Abstract
The RgpA-Kgp proteinase-adhesin complexes are a primary virulence factor of Porphyromonas gingivalis, a major pathogen in the development of chronic periodontitis. The RgpA-Kgp complexes have been suggested to bias the immune response to a Th2 phenotype in disease by hydrolysis of Th1 cytokines. Here, we show that the RgpA-Kgp complexes hydrolyze and inactivate interleukin-4 (IL-4) and IL-5 under physiologically relevant conditions. Using the IL-4/IL-5-dependent TF1.8 T-cell line, it was found that at equimolar ratios of cytokine to RgpA-Kgp complexes, IL-4 and IL-5 were inactivated in the culture medium. The inactivation of IL-4 and IL-5 was RgpA-Kgp concentration dependent, as at an enzyme-to-cytokine molar ratio of 1:8, the bioactivity of the cytokines was greater than at the higher concentration of RgpA-Kgp of 1:1. Furthermore, inactivation of the cytokines by the RgpA-Kgp complexes was time dependent, as longer preincubation times resulted in lower cytokine activity. IL-5 was found to be slightly more resistant to inactivation than IL-4. Mass spectrometric analyses of IL-4 and IL-5 showed that hydrolysis by RgpA-Kgp complexes was C terminal to Arg and Lys residues of the cytokines. The peptides released indicated that the regions of IL-4 and IL-5 important for bioactivity were being hydrolyzed in the first 15 min of incubation. The ability of the RgpA-Kgp complexes to degrade Th2 cytokines may contribute to immune dysregulation and may play a role in the pathology of chronic periodontitis.
The RgpA-Kgp proteinase-adhesin complexes (RgpA-Kgp complexes) of the periodontal pathogen Porphyromonas gingivalis have been suggested to play a major role in the dysregulation of the host immune response (22). The ability of the RgpA-Kgp complexes to dysregulate the immune response has been linked to their Arg (RgpA)- and Lys (Kgp)-specific proteolytic activities, which have been shown to degrade a range of host proteins (11, 12, 22, 29, 35, 42). The Arg- and Lys-specific proteinases have been reported to hydrolyze complement factors C5 and C3 to release the chemotactic factors C5a and C3a, respectively, which have been suggested to enhance the inflammatory response by attracting neutrophils to the site of infection (37). Furthermore, the proteinases degrade C3b and C5b, thereby reducing the opsonization of the bacteria. In addition, the proteinases have been shown to degrade immunoglobulin G (IgG), IgA, IgM, IgD, and IgE and the T-cell surface markers CD4 and CD8, which decrease T-cell proliferation (5, 6, 8).
A number of cytokines have also been reported to be degraded by the Arg- and Lys-specific proteinases of the RgpA-Kgp complexes. Tumor necrosis factor alpha (TNF-α) was demonstrated to be degraded at low concentrations of the purified Arg-specific and Lys-specific proteinases (enzyme-to-substrate [E:S] ratios of 1:25 and 1:100, respectively) over a time course of 8 h, thus contributing to the impairment of polymorphonuclear leukocyte function (4). Mikolajczyk-Pawlinska et al. (15) have shown that interleukin-8 (IL-8) is degraded at low concentrations of purified Arg- and Lys-specific proteinases (an E:S molar ratio of 1:550), where complete degradation was observed within 10 h. Studies using Western blot analysis demonstrated that the RgpA and Kgp proteinases degraded gamma interferon (IFN-γ) and IL-12 and that CD4+ T cells incubated with IL-12 that had been preexposed to the proteinases secreted less IFN-γ (44, 45). Yun et al. (44, 45) concluded from these studies that degradation of these Th1 cytokines (IL-12 and IFN-γ) biases the immune response to a Th2 cytokine response and speculated that disease (chronic periodontitis) was therefore associated with a Th2 cytokine response.
