosine deaminase and related metal-dependent hydrolases | −2.056 | |
| APL_1863 | Predicted glycosyltransferase | −2.027 | |
| APL_1956 | Hypothetical protein | −2.000 | |
| APL_0936 | Putative integral membrane protein | −1.829 | |
| Energy metabolism | |||
| APL_1849 | lldD | l-Lactate dehydrogenase (FMN dependent) and related alpha-hydroxy acid dehydrogenases | −3.284 |
| APL_0592 | guaA | GMP synthase | −2.629 |
| APL_1219 | fldA | Flavodoxin | −2.276 |
| Transport and binding proteins: cations and iron | |||
| APL_1047 | hgbA | Hemoglobin-binding protein A precursor | −9.346 |
| APL_1571 | tonB1 | Periplasmic energy transducing protein TonB1 | −5.319 |
| APL_1952 | Outer membrane receptor protein, mostly Fe transport | −3.517 | |
| APL_0077 | exbD2 | Energy transducing protein ExbD2 | −3.499 |
| APL_1953 | Outer membrane receptor protein, mostly Fe transport | −2.874 | |
| APL_0670 | Putative Fe2+/Pb2+ permease | −2.406 | |
| APL_1048 | hugZ | Heme utilization protein | −2.387 |
| APL_0272 | yfeA | Iron (chelated) ABC transporter, periplasmic binding protein | −2.339 |
| APL_0271 | yfeB | putative chelated iron transport system ATP-binding protein | −2.251 |
| APL_0714 | Putative ABC-type enterochelin transport system, periplasmic component | −2.178 | |
| APL_1954 | Outer membrane receptor proteins, mostly Fe transport | −2.163 | |
| APL_0715 | COG4606: ABC-type enterochelin transport system, permease component | −1.993 | |
| APL_1955 | Outer membrane receptor proteins, mostly Fe transport | −1.891 | |
| APL_0717 | COG4604: ABC-type enterochelin transport system, ATPase component | −1.760 | |
| APL_0127 | yfeD | Putative iron transport system membrane protein | −1.718 |
| Transport and binding proteins: others | |||
| APL_0300 | tolQ | Colicin transport protein | −2.584 |
| APL_1392 | ptnC | Mannose permease component IIC | −2.135 |
| APL_1393 | ptnD | Mannose permease component IID | −2.043 |
| APL_1584 | cpxB | Capsule polysaccharide export inner membrane protein | −1.863 |
| APL_1585 | cpxA | Capsule polysaccharide export ATP-binding protein | −1.572 |
| APL_1583 | cpxC | Capsule polysaccharide export inner membrane protein | −1.549 |
| Transcription | |||
| APL_0638 | nusA | Transcription elongation factor | −3.997 |
| APL_0577 | pnp | Polyribonucleotide nucleotidyltransferase | −2.375 |
| APL_0757 | rnb | Exoribonuclease II | −1.665 |
| APL_0543 | rnc | RNase III | −1.539 |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| APL_0148 | rnr1 | Ribonucleotide reductase, alpha subunit | −2.556 |
| APL_0593 | guaB | Inosine-5′-monophosphate dehydrogenase | −2.474 |
| APL_0147 | Ribonucleotide reductase, beta subunit | −2.438 | |
| APL_0255 | gpt | Xanthine phosphoribosyltransferase | −2.316 |
| APL_0661 | purK | Phosphoribosylaminoimidazole carboxylase ATPase subunit | −2.091 |
| APL_1172 | purD | Phosphoribosylamine-glycine ligase | −1.883 |
| APL_2018 | purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | −1.717 |
| APL_0834 | udk | Uridine kinase | −1.396 |
| Protein fate | |||
| APL_1507 | tig | FKBP-type peptidyl-prolyl cis-trans isomerase (trigger factor) | −3.849 |
| APL_0364 | Ssa1 | Autotransporter serine protease | −2.279 |
| Protein synthesis | |||
| APL_1718 | rplK | 50S ribosomal protein L11 | −5.061 |
| APL_0639 | infB | Translation initiation factor 2 (IF-2) | −4.243 |
| APL_1774 | rplF | Ribosomal protein L6 | −3.954 |
| APL_1720 | rplJ | Ribosomal protein L10 | −3.833 |
| APL_1558 | rpsT | 30S ribosomal protein S20 | −3.720 |
| APL_1765 | rplV | 50S ribosomal protein L22 | −3.039 |
| APL_0566 | rpsB | 30S ribosomal protein S2 | −2.842 |
| APL_1400 | rpsG | 30S ribosomal protein S7 | −2.714 |
| APL_0487 | rplY | 50S ribosomal protein L25 (general stress protein Ctc) | −2.364 |
| APL_1762 | rplW | 50S ribosomal protein L23 | −2.330 |
| APL_1763 | rplB | 50S ribosomal protein L2 | −2.265 |
| APL_1761 | rplD | 50S ribosomal protein L4 | −2.254 |
| APL_0223 | infC | Translation initiation factor 3 (IF-3) | −2.192 |
| APL_1773 | rpsH | 30S ribosomal protein S8 | −2.100 |
| APL_1775 | rplR | 50S ribosomal protein L18 | −2.020 |
| APL_0399 | ksgA | Dimethyladenosine transferase (rRNA methylation) | −1.950 |
| APL_1401 | rpsL | 30S ribosomal protein S12 | −1.914 |
| APL_1972 | rpmG | 50S ribosomal protein L33 | −1.902 |
| APL_0040 | yhbZ | Hypothetical GTP-binding protein | −1.721 |
| APL_1228 | infA | Translation initiation factor 1 | −1.716 |
| APL_0678 | efp | Translation elongation factor P | −1.708 |
| APL_1781 | rpsM | 30S ribosomal protein S13 | −1.705 |
| APL_1383 | tRNA (guanine-N(7)-)-methyltransferase | −1.688 | |
| APL_0641 | truB | tRNA pseudouridine synthase B | −1.660 |
| APL_1476 | tyrS | Tyrosyl-tRNA synthetase | −1.646 |
| APL_1789 | rplS | 50S ribosomal protein L19 | −1.521 |
| Cellular processes | |||
| APL_0669 | Predicted iron-dependent peroxidase | −2.630 | |
| APL_1443 | apxIB | Toxin RTX-I translocation ATP-binding protein | −1.753 |
| Cell envelope | |||
| APL_0933 | ompP1 | Putative long-chain-fatty-acid transport protein precursor | −8.199 |
| APL_0651 | galU | UTP-glucose-1-phosphate uridylyltransferase | −2.114 |
| APL_1989 | Predicted membrane protein | −1.951 | |
| APL_1071 | Putative xanthine/uracil permease | −1.817 | |
| Fatty acid and phospholipid metabolism | |||
| APL_1864 | accB | Biotin carboxyl carrier protein of acetyl-CoA carboxylaseb | −2.570 |
| APL_1865 | accC | Biotin carboxylase | −2.444 |
| APL_1889 | fabA | 3-Hydroxydecanoyl-(acyl carrier protein) dehydratase | −2.426 |
| APL_1385 | plsX | Fatty acid/phospholipid biosynthesis enzyme PlsX | −1.922 |
| Amino acid biosynthesis | |||
| APL_0194 | aroK | Shikimate kinase | −2.401 |
| APL_1499 | thrC | Threonine synthase | −2.083 |
| DNA metabolism | |||
| APL_0190 | fis | Factor for inversion stimulation (Fis), transcriptional activator | −2.209 |
| APL_0074 | recR | Recombinational DNA repair protein (RecF pathway) | −1.634 |
| Central intermediary metabolism | |||
| APL_1508 | Putative rhodanese-related sulfurtransferase | −2.860 | |
| APL_0175 | dksA | DnaK suppressor protein | −1.845 |
| APL_0349 | glgA | Glycogen synthase | −1.505 |
Eighty-eight genes.