Patients with chronic periodontitis have been shown to express elevated levels of proinflammatory cytokines, including IL-1, IL-6, and TNF-α, in gingival tissues and gingival crevicular fluid (13, 30, 31). These studies and a number of others (20, 22, 36) have suggested that a Th1 inflammatory immune response is associated with chronic periodontitis progression, whereas an anti-inflammatory Th2 immune response is not associated with progression of disease (20, 22, 36). Houri-Haddad et al. (7), utilizing a mouse subcutaneous-lesion model, have demonstrated that the production of IFN-γ is associated with P. gingivalis-induced inflammation, whereas IFN-γ-deficient (IFN-γ−/−) mice exhibited decreased local inflammation and produced a protective IgG1 antibody response to P. gingivalis. Additionally, both IFN-γ and IL-6 have been reported to contribute to P. gingivalis-induced bone loss in mice, as animals lacking IFN-γ and IL-6 exhibited no bone loss after intraoral challenge with P. gingivalis (2).
A range of inflammatory cytokines have been reported to be induced by P. gingivalis in both in vitro and in vivo studies. Stimulation of human epithelial cells and fibroblasts with P. gingivalis cells in vitro resulted in an upregulation of the proinflammatory cytokines IL-1β, IL-6, IL-8, and TNF-α (1). In a similar study, a proinflammatory cytokine, IL-8, was also demonstrated to be upregulated after stimulation of human gingival fibroblasts with the P. gingivalis Arg-specific proteinase in vitro (24). IL-17, another proinflammatory cytokine, has been found in periodontal lesions, indicating it may play a role in Th1 modulation (38). These data suggest that P. gingivalis and its virulence factors upregulate a proinflammatory cytokine response during infection and are in conflict with the suggestion by Yun et al. (44, 45) that the RgpA and Kgp proteinases preferentially degrade Th1 cytokines.
In this paper, we investigate the degradation and bioavailability of the anti-inflammatory (Th2) cytokines IL-4 and IL-5 in the presence of the RgpA-Kgp complexes of P. gingivalis by utilizing the IL-4/IL-5-dependent TF1.8 T-cell line and mass spectrometric analysis.
MATERIALS AND METHODS
Isolation and purification of the RgpA-Kgp proteinase-adhesin complexes.
P. gingivalis strain W50 cells were grown as previously described (20), and the RgpA-Kgp complexes were isolated, purified, and analyzed as described by Pathirana et al. (25).
IL-4 and IL-5 stimulation of the TF1.8 T-cell line after incubation with RgpA-Kgp complexes.
Human recombinant IL-4 (catalog no. 204-IL/CF; R&D Systems, Minneapolis, MN) or human recombinant IL-5 (catalog no. 205-IL/CF; R&D Systems, Minneapolis, MN) was added to the culture medium at a concentration of 0.01 to 20.00 ng/ml in 96-well microtiter plates or was preincubated with the RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:1 or 1:8 in phosphate-buffered saline (PBS) (0.15 M NaCl, 0.01 M Na2HPO4, 1.5 mM KH2PO4, 3 mM KCl) at 37°C before being added to the culture medium. Aliquots of the incubation mixtures containing 4 ng of IL-4 or IL-5 and RgpA-Kgp complexes were taken at 1, 4, 8, and 24 h and then serially diluted in 96-well microtiter plates with AIM-V medium (Gibco) supplemented with 2 mM l-glutamine, 10 mM HEPES, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate (supplied by JRH Biosciences, Parkville, Australia), and 0.1 mM monothioglycerol (Sigma, Australia). The zero time point involved adding the Rgp-Kgp complexes to the culture medium at the same time as the cytokines. TF1.8 T cells cultured in AIM-V medium were added to each well at a concentration of 2.5 × 104 cells/well in a total culture volume of 100 μl and incubated for 48 h at 37°C in an atmosphere of 5% CO2 in air. T-cell proliferation was measured using [3H]thymidine incorporation as previously described (20). Data are expressed as the stimulatory index (SI) ± standard deviation (SD), where the SI is calculated as the test counts per minute divided by the negative control counts per minute.