CoA, coenzyme A.
TABLE 4.
A. pleuropneumoniae genes which are upregulated during adherence to SJPL cellsa
| Locus tag | Gene | Description | Fold change |
|---|---|---|---|
| Hypothetical/unclassified/unknown | |||
| APL_0568 | Hypothetical membrane protein | 3.083 | |
| APL_1459 | Hypothetical protein | 2.882 | |
| APL_1690 | Predicted periplasmic/secreted protein | 2.609 | |
| APL_1471 | Putative sugar transferase | 2.450 | |
| APL_1380 | Uncharacterized conserved protein | 2.310 | |
| APL_2002 | Hypothetical protein | 2.301 | |
| APL_0750 | MscS family protein | 2.181 | |
| APL_1575 | Hypothetical protein | 1.940 | |
| APL_1103 | Predicted inner membrane protein | 1.844 | |
| APL_1854 | pqiA | Uncharacterized paraquat-inducible protein A | 1.746 |
| APL_0217 | Hypothetical protein | 1.640 | |
| Biosynthesis of cofactors | |||
| APL_0776 | ispE | 4-Diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate synthase | 2.913 |
| Energy metabolism | |||
| APL_1849 | lldD | l-Lactate dehydrogenase | 6.756 |
| APL_1820 | rpe | d-Ribulose-phosphate-3 epimerase | 6.687 |
| APL_1191 | namA | NADPH dehydrogenase | 4.524 |
| APL_1331 | hyaA | Hydrogenase 2 small chain precursor | 3.366 |
| APL_1685 | fucK | l-Fuculokinase | 3.195 |
| APL_1689 | fucO | Probable alcohol dehydrogenase, class IV | 2.887 |
| APL_1684 | fucI | l-Fucose isomerase | 2.812 |
| APL_1333 | hybB | Putative Ni/Fe hydrogenase 2 b-type cytochrome component | 2.796 |
| APL_1019 | kdgK | 2-Dehydro-3-deoxygluconokinase | 2.334 |
| APL_0896 | fdhE | Formate dehydrogenase accessory protein FdhE | 2.007 |
| Transport and binding proteins: cations and iron | |||
| APL_1793 | fecE | Fe(III) dicitrate ABC transporter, ATP-binding protein | 6.113 |
| APL_1955 | Outer membrane receptor proteins, mostly Fe transport | 3.271 | |
| APL_1981 | corA | Magnesium transport protein CorA | 3.001 |
| APL_0077 | exbD2 | Energy transducing protein ExbD2 | 2.346 |
| Transport and binding proteins: others | |||
| APL_0066 | dppC | Dipeptide transport system, permease components | 12.073 |
| APL_0870 | Putative C4-dicarboxylate transporter | 6.308 | |
| APL_1713 | Putative oligopeptide transporter | 4.639 | |
| APL_0191 | Predicted Na+-dependent transporters of the SNF family | 4.638 | |
| APL_0309 | yheS | Putative ABC transporter ATP-binding protein YheS | 2.331 |
| APL_1848 | cysA | Sulfate/thiosulfate import ATP-binding protein CysA | 1.938 |
| APL_1249 | sapF | Peptide transport system ATP-binding protein SapF | 1.858 |
| APL_0260 | modC | Molybdenum import ATP-binding protein ModC | 1.799 |
| APL_0582 | sotB | Putative efflux transporter | 1.773 |
| Regulatory functions | |||
| APL_1099 | Organic radical activating enzymes | 7.175 | |
| APL_1962 | hflX | GTP-binding protein HflX | 2.005 |
| Transcription | |||
| APL_0176 | pcnB | Putative poly(A) polymerase | 2.566 |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| APL_0775 | prsA | Ribose-phosphate pyrophosphokinase | 8.463 |
| APL_0425 | purF | Amidophosphoribosyltransferase | 1.812 |
| Protein fate | |||
| APL_1742 | srp54 | Signal recognition particle protein (sigma 54 like) | 6.861 |
| APL_1905 | dnaJ | Chaperone protein DnaJ | 3.383 |
| APL_1330 | hypF | Carbamoyltransferase HypF | 2.080 |
| Protein synthesis | |||
| APL_0034 | engD | GTP-dependent nucleic acid-binding protein EngD | 17.924 |
| APL_0575 | deaD | Superfamily II DNA and RNA helicases | 6.254 |
| APL_0487 | rplY | 50S ribosomal protein L25 (general stress protein Ctc) | 3.429 |
| APL_1325 | Putative 2-methylthioadenine synthetase | 2.357 | |
| APL_1112 | rumA | 23S rRNA (uracil-5-)-methyltransferase RumB | 2.145 |
| APL_0853 | Methionine synthase II (cobalamin independent) | 2.078 | |
| Cellular processes | |||
| APL_1922 | pgaB | Biofilm PGA synthesis lipoprotein PgaB precursor | 7.257 |
| APL_0011 | ftsL | Cell division protein FtsL | 3.349 |
| APL_1923 | pgaC | Biofilm PGA synthesis N-glycosyltransferase PgaC | 2.454 |
| Cell envelope | |||
| APL_0551 | tadB | Tight adherence protein B | 2.453 |
| APL_1841 | murI | Glutamate racemase | 2.340 |
| APL_0016 | murD | UDP-N-acetylmuramoy-l-alanine-d-glutamate synthetase | 2.113 |
| APL_1598 | mrdB | Rod-shape determining protein | 2.096 |
| APL_0419 | lgtF | UDP-glucose-lipooligosaccharide glucosyltransferase | 1.929 |
| APL_1554 | wecA | Putative undecaprenyl-phosphate α-N-acetylglucosaminyl 1-phosphate transferase | 1.577 |
| APL_0555 | rcpA | Rough colony protein A | 1.559 |
| Fatty acids and phospholipids metabolism | |||
| APL_1864 | accB | Biotin carboxyl carrier protein of acetyl-CoA carboxylaseb | 8.665 |
| APL_1865 | accC | Biotin carboxylase | 4.016 |
| APL_0887 | fadI | 3-ketoacyl-CoA thiolase | 3.675 |
| Amino acids biosynthesis | |||
| APL_0320 | metC | Cystathionine beta-lyase | 9.278 |
| APL_0099 | ilvG | Acetolactate synthase isozyme II large subunit | 6.897 |
| APL_1452 | serA | d-3-Phosphoglycerate dehydrogenase | 3.896 |
| APL_0469 | trpB | Tryptophan synthase beta chain | 3.274 |
| APL_0139 | leuC | 3-Isopropylmalate dehydratase large subunit 2 | 3.253 |
| APL_1951 | proA | Gamma-glutamyl phosphate reductase | 2.846 |
| APL_1147 | trpG | Putative anthranilate synthase component II | 2.723 |
| APL_0249 | thrB | Homoserine kinase | 2.459 |
| APL_1125 | Putative cysteine desulfurase | 2.068 | |
| DNA metabolism | |||
| APL_1196 | Type I site-specific restriction-modification system, R subunit | 3.251 | |
| APL_1194 | Type I restriction-modification system, methyltransferase subunit | 2.816 | |
| APL_1146 | rmuC | DNA recombination protein RmuC homolog | 2.462 |
| APL_0874 | holA | DNA polymerase III, delta subunit | 2.022 |
| APL_0287 | hsdM | Putative type I restriction-modification system, methyltransferase subunit | 2.005 |
| Central intermediary metabolism | |||
| APL_2045 | sseA | Probable thiosulfate sulfurtransferase | 1.942 |
| APL_1843 | cysJ | Sulfite reductase [NADPH] flavoprotein alpha component | 1.780 |
Seventy-nine genes.
CoA, coenzyme A.
TABLE 5.
A. pleuropneumoniae genes which are downregulated during adherence to SJPL cellsa
| Locus tag | Gene | Description | Fold change |
|---|---|---|---|
| Hypothetical/unclassified/unknown | |||
| APL_1887 | Esterase domain-containing protein | −6.273 | |
| APL_1284 | Putative DNA-binding protein | −3.076 | |
| APL_0049 | Hypothetical protein | −2.982 | |
| APL_0970 | Hypothetical protein | −2.980 | |
| APL_1100 | Hypothetical protein | −2.928 | |
| APL_1437 | Hypothetical protein | −2.751 | |
| APL_0116 | Hypothetical protein | −2.566 | |
| APL_0389 | ompP4 | Lipoprotein E precursor, predicted secreted acid phosphatase | −2.509 |
| APL_0704 | Potential type III restriction enzyme | −2.293 | |
| APL_1365 | Hly | Hypothetical protein | −2.174 |
| APL_0110 | Hypothetical protein | −2.144 | |
| APL_1396 | Hypothetical protein | −2.125 | |
| APL_0756 | Hypothetical protein | −2.098 | |
| APL_0889 | Hypothetical protein | −1.757 | |
| Energy metabolism | |||
| APL_0434 | gapA | Glyceraldehyde-3-phosphate dehydrogenase | −4.981 |
| APL_0892 | fdxG | Formate dehydrogenase, nitrate-inducible, major subunit | −4.853 |
| APL_0771 | lpdA | Dihydrolipoyl dehydrogenase | −4.765 |
| APL_1379 | ccp | Cytochrome c peroxidase | −4.447 |
| APL_0983 | tktA | Transketolase 2 | −3.790 |
| APL_0894 | fdxH | Formate dehydrogenase, iron-sulfur subunit | −3.627 |
| APL_1251 | pgk | 3-Phosphoglycerate kinase | −3.625 |
| APL_1450 | fbp | Fructose-1,6-bisphosphatase | −3.315 |
| APL_0181 | gloA | Lactoylglutathione lyase | −3.046 |
| APL_1925 | tpiA | Triosephosphate isomerase | −3.023 |
| APL_1091 | aspA | Aspartate ammonia-lyase | −2.722 |
| APL_0688 | torZ | Trimethylamine-N-oxide reductase precursor | −2.687 |
| APL_1011 | adh2 | Aldehyde-alcohol dehydrogenase 2 | −2.642 |
| APL_0772 | aceF | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) | −2.620 |
| APL_1137 | pgi | Glucose-6-phosphate isomerase | −2.595 |
| APL_0486 | maeA | NADP-dependent malic enzyme | −2.416 |
| APL_1526 | frdD | Fumarate reductase subunit D | −2.267 |
| APL_0101 | nrfB | Nitrate reductase, cytochrome-c-type protein | −2.231 |
| APL_1250 | fba | Fructose bisphosphate aldolase | −2.124 |
| Transport and binding proteins: others | |||
| APL_1620 | cbiO | Predicted ABC-type cobalt transport, ATPase component | −2.108 |
| APL_0447 | lctP | Putative l-lactate permease | −2.068 |
| APL_0719 | Putative phosphate permeases | −1.816 | |
| APL_0791 | Transmembrane transport protein-permease | −1.702 | |
| Regulatory functions | |||
| APL_0656 | hlyX | Regulatory protein HlyX | −2.727 |
| APL_0629 | cpxR | Transcriptional regulatory protein CpxR | −2.367 |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| APL_0769 | ushA | UshA precursor | −3.470 |
| APL_0646 | cpdB | 2′,3′-Cyclic-nucleotide 2′-phosphodiesterase precursor | −2.778 |
| APL_1014 | deoD | Purine-nucleoside phosphorylase | −2.247 |
| Protein fate | |||
| APL_1154 | Putative zinc protease | −2.401 | |
| APL_1456 | slyD | FkbP-type peptidyl-prolyl cis-trans isomerase | −2.313 |
| Protein synthesis | |||
| APL_0740 | rpsA | 30s ribosomal protein S1 | −1.608 |
| Cellular processes | |||
| APL_1445 | apxIC | RTX-I toxin-activating lysine-acyltransferase ApxIC | −4.117 |
| APL_0956 | apxIIA | RTX-II toxin determinant A | −4.052 |
| APL_0004 | sodC | Superoxide dismutase (Cu/Zn) precursor | −3.193 |
| Cell envelope | |||
| APL_1494 | ftpA | COG0783: DNA-binding ferritin-like protein (oxidative damage protectant) | −4.223 |
| APL_0652 | manB | Phosphomannomutase | −3.375 |
| Central intermediary metabolism | |||
| APL_0645 | ackA | Acetate kinase | −3.190 |
| APL_1899 | ppa | Inorganic pyrophosphatase | −2.207 |
Fifty-two genes.