Analysis of degradation of IL-4 and IL-5 by the RgpA-Kgp complexes using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
RgpA-Kgp complexes were added to 10 μM of either IL-4 or IL-5 in PBS or AIM-V medium at an enzyme-to-cytokine molar ratio of 1:8. After incubation at 37°C for 0 min, 15 min, 30 min, 1 h, 4 h, 8 h, and 24 h, 1-μl aliquots of the reaction mixture were removed and acidified with a final concentration of 1% formic acid to stop further enzymatic degradation. Samples containing the RgpA-Kgp complexes or IL-4 or IL-5 alone were also incubated at 37°C and prepared as controls. Equal volumes (0.3 μl) of sample solution and matrix solution (α-cyano-4-hydroxycinnamic acid saturated in 33% acetonitrile aqueous solution containing 0.1% trifluoroacetic acid) were mixed on a ground-steel MALDI target (Bruker Daltonics, Bremen, Germany). After being air dried at room temperature, the spots were washed twice with 5 μl of cold 0.1% trifluoroacetic acid and air dried completely before being analyzed in an Ultraflex MALDI TOF/TOF mass spectrometer with a LIFT II upgrade (Bruker Daltonics, Bremen, Germany). MALDI-TOF/TOF MS spectra were acquired in linear or reflector-positive mode from 200 laser shots, using an N2 laser producing laser irradiation at a λ of 337 nm. Constant laser power was used in each acquisition method to facilitate direct comparison of samples within the same mass range. Compass 1.1 software (Bruker Daltonics, Bremen, Germany), including flexControl 2.4 and BioTools 3.0, was used for instrument control, spectrum annotation, and analysis.
Statistical analysis.
The TF1.8 T-cell proliferation data (counts per minute) were log10 transformed, and normal distribution of the data was confirmed using Levene's test of homogeneity of variances. The transformed proliferation data were statistically analyzed using a one-way analysis of variance and Dunnett's T3 test (17).
RESULTS
Bioactivity of IL-4 and IL-5 in the presence of P. gingivalis W50 RgpA-Kgp complexes.
The bioactivity of IL-4 and IL-5 in the presence of the RgpA-Kgp complexes was evaluated by utilizing the TF1.8 T-cell line, which is dependent upon IL-4 or IL-5 for cell growth. To determine whether the RgpA-Kgp complexes affected the bioactivity of IL-4 or IL-5, RgpA-Kgp complexes were added to the culture medium at an enzyme-to-cytokine molar ratio of 1:1 (Fig. 1) or 1:8 (Fig. 2 and 3). The addition of the RgpA-Kgp complexes at both molar ratios (1:1 and 1:8) significantly reduced (P < 0.05) TF1.8 T-cell proliferation compared with the controls not containing the RgpA-Kgp complexes (Fig. 1, 2, and 3). The effect of the RgpA-Kgp complexes was dose dependent, as the lower concentration (enzyme-to-cytokine ratio, 1:8) produced less reduction in the stimulation of the TF1.8 T cells.
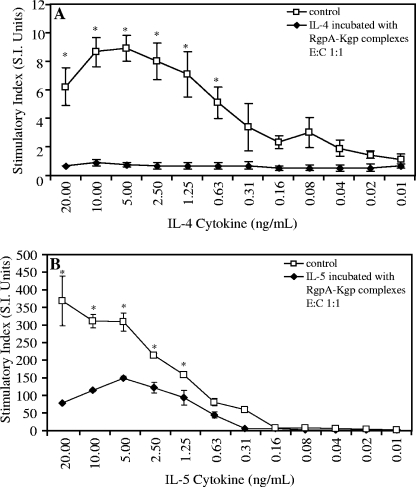
FIG. 1.