The genes that showed the highest level of upregulation during planktonic growth belonged to the “Energy metabolism” functional class, and this class was also the most affected, with 24 out of the 82 upregulated genes. Surprisingly, most of these genes are involved in anaerobic respiration. Various enzymes involved in anaerobic respiration using alternative electron acceptors were upregulated: genes encoding subunits of the formate dehydrogenase (fdxG, and fdnHI) and nitrate reductase (nrfABC), which are essential for anaerobic respiration on nitrate (35, 62), and genes encoding subunits of the fumarate reductase (frdACD), which allows fumarate to serve as a terminal electron acceptor (10), were all upregulated. Furthermore, the genes pgi, fbp, and pykA, which encode three enzymes involved at various steps of glycolysis, glucose-6-phosphate isomerase, fructose-1,6-bisphosphatase, and pyruvate kinase, respectively, showed an increase in transcription. Two dehydrogenases, alcohol dehydrogenase I (encoded by adhI) and malate:quinone oxidoreductase (encoded by mqo), are also involved in anaerobic respiration, the latter being controlled by the ArcA-ArcB two-component system (59). The genes aspA and dmsA were also upregulated. The “Transport and binding proteins” class was the second most affected, with 12 upregulated genes. Genes involved in the transport of l-lactate (lctP), formate and nitrite (yrhG), sucrose (ptsB), and glycerol (glpT) were all induced, as was the gene modA, which encodes a periplasmic protein involved in the ABC transport of molybdate (25). The gene APL_0443, coding for a putative autotransporter adhesin, showed a 1.9-fold induction.
Genes downregulated during planktonic growth mostly belonged to the “Protein synthesis” and “Transport and binding proteins: cations and iron” functional classes. The hgbA hemoglobin receptor gene and its hugZ heme utilization protein cotranscript were downregulated, and other well-established iron acquisition-related genes included tonB1 and exbD2. These genes are cotranscribed with other members of the TonB1 (exbB1, exbD1, tbpA, and tbpB) and TonB2 (exbB2 and exbD2) energy transduction system (7), but these were not identified in our study. At this time, the genes exbB1, exbD1, and tbpB are not present on AppChip1, and the gene tbpA was not downregulated. The genes exbB2 and tonB2, however, exhibit a twofold average downregulation, but variations between chips might have caused these to be ignored by our very stringent analysis parameter (false discovery rate = 0%). A high number of genes that were identified for the first time in A. pleuropneumoniae in our previous transcript profiling experiment under iron restriction (13) were also downregulated. The open reading frames APL_1952 to APL_1955, which code for a putative second hemoglobin receptor system and are likely transcriptionally linked, were repressed, as were the genes APL_0714-APL_0715-APL_0717, encoding a putative ABC-type siderophore transport system, and the genes yfeABD, likely responsible for the ABC-like periplasmic binding protein-dependent transport of chelated iron and possibly manganese. The genes cpxABC, coding for the capsule polysaccharide ABC-type export system, were also all downregulated, along with the gene ssa1, encoding a putative autotransporter serine protease.
Interestingly, some genes potentially involved in adhesion and biofilm biosynthesis were upregulated during adherence to SJPL cells. The genes rcpA and tadB, which belong to a large operon of 14 genes, were upregulated, as were the genes pgaBC, involved in poly-β-1,6-N-acetyl-d-glucosamine biofilm biosynthesis. A small number of genes involved in iron acquisition were also upregulated, the most notable being fecE and APL_1955. Once again, genes involved in anaerobic respiration were shown to be upregulated. Other enzyme genes coding for hydrogenases (hyaA and hybB) or dehydrogenases (lldD and fdhE) involved in energy metabolisms also showed upregulation. The gene fucO, essential for the anaerobic degradation of fucose, and the genes fucI and fucK, (11), involved in the general fucose degradation pathway, were also upregulated.
Only 52 genes were identified as downregulated, and most of them belong to the “Energy metabolism” functional class. The six enzymes which catalyze the first six steps of glycolysis (encoded by gapA, pgk, fbp, tpiA, pgi, and fba) were downregulated, as was the gene maeA, responsible for the first step of gluconeogenesis, and the gene tktA, which links glycolysis to the pentose-phosphate pathway. hlyX, coding for the A. pleuropneumoniae FNR anaerobic global regulator homolog, was repressed 2.72-fold. The toxin genes apxIC and apxIIA also showed downregulation during adhesion to SJPL cells.
Adherence and invasion of A. pleuropneumoniae and other members of the Pasteurellaceae.
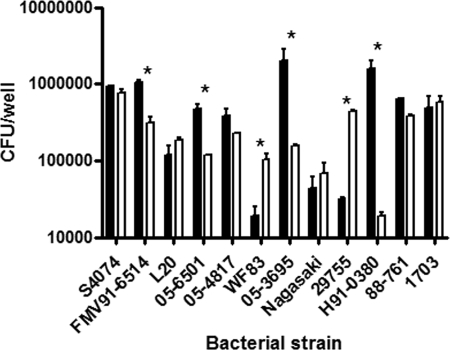
Other serotypes of A. pleuropneumoniae and different swine-colonizing members of the Pasteurellaceae were tested using the adherence models. Differences in adherence were observed between strains for a given cell line and between the cell lines for a given strain (Fig. 6). We noticed that the field strains of A. pleuropneumoniae adhered significantly more to the cell lines than the reference strain of the same serotype. We also noticed that the level of adherence to a given cell line was strain dependent. Following the observation that all members of the Pasteurellaceae tested adhered to the cell lines, invasion tests were performed. A. pleuropneumoniae S4074 did not invade either cell line in our infection model, while the other members of the Pasteurellaceae tested showed invasion. H. parasuis showed the highest level of invasion, although at a reduced level compared to invasion seen with endothelial cells (Fig. 7) (60).
FIG. 6.