Bioactivity of human recombinant IL-4 and IL-5 in the presence of P. gingivalis W50 RgpA-Kgp complexes at a 1:1 enzyme-to-cytokine (E:C) molar ratio. A human T-cell line (TF 1.8), dependent on IL-4 or IL-5 for growth, was cultured in the presence of human recombinant IL-4 (A) or human recombinant IL-5 (B), together with RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:1 or without the RgpA-Kgp complexes (control). The cells were grown for 48 h, and cell growth was detected by utilizing [3H]thymidine incorporation. The data are expressed as the SI and are the average of triplicate assays ± SD. *, P < 0.05. Control growth is significantly greater than growth in the presence of the RgpA-Kgp complexes.
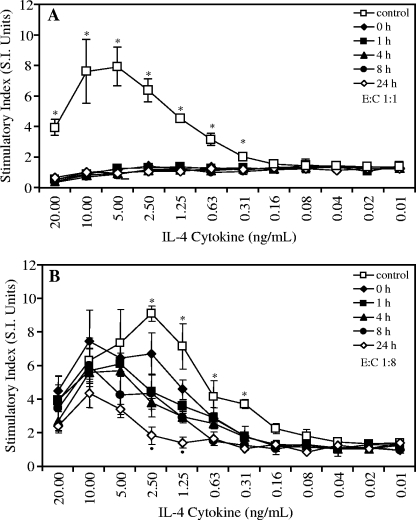
FIG. 2.
Time course analysis of bioactivity of human recombinant IL-4 after incubation with the RgpA-Kgp complexes at a 1:1 enzyme-to-cytokine (E:C) molar ratio (A) and at a 1:8 enzyme-to-cytokine molar ratio (B). A human T-cell line (TF 1.8), dependent on IL-4 or IL-5 for growth, was grown in the presence of human recombinant IL-4 that had been previously incubated with the RgpA-Kgp complexes for 0, 1, 4, 8, and 24 h and compared with growth in IL-4 without incubation with the RgpA-Kgp complexes (control). The cells were grown for 48 h, and cell growth was detected by utilizing [3H]thymidine incorporation. The data are expressed as the SI and are the average of triplicate assays ± SD. (A) *, P < 0.05; the control is significantly higher than all other incubation times. (B) *, P < 0.05; the control is significantly higher than all other incubation times. •, P < 0.05; 24-h incubation was significantly lower than 0 h.
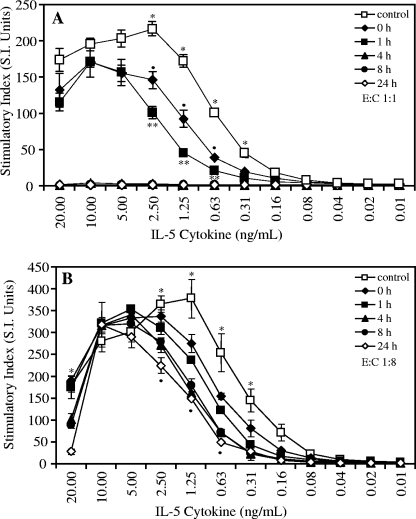
FIG. 3.
Time course analysis of bioactivity of human recombinant IL-5 after incubation with the RgpA-Kgp complexes at a 1:1 enzyme-to-cytokine (E:C) molar ratio (A) and at a 1:8 enzyme-to-cytokine molar ratio (B). A human T-cell line (TF 1.8), dependent on IL-4 or IL-5 for growth, was grown in the presence of human recombinant IL-5 that had been previously incubated with the RgpA-Kgp complexes for 0, 1, 4, 8, and 24 h and compared with growth in IL-4 without incubation with the RgpA-Kgp complexes (control).The cells were grown for 48 h, and cell growth was detected by utilizing [3H]thymidine incorporation. The data are expressed as the SI and are the average of triplicate assays ± SD. (A) *, P < 0.05; the control is significantly higher than all incubation times. •, P < 0.05; 0-h incubation is significantly higher than 1-h incubation. **, P < 0.05; 0- and 1-h incubations are significantly higher than 4-, 8-, and 24-h incubations. (B) *, P < 0.05; the control is significantly higher than all other incubation times. •, P < 0.05; 4-, 8-, and 24-h incubations are significantly lower than 0- and 1-h incubations.
IL-4 stimulation of TF1.8 T cells after preincubation with RgpA-Kgp complexes.
IL-4 was preincubated with RgpA-Kgp complexes at E:S molar ratios of 1:1 and 1:8 for 1, 4, 8, and 24 h (Fig. 2). A zero time point was included in all experiments as described above. Aliquots from each IL-4/RgpA-Kgp preincubation were serially diluted in AIM-V medium and used to stimulate TF1.8 T cells for 48 h, and the bioactivity was compared with that of IL-4 without RgpA-Kgp complexes.
The effect of preincubation of IL-4 with RgpA-Kgp at an enzyme-to-cytokine molar ratio of 1:1 on the stimulation of TF1.8 T cells is shown in Fig. 2A. No proliferation was observed for the TF1.8 T cells that were incubated with IL-4 that had been preincubated with RgpA-Kgp complexes at any of the preincubation times tested. The same results were obtained whether the cytokine and RgpA-Kgp complexes were preincubated in PBS or AIM-V medium. The effect of preincubation of IL-4 with the RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:8 on the stimulation of TF1.8 T cells is shown in Fig. 2B. A significant reduction (P < 0.05) in TF1.8 T-cell proliferation with IL-4 preincubated with RgpA-Kgp complexes was observed for all preincubation times; however, the reduction in stimulation of growth was less at the lower enzyme concentration (enzyme-to-cytokine ratio, 1:8).
IL-5 stimulation of TF1.8 T cells after preincubation with the RgpA-Kgp complexes.
IL-5 was preincubated with the RgpA-Kgp complexes at enzyme-to-cytokine molar ratios of 1:1 and 1:8 for 1 h, 4 h, 8 h, and 24 h (Fig. 3). A zero time point was included in all experiments as described above. After preincubation, aliquots from each time point were serially diluted in AIM-V medium and used to stimulate TF1.8 T cells for 48 h, and the bioactivity was compared with that of IL-5 without RgpA-Kgp complexes.
The effect of preincubation of IL-5 with the RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:1 on the stimulation of TF1.8 T cells is shown in Fig. 3A. Stimulation of the TF1.8 T cells with IL-5 preincubated with RgpA-Kgp complexes was significantly reduced for all preincubation times (P < 0.05). No proliferation of TF1.8 T cells was observed when IL-5 was preincubated with RgpA-Kgp complexes for 4, 8, and 24 h.
The TF1.8 T cells were also stimulated with IL-5 that had been preincubated with the RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:8 (Fig. 3B). Preincubation of IL-5 with RgpA-Kgp complexes resulted in significantly reduced proliferation of the TF1.8 T cells for all preincubation times (P < 0.05); however, the level of reduction of T-cell stimulation by the RgpA-Kgp complexes was substantially less at the lower enzyme concentration (enzyme-to-cytokine ratio, 1:8).
Analysis of the degradation of IL-4 and IL-5 by the RgpA-Kgp complexes using MALDI-TOF/TOF MS.
To investigate the degradation of IL-4 and IL-5 by the RgpA-Kgp complexes, IL-4 and IL-5 cytokines were incubated with the RgpA-Kgp complexes at an enzyme-to-cytokine molar ratio of 1:8. Samples were taken at sequential time points and analyzed by MALDI-TOF/TOF MS, and the degradation products were identified by peptide mass fingerprinting. Incubation of IL-4 cytokine with the RgpA-Kgp complexes revealed that hydrolysis of IL-4 was observed at the first time point of 15 min (Table 1). The hydrolysis occurred C terminal to the Arg and Lys residues of the cytokine. After 15 min of incubation, IL-4 was hydrolyzed at the carboxyl side of six Arg residues and six Lys residues to yield peptides 1 to 9 (Table 1). Nine of these hydrolysis sites (peptides 2 to 8) are located in the center of the IL-4 amino acid sequence, whereas the hydrolyzed peptides 1 and 9 (Table 1) were at the N terminus and near the C terminus of the cytokine, respectively, resulting from hydrolysis at three Lys residues. After 30 min of incubation, the carboxyl side of residue Arg116 was hydrolyzed, resulting in the release of peptide 10 (104EANQSTLENFLER116). Incubation for 1 h resulted in hydrolysis at the carboxyl side of residue Arg122, producing peptide 11 (123EKYSKCSS130), the C-terminal peptide. Two predicted peptide fragments, from a theoretical tryptic digest of IL-4 with an m/z value of over 600, Thr14-Arg48 and Asn90-Lys103, were not observed. The peptide fragments identified by peptide mass fingerprinting gave 58% sequence coverage of IL-4.