Adherence of 12 members of the Pasteurellaceae to the SJPL (filled bars) or NPTr (empty bars) cell line after 3 h of incubation. The strains include A. pleuropneumoniae serotype 1 S4074 and FMV91-6514, A. pleuropneumoniae serotype 5b L20 and 05-6501, A. pleuropneumoniae serotype 5a 05-4817, A. pleuropneumoniae serotype 7 WF83 and 05-3695, H. parasuis serotype 5 Nagasaki and 29755, A. suis serotype O2/K2 H91-0380, and P. multocida capsular type A 88-761 and capsular type D 1703. Asterisks represent statistical differences (P < 0.05) in adherence of the given strain between the two cell lines.
FIG. 7.
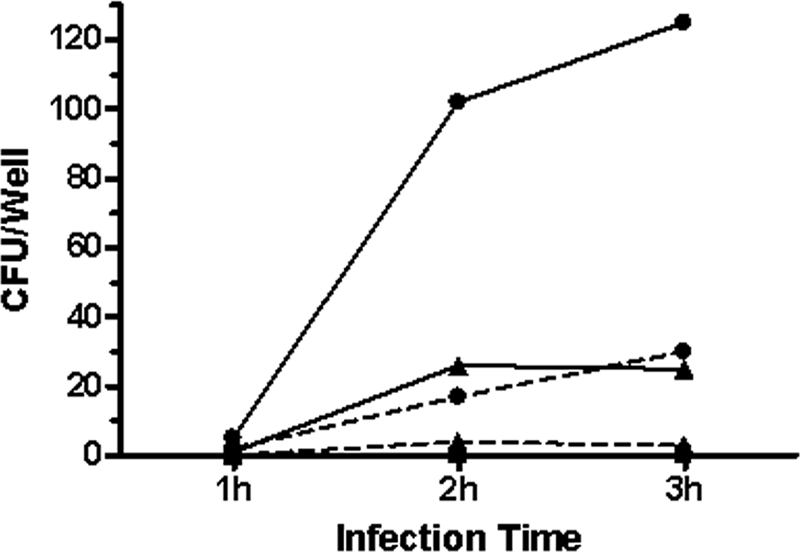
Invasion by A. pleuropneumoniae S4074 (▪), H. parasuis Nagasaki (▴), and H. parasuis 29755 (•) of SJPL (full line) or NPTr (dashed line) cells from 1 h to 3 h.
DISCUSSION
Using immortalized porcine lung and tracheal epithelial cells, we were able to study the host-pathogen interactions of A. pleuropneumoniae. In our models, A. pleuropneumoniae provoked cell death very rapidly through necrosis and not apoptosis. The presence of this bacterium causes many changes in the protein profiles of both epithelial cell lines. Indeed, using an antibody microarray as a rapid screening for differential protein expression, we were able to direct our efforts toward the NF-κB pathway, since numerous differentially expressed proteins were implicated in the NF-κB pathway, including IKKα and IKKβ. NF-κB consists of a homo- or heterodimer composed of the five mammalian Rel proteins, p65, c-Rel, p50, p52, and RelB (24), with the p50/p65 heterodimer being the most abundant and active of the NF-κB complexes (2). Out of the five subunits, only p65 (RelA), RelB, and c-Rel were found to contain the C-terminal transactivation domains essential for gene activation. In contrast, p50 and p52 do not possess the transactivation domains and therefore cannot act as transcriptional activators by themselves (39). Additionally, p50 and p52 are synthesized as precursor proteins that belong to the IκB family known as inhibitors of NF-κB, and homo- or heterodimers of p50 and p52 were also reported to repress κB site-dependent transcription in vivo (39). Interestingly, the p50 subunit was found to be induced in the SJPL cells but not in the NPTr cells after 3 h of incubation with A. pleuropneumoniae S4074, and inversely, the p65 subunit was induced in the NPTr cells only. It should be noted that the absence of detection of either p50 in the NPTr cells or p65 in the SJPL cells is not due to a weak bacterium-induced expression but is most probably due to the incapacity of the cell line to express the protein, since no basal level of expression was observed under unstimulated conditions for the subunits p50 in the NPTr cells and p65 in the SJPL cells. Those results suggest that in the absence of p65, inactive p50/p50 homodimers are more likely to form in the SJPL cells. The absence of IL-8 production by the SJPL cells might be explained by the weak NF-κB induction observed in the EMSAs but certainly correlates with the absence of the p65 subunit, necessary for binding to the IL-8 promoter. Previous studies have also shown that the binding affinity of p50 for the human IL-8 promoter is weak compared to the binding of the p65 subunit (38). Different pathways can activate NF-κB, the most frequent in gram-negative bacterial infection being the classical pathway through Toll-like receptor activation by LPS (44). We demonstrated that this is the case for the SJPL cells but not for the NPTr cells. A possibility is that an alternative pathway for NF-κB activation was used in the NPTr cells where IKKα homodimers are activated instead of the IKKβ in the classical pathway, leading to NF-κB2/p100 phosphorylation. This is a possibility, since this modification creates the production of p52 (44), a subunit which seems to be present in the stimulated NPTr cells, as seen in the EMSA, where a band slightly higher than p50 was detected, and since IKKα was upregulated in the NPTr antibody microarray. This pathway is generally triggered by TNF receptor family members, such as LTβR, BAFF-R, CD40, and CD30 (44). Additional experiments are necessary, however, to confirm this theory.
The presence of the epithelial cells stimulated differential expression of many A. pleuropneumoniae genes. Although it was shown previously, with the evidence of a putative involvement in virulence of genes dmsA and aspA, that genes involved in anaerobic respiration might in fact be essential for full virulence of A. pleuropneumoniae in the host (3, 5, 30), it is still unclear why genes involved in anaerobic respiration are upregulated under our experimental conditions. While such a metabolic switch might be important in vivo to adapt to the lack of oxygen in the deep lung tissues, it should not be necessary in our experimental setup unless this apparent aerobic/anaerobic shift is controled by a host cell-associated factor rather than by oxygen sensors. In fact, it is worth noting that this metabolic shift does not seem to be complete, since genes involved in aerobic respiration are not downregulated. The upregulation of the gene sodA, coding for a cytoplasmic Mn superoxide dismutase (21), also seems to indicate that aerobic respiration is not stopped, since these cytoplasmic superoxide dismutases are specifically useful in removing superoxide anions generated during the course of aerobic respiration (54).