TABLE 1.
Sequences of identified fragments of human recombinant IL-4 and IL-5 after incubation with the RgpA-Kgp complexes
| Peptide no. | Sequence of identified fragmenta | Calculated MH+ | Observed MH+ | Time peptide first observed |
|---|---|---|---|---|
| IL-4 | ||||
| 1 | 1MHKCDITLQEIIK13 | 1571.83 | 1571.89 | 15 min |
| 2 | R48↓AATVLRQFYSHHEK62 | 1686.87 | 1686.91 | 15 min |
| 3 | R54↓QFYSHHEK62 | 1075.50 | 1075.51 | 15 min |
| 4 | R54↓QFYSHHEKDTR65 | 1447.67 | 1447.71 | 15 min |
| 5 | K62↓DTRCLGATAQQFHR76 | 1603.78 | 1603.82 | 15 min |
| 6 | R65↓CLGATAQQFHR76 | 1231.60 | 1231.63 | 15 min |
| 7 | K78↓QLIRFLK85 | 917.60 | 917.59 | 15 min |
| 8 | R82↓FLKRLDR89 | 947.58 | 947.57 | 15 min |
| 9 | K103↓EANQSTLENFLERLK118 | 1791.92 | 1791.96 | 15 min |
| 10 | K103↓EANQSTLENFLER116 | 1550.75 | 1550.86 | 30 min |
| 11 | R122↓EKYSKCSS130 | 931.42 | 931.46 | 1 h |
| IL-5 | ||||
| 12 | 1IPTEIPTSALVK12 | 1268.75 | 1268.80 | 15 min |
| 13 | 1IPTEIPTSALVKETLALLSTHR22 | 2390.37 | 2390.36 | 15 min |
| 14 | K12↓ETLALLSTHR22 | 1140.64 | 1140.67 | 15 min |
| 15 | R22↓TLLIANETLR32 | 1143.67 | 1143.69 | 15 min |
| 16 | R32↓IPVPVHK39 | 789.50 | 789.58 | 15 min |
| 17 | K39↓NHQLCTEEIFQGIGTLESQTVQGGTVER67 | 3074.49 | 3074.43 | 15 min |
| 18 | R67↓LFKNLSLIK76 | 1075.69 | 1075.67 | 15 min |
| 19 | R67↓LFKNLSLIKKYIDGQKK84 | 2036.23 | 2036.03 | 15 min |
| 20 | K70↓NLSLIKKYIDGQK83 | 1519.88 | 1519.88 | 15 min |
| 21 | K76↓KYIDGQKKKCGEER90 | 1681.87 | 1681.88 | 15 min |
| 22 | K83↓KKCGEER90 | 849.43 | 849.42 | 15 min |
| 23 | K83↓KKCGEERRR92 | 1161.63 | 1161.61 | 15 min |
| 24 | K83↓KKCGEER90 | |||
| | | 3922.33 | 3922.26 | 15 min | |
| K39↓NHQLCTEEIFQGIGTLESQTVQGGTVER67 | ||||
| 25 | K77↓YIDGQKKKCGEER90 | |||
| | | 4627.11 | 4627.15 | 15 min | |
| K39↓NHQLCTEEIFQGIGTLESQTVQGGTVER67 | ||||
| 26 | K76↓KYIDGQKKKCGEER90 | |||
| | | 4755.28 | 4755.28 | 15 min | |
| K39↓NHQLCTEEIFQGIGTLESQTVQGGTVER67 | ||||
| 27 | K77↓YIDGQKKKCGEER90 | 1553.77 | 1553.89 | 1 h |
| 28 | R91↓RVNQFLDYLQEFLGVMNTEWIIES115 | 2944.46 | 2944.57 | 1 h |
↓ indicates point of cleavage.