A gene with possible involvement in virulence was also identified. Locus tag APL_0443 is described as an autotransporter adhesin. This protein shows a region of high homology with the A. actinomycetemcomitans extracellular matrix protein adhesin A (EmaA), an oligomeric autotransporter with a YadA domain (57), and the putative Mannheimia haemolytica Hsf protein (40). In Haemophilus influenzae serotype b, Hsf (Haemophilus surface fibrils) is considered the major nonpilus adhesin (55) and was found to be associated with adherence to human epithelial cells (6, 26). Whether this putative A. pleuropneumoniae Hsf protein has these properties remains to be seen, but the upregulation of this gene during planktonic life over SJPL cells might hint to a possible role in the initial steps of A. pleuropneumoniae adhesion during infection.
The fact that iron, in DMEM, is available only in the form of ferric nitrate (12) might explain why iron acquisition systems are more expressed in cell-free DMEM than during planktonic growth. Experiments conducted in our laboratory have shown that ferric nitrate cannot support growth of A. pleuropneumoniae in an ethylenediamine dihydroxyphenyl acetic acid iron-restricted medium (13). Lysis of SJPL cells, leading to the release of the intracellular content of those cells in the medium, supplies the planktonic bacteria with more readily available sources of iron, for example, iron-sulfur clusters in the catalytic core of enzymes and ferritin-bound ferric iron.
Some genes with possible involvement in virulence also came up as downregulated during planktonic growth. Downregulation of the cpxABC operon during planktonic growth over SJPL cells might indicate that when in contact with host cells, A. pleuropneumoniae might wear a thinner polysaccharide layer in order to unmask some adhesins. Surprisingly, this downregulation of the cpx operon was not seen during adhesion to SJPL cells. Upon verification of changes during adhesion, only cpxC showed a low level of downregulation (−1.24), although this was not statistically significant. Repression of the gene ssa1 was surprising, since this gene, also termed aasP, was shown to be expressed in vivo during the chronic stage of the disease (3). However, this gene was also shown to be iron responsive, as indicated by its upregulation during iron restriction (13). It might therefore simply follow the same trend as other iron-responsive genes which were downregulated during planktonic growth.
Our main focus, when looking at overexpressed genes during adherence to porcine lung epithelial cells, was to search for new potential adhesins. The genes tadB and rcpA are part of a large operon which, in A. actinomycetemcomitans, is composed of 14 genes (36) and mediates nonspecific adhesion to solid surfaces, whether they are biological surfaces or not (16). The genetic organization of the A. pleuropneumoniae tad locus is identical to that for A. actinomycetemcomitans (58). Although it is suspected that the tad genes might be transcribed as an operon, only two genes were identified as upregulated in our study. The 12 other genes are present on the microarray but are not significantly induced. Expression of the tad genes is responsible for the rough colony phenotype of A. actinomycetemcomitans, but smooth variants often arise after continued passage on rich medium (49) since mutations often appear in the promoter region of the gene flp-1 (58). We suspect that this might also be the case for A. pleuropneumoniae, since most field isolates exhibit this rough colony phenotype while the reference strains are often smooth colony variants. As is the case for A. actinomycetemcomitans, the Tad proteins might play an important role for the colonization of the respiratory tract by A. pleuropneumoniae, but this will have to be further investigated. Other genes possibly involved in adhesion were also upregulated during adhesion to SJPL cells. The genes pgaB and pgaC are both involved in poly-β-1,6-N-acetyl-d-glucosamine (PGA) biofilm formation. A pgaABCD cluster is present in the App5b L20 genome, and the gene pgaC has been shown to be present in 15 reference strains (29). These results are interesting since the only components that have been clearly shown to be involved in A. pleuropneumoniae adhesion to lung surfaces to date are LPS (1, 8, 34, 45, 46).
The gene hlyX was downregulated during adhesion to SJPL cells. This gene, which encodes the A. pleuropneumoniae FNR anaerobic global regulator homolog, was shown to be important for the colonization and persistence of A. pleuropneumoniae in the respiratory tracts of swine (5). The repression of hlyX probably explains the repression of aspA, which is presumably regulated by HlyX, as well as the downregulation of a few other genes linked with anaerobic respiration (fdxG, torZ, nrfB, and frdD). Genes putatively regulated by HlyX have been shown to be induced by bronchoalveolar lavage fluid from infected pigs (30), and it is possible that hlyX expression follows the same pattern. Also, putative HlyX-regulated genes were upregulated during planktonic growth over SJPL cells.
ApxI and ApxII have been shown to be major virulence factors in A. pleuropneumoniae. Not much is known about transcriptional regulation of those toxins in A. pleuropneumoniae. Studies have shown that levels of oxygen do not influence the levels of ApxI and ApxII (33) and that the iron response regulator Fur seems to have variable effects depending on the calcium concentration in the culture medium (28). Under high calcium concentrations, Fur seemed to act as an activator of the apxI operon, while it seemed to act as a repressor under low calcium concentrations. A previous microarray study conducted under iron restriction showed that Fur does have an effect on ApxI transcription (13). One would normally expect these toxins to be induced under conditions mimicking the in vivo environment, mostly after contact with epithelial cells. Downregulation of the genes apxIC and apxIIA was therefore intriguing. Perhaps smaller concentrations of RTX toxins are required when the bacteria are in close proximity to host cells, leading to the downregulation of the toxins following adherence.
Adherence is seen in both models for all A. pleuropneumoniae strains and serotypes tested. It is interesting to note that field strains adhere more to the cell lines than the reference strain of the same serotype. No invasion is noticed for A. pleuropneumoniae, even though close relatives, such as A. actinomycetemcomitans and H. parasuis, are known to be invasive (17, 42, 60).
Overall these results showed the efficacy of the models and allowed us to gain a great amount of knowledge of A. pleuropneumoniae host-pathogen interactions. Indeed, interaction of A. pleuropneumoniae with host epithelial cells seems to involve complex cross talk which results in the regulation of various bacterial genes. Many virulence genes were upregulated, including genes coding for the putative adhesins Hsf and poly-β-1,6-N-acetyl-d-glucosamine, while capsular polysaccharide-associated genes were downregulated, possibly exposing adhesins usually hidden by a thick capsule. Incubation with A. pleuropneumoniae then led, for both cell lines, to the induction of NF-κB. This is done through the activation of a Toll receptor for the SJPL cells but through an alternative pathway for the NPTr cells. The NPTr cells then secrete IL-8, which is known to attract neutrophils to the infection site, while the SJPL cells do not due to the absence of the p65 subunit of NF-κB. These models are a biologically relevant tool for studying porcine respiratory tract pathogens which could be further used, in the future, to evaluate the effect of a preinfection with agents such as mycoplasmas and viruses, often present with bacterial pathogens under field conditions.