Incubation of IL-5 with the RgpA-Kgp complexes also revealed degradation at the first time point of 15 min (Table 1), with all digestion products observed within 1 h of incubation. After 15 min of incubation, IL-5 was hydrolyzed at the carboxyl side of five Arg residues and six Lys residues to yield peptides 12 to 23 (Table 1). In addition, three disulfide-linked peptides (linked through Cys44 and Cys86) were observed to be released from IL-5 in the first 15 min of hydrolysis (Table 1). After 1 h of incubation, peptides 27 and 28 from the C-terminal end of IL-5 were also observed. The peptide fragments identified by peptide mass fingerprinting gave complete sequence coverage of IL-5.
DISCUSSION
IL-4 and IL-5 are both Th2 anti-inflammatory cytokines that regulate a number of different cell types and their functions. IL-4 is a 14-kDa molecule primarily produced by T cells that has a range of functions, including the costimulation of B cells, T cells, and mast cells (3, 23, 27, 28), inducing antibody isotype switching to IgE, in mice to IgG1 (26), and in humans to IgG4 (18). IL-5 exists as a 34-kDa homodimer and is primarily produced by activated T cells (40); it regulates the production of eosinophils (32, 34, 43). In this study, IL-4 and IL-5 were both found to be degraded and inactivated by the major virulence factor of P. gingivalis, the RgpA-Kgp complexes, under physiologically relevant conditions. The studies were conducted with the TF1.8 T-cell line cultured in AIM-V medium containing, inter alia, 2% (wt/vol) serum albumin. This culture system reflects the environment of the interstitial fluid and physiologically relevant cytokine concentrations. Recently, O'Brien-Simpson et al. (21) have demonstrated the presence of the RgpA-Kgp complexes in gingival tissue from diseased sites in periodontitis patients. Thus, taken together, these results are consistent with the diffusion of the RgpA-Kgp complexes from subgingival dental plaque into the interstitial fluid of gingival tissue, resulting in the degradation and inactivation of at least some of the IL-4 and IL-5 present.
The effect of RgpA-Kgp on IL-4 and IL-5 was dependent on the RgpA-Kgp enzyme concentration and on the time of preincubation, which is consistent with degradation of the cytokines being the major mechanism of inactivation. This may indicate that the RgpA-Kgp complexes in periodontal tissues, even at low concentrations, are able to degrade the Th2 cytokines IL-4 and IL-5, thereby contributing to the disruption of the anti-inflammatory immune responses. Utilizing mass spectrometric analysis, hydrolysis of IL-4 at certain Arg and Lys residues within 15 min was confirmed, indicating that both the RgpA and Kgp proteinases were involved in cleavage of the cytokine. The IL-4 residues Glu10, Arg89, Arg122, Tyr125, and Ser126 have been reported to be critical for IL-4 receptor binding and transmembrane signaling (9, 10, 41). Within 15 min, the RgpA-Kgp complexes had hydrolyzed IL-4 to produce peptides containing Glu10 and Arg89 (peptides 1 and 8), and within 1 h, Arg122 was hydrolyzed to release peptide 11 containing Tyr125 and Ser126. This hydrolysis of the cytokine separated the critical residues required for IL-4 binding and signaling, which would account for the loss of IL-4 bioactivity.