Supplementary Material
Acknowledgments
This work was supported by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (DGPIN0003428) to M. Jacques and by a team grant from the FQRNT (2006-PR-106088) to M. Jacques and M. Gottschalk.
We thank M. Ferrari for the NPTr cell line and R. Webster for the SJPL cell line. We also acknowledge the contribution of Isabelle Gaucher and Geneviève Pelletier-Jacques.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 12 January 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abul-Milh, M., S. E. Paradis, J. D. Dubreuil, and M. Jacques. 1999. Binding of Actinobacillus pleuropneumoniae lipopolysaccharides to glycosphingolipids evaluated by thin-layer chromatography. Infect. Immun. 674983-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeuerle, P. A., and T. Henkel. 1994. Function and activation of NF-κB in the immune system. Annu. Rev. Immunol. 12141-179. [DOI] [PubMed] [Google Scholar]
- 3.Baltes, N., F. F. Buettner, and G. F. Gerlach. 2007. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Vet. Microbiol. 123110-121. [DOI] [PubMed] [Google Scholar]
- 4.Baltes, N., and G. F. Gerlach. 2004. Identification of genes transcribed by Actinobacillus pleuropneumoniae in necrotic porcine lung tissue by using selective capture of transcribed sequences. Infect. Immun. 726711-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltes, N., M. N′Diaye, I. D. Jacobsen, A. Maas, F. F. Buettner, and G. F. Gerlach. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect. Immun. 734614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barenkamp, S. J., and J. W. St Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 191215-1223. [DOI] [PubMed] [Google Scholar]
- 7.Beddek, A. J., B. J. Sheehan, J. T. Bosse, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2004. Two TonB systems in Actinobacillus pleuropneumoniae: their roles in iron acquisition and virulence. Infect. Immun. 72701-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belanger, M., D. Dubreuil, J. Harel, C. Girard, and M. Jacques. 1990. Role of lipopolysaccharides in adherence of Actinobacillus pleuropneumoniae to porcine tracheal rings. Infect. Immun. 583523-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchette, J., P. Pouliot, and M. Olivier. 2007. Role of protein tyrosine phosphatases in the regulation of interferon-γ-induced macrophage nitric oxide generation: implication of ERK pathway and AP-1 activation. J. Leukoc. Biol. 81835-844. [DOI] [PubMed] [Google Scholar]
- 10.Cecchini, G., I. Schroder, R. P. Gunsalus, and E. Maklashina. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553140-157. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y. M., Y. Zhu, and E. C. Lin. 1987. The organization of the fuc regulon specifying L-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol. Gen. Genet. 210331-337. [DOI] [PubMed] [Google Scholar]
- 12.Conrad, D. R. 2007. Ferric and ferrous iron in cell culture. Sigma-Aldrich Co., St. Louis, MO. http://www.sigmaaldrich.com/life-science/cell-culture/learning-center/media-expert/iron.html.
- 13.Deslandes, V., J. H. Nash, J. Harel, J. W. Coulton, and M. Jacques. 2007. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enriquez-Verdugo, I., A. L. Guerrero, J. J. Serrano, D. Godinez, J. L. Rosales, V. Tenorio, and M. de la Garza. 2004. Adherence of Actinobacillus pleuropneumoniae to swine-lung collagen. Microbiology 1502391-2400. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, M., A. Scalvini, M. N. Losio, A. Corradi, M. Soncini, E. Bignotti, E. Milanesi, P. Ajmone-Marsan, S. Barlati, D. Bellotti, and M. Tonelli. 2003. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J. Virol. Methods 107205-212. [DOI] [PubMed] [Google Scholar]
- 16.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 441063-1076. [DOI] [PubMed] [Google Scholar]
- 17.Fives-Taylor, P., D. Meyer, and K. Mintz. 1995. Characteristics of Actinobacillus actinomycetemcomitans invasion of and adhesion to cultured epithelial cells. Adv. Dent. Res. 955-62. [DOI] [PubMed] [Google Scholar]
- 18.Foote, S. J., J. T. Bosse, A. B. Bouevitch, P. R. Langford, N. M. Young, and J. H. Nash. 2008. The complete genome sequence of Actinobacillus pleuropneumoniae L20 (serotype 5b). J. Bacteriol. 1901495-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey, J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3257-261. [DOI] [PubMed] [Google Scholar]
- 20.Frey, J., R. Kuhn, and J. Nicolet. 1994. Association of the CAMP phenomenon in Actinobacillus pleuropneumoniae with the RTX toxins ApxI, ApxII and ApxIII. FEMS Microbiol. Lett. 124245-251. [DOI] [PubMed] [Google Scholar]
- 21.Fridovich, I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 6497-112. [DOI] [PubMed] [Google Scholar]
- 22.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27311-327. [DOI] [PubMed] [Google Scholar]
- 23.Gelfanova, V., E. J. Hansen, and S. M. Spinola. 1999. Cytolethal distending toxin of Haemophilus ducreyi induces apoptotic death of Jurkat T cells. Infect. Immun. 676394-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16225-260. [DOI] [PubMed] [Google Scholar]
- 25.Grunden, A. M., and K. T. Shanmugam. 1997. Molybdate transport and regulation in bacteria. Arch. Microbiol. 168345-354. [DOI] [PubMed] [Google Scholar]
- 26.Hallstrom, T., E. Trajkovska, A. Forsgren, and K. Riesbeck. 2006. Haemophilus influenzae surface fibrils contribute to serum resistance by interacting with vitronectin. J. Immunol. 177430-436. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins, P. A., and S. Sriskandan. 2005. Mammalian Toll-like receptors: to immunity and beyond. Clin. Exp. Immunol. 140395-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, Y. M., N. Chin, C. F. Chang, and Y. F. Chang. 2003. Cloning and characterization of the Actinobacillus pleuropneumoniae fur gene and its role in regulation of ApxI and AfuABC expression. DNA Seq. 14169-181. [DOI] [PubMed] [Google Scholar]
- 29.Izano, E. A., I. Sadovskaya, E. Vinogradov, M. H. Mulks, K. Velliyagounder, C. Ragunath, W. B. Kher, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen, I., J. Gerstenberger, A. D. Gruber, J. T. Bosse, P. R. Langford, I. Hennig-Pauka, J. Meens, and G. F. Gerlach. 2005. Deletion of the ferric uptake regulator Fur impairs the in vitro growth and virulence of Actinobacillus pleuropneumoniae. Infect. Immun. 733740-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsen, I. D., J. Meens, N. Baltes, and G. F. Gerlach. 2005. Differential expression of non-cytoplasmic Actinobacillus pleuropneumoniae proteins induced by addition of bronchoalveolar lavage fluid. Vet. Microbiol. 109245-256. [DOI] [PubMed] [Google Scholar]