IL-5, another anti-inflammatory cytokine, is often, but not always, coexpressed with IL-4 (33). Mass spectrometric analysis showed that IL-5 was degraded by the RgpA-Kgp complexes involving both proteinases, with all possible peptide fragments identified within 1 h. Although hydrolysis of IL-5 began within 15 min, it is apparent from the TF1.8 T-cell proliferation data that IL-5 was not as readily inactivated by RgpA-Kgp as IL-4. The cysteine residues (Cys44 and Cys86) of IL-5 have been previously reported to stabilize the IL-5 homodimer by the formation of a disulfide bond, and the homodimer structure is essential for biological function (14, 16, 39). Interestingly, disulfide bond formation between Cys44 and Cys86 was confirmed in this study by the identification of peptides 24 to 26 (Table 1). Furthermore, recombinant IL-5, which was used in this study, has been reported to be a glycoprotein (16). This may indicate that the homodimer structure of IL-5 and/or the glycosylation helps to protect the cytokine from rapid degradation by the complexes, which could explain the slightly greater resistance of the cytokine to inactivation. Peptides containing Cys44 and Cys86 not involved in a disulfide bond (peptides 17 and 21 to 27) (Table 1) were released by RgpA-Kgp, indicating that not all the IL-5 existed in the disulfide form. Furthermore, the full sequence coverage from the mass spectrometric analysis indicated that not all the IL-5 was glycosylated. The hydrolysis of IL-5 shown by the mass spectrometric analysis would account for the ultimate loss of TF1.8 T-cell proliferation observed with the RgpA-Kgp complexes at a 1:1 molar ratio.
A number of studies have reported that an anti-inflammatory cytokine response does not result in progression of chronic periodontitis, whereas an inflammatory response is associated with progression and severity of disease. Patients with chronic periodontitis are reported to express elevated levels of inflammatory cytokines in diseased gingival tissue in comparison to healthy tissue (13). In an experimentally induced model of chronic periodontitis in mice, T cells from mice intraorally challenged with P. gingivalis were shown to have a proinflammatory (Th1) cytokine profile, whereas immunization with the RgpA-Kgp complexes of P. gingivalis, which induced protection, resulted in a predominant Th2 (IL-4) cytokine response (20). In another study, mice that were polarized to produce a Th1 response showed exacerbated levels of disease upon challenge with P. gingivalis, in contrast to Th2-polarized mice, which experienced less disease (36). These studies are part of a larger body of work suggesting that an anti-inflammatory cytokine response is not associated with progression of periodontitis whereas an inflammatory response is associated with disease progression. In the current study, the results show that IL-4 and IL-5 are inactivated by the RgpA-Kgp complexes under physiologically relevant conditions and that both cytokines are degraded by the RgpA-Kgp proteinases. IL-4 plays a crucial role in the host anti-inflammatory responses, as it promotes the production of the antibodies (mouse IgG1 and human IgG4) that are reported to induce protection against disease (19, 20). Our studies and those of other research groups suggest that both anti-inflammatory and proinflammatory cytokines are degraded and inactivated by the proteinases of the RgpA-Kgp complexes, thereby possibly contributing to the dysregulation of the host immune response rather than polarizing it to one response or another. This dysregulation is likely to be localized to the site of infection, where there is a high level of pathogens in subgingival plaque and hence secreted proteinases in the subjacent tissues (21). In the tissues, IL-4 and IL-5, along with other cytokines, may be rapidly degraded and their bioavailability reduced. Immune dysregulation in chronic periodontitis has been proposed previously (13, 30, 31), and the results of this study support that proposal.
Acknowledgments
This project was supported by the Australian National Health and Medical Research Council (project no. 454475).
There are no conflicts of interest.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Andrian, E., D. Grenier, and M. Rouabhia. 2004. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect. Immun. 724689-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, P. J., M. Dixon, R. T. Evans, L. Dufour, E. Johnson, and D. C. Roopenian. 1999. CD4+ T Cells and the proinflammatory cytokines gamma interferon and interleukin-6 Contribute to alveolar bone loss in mice. Infect. Immun. 672804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, S. C., G. Sellge, A. Lorentz, W. Sebald, R. Raab, and M. P. Manns. 1999. IL-4 enhances proliferation and mediator release in mature human mast cells