- 32.Jacques, M., J. Labrie, F. St Michael, A. D. Cox, M. A. Paradis, C. P. Dick, C. Klopfenstein, A. Broes, N. Fittipaldi, and M. Gottschalk. 2005. Isolation of an atypical strain of Actinobacillus pleuropneumoniae serotype 1 with a truncated lipopolysaccharide outer core and no O-antigen. J. Clin. Microbiol. 433522-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarma, E., G. Corradino, and L. B. Regassa. 2004. Anaerobiosis, growth phase and Actinobacillus pleuropneumoniae RTX toxin production. Microb. Pathog. 3729-33. [DOI] [PubMed] [Google Scholar]
- 34.Jeannotte, M. E., M. Abul-Milh, J. D. Dubreuil, and M. Jacques. 2003. Binding of Actinobacillus pleuropneumoniae to phosphatidylethanolamine. Infect. Immun. 714657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, R. W., A. Lamont, and P. B. Garland. 1980. The mechanism of proton translocation driven by the respiratory nitrate reductase complex of Escherichia coli. Biochem. J. 19079-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9429-437. [DOI] [PubMed] [Google Scholar]
- 37.Kato, S., N. Sugimura, K. Nakashima, T. Nishihara, and Y. Kowashi. 2005. Actinobacillus actinomycetemcomitans induces apoptosis in human monocytic THP-1 cells. J. Med. Microbiol. 54293-298. [DOI] [PubMed] [Google Scholar]
- 38.Kunsch, C., R. K. Lang, C. A. Rosen, and M. F. Shannon. 1994. Synergistic transcriptional activation of the IL-8 gene by NF-kappa B p65 (RelA) and NF-IL-6. J. Immunol. 153153-164. [PubMed] [Google Scholar]
- 39.Lernbecher, T., U. Muller, and T. Wirth. 1993. Distinct NF-kappa B/Rel transcription factors are responsible for tissue-specific and inducible gene activation. Nature 365767-770. [DOI] [PubMed] [Google Scholar]
- 40.Lo, R. Y. 2001. Genetic analysis of virulence factors of Mannheimia (Pasteurella) haemolytica A1. Vet. Microbiol. 8323-35. [DOI] [PubMed] [Google Scholar]
- 41.Martellini, A., P. Payment, and R. Villemur. 2005. Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Res. 39541-548. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, D. H., J. E. Lippmann, and P. M. Fives-Taylor. 1996. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect. Immun. 642988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, D. H., P. K. Sreenivasan, and P. M. Fives-Taylor. 1991. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect. Immun. 592719-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikori, M. 2005. Classical and alternative NF-kB activation pathways and their roles in lymphoid malignancies. J. Clin. Exp. Hematop. 4515-24. [Google Scholar]
- 45.Paradis, S. E., D. Dubreuil, S. Rioux, M. Gottschalk, and M. Jacques. 1994. High-molecular-mass lipopolysaccharides are involved in Actinobacillus pleuropneumoniae adherence to porcine respiratory tract cells. Infect. Immun. 623311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paradis, S. E., J. D. Dubreuil, M. Gottschalk, M. Archambault, and M. Jacques. 1999. Inhibition of adherence of Actinobacillus pleuropneumoniae to porcine respiratory tract cells by monoclonal antibodies directed against LPS and partial characterization of the LPS receptors. Curr. Microbiol. 39313-0320. [DOI] [PubMed] [Google Scholar]
- 47.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramjeet, M., V. Deslandes, F. St Michael, A. D. Cox, M. Kobisch, M. Gottschalk, and M. Jacques. 2005. Truncation of the lipopolysaccharide outer core affects susceptibility to antimicrobial peptides and virulence of Actinobacillus pleuropneumoniae serotype 1. J. Biol. Chem. 28039104-39114. [DOI] [PubMed] [Google Scholar]
- 49.Rosan, B., J. Slots, R. J. Lamont, M. A. Listgarten, and G. M. Nelson. 1988. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol. Immunol. 358-63. [DOI] [PubMed] [Google Scholar]
- 50.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34374-378. [DOI] [PubMed] [Google Scholar]
- 51.Schaller, A., R. Kuhn, P. Kuhnert, J. Nicolet, T. J. Anderson, J. I. MacInnes, R. P. Segers, and J. Frey. 1999. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 1452105-2116. [DOI] [PubMed] [Google Scholar]
- 52.Seo, S. H., O. Goloubeva, R. Webby, and R. G. Webster. 2001. Characterization of a porcine lung epithelial cell line suitable for influenza virus studies. J. Virol. 759517-9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheehan, B. J., J. T. Bosse, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2003. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 713960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheehan, B. J., P. R. Langford, A. N. Rycroft, and J. S. Kroll. 2000. [Cu, Zn]-Superoxide dismutase mutants of the swine pathogen Actinobacillus pleuropneumoniae are unattenuated in infections of the natural host. Infect. Immun. 684778-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 1786281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Straw, B. E. 2006. Diseases of swine, 9th ed. Blackwell Publishing, Ames, IA.
- 57.Tang, G., T. Ruiz, R. Barrantes-Reynolds, and K. P. Mintz. 2007. Molecular heterogeneity of EmaA, an oligomeric autotransporter adhesin of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Microbiology 1532447-2457. [DOI] [PubMed] [Google Scholar]
- 58.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. 5363-375. [DOI] [PubMed] [Google Scholar]
- 59.van der Rest, M. E., C. Frank, and D. Molenaar. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J. Bacteriol. 1826892-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanier, G., A. Szczotka, P. Friedl, S. Lacouture, M. Jacques, and M. Gottschalk. 2006. Haemophilus parasuis invades porcine brain microvascular endothelial cells. Microbiology 152135-142. [DOI] [PubMed] [Google Scholar]
- 61.Van Overbeke, I., K. Chiers, G. Charlier, I. Vandenberghe, J. Van Beeumen, R. Ducatelle, and F. Haesebrouck. 2002. Characterization of the in vitro adhesion of Actinobacillus pleuropneumoniae to swine alveolar epithelial cells. Vet. Microbiol. 8859-74. [DOI] [PubMed] [Google Scholar]
- 62.Wang, H., and R. P. Gunsalus. 2003. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 1855076-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., J. M. Tennent, A. Ingham, G. Beddome, C. Prideaux, and W. P. Michalski. 2000. Identification of type 4 fimbriae in Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 18915-18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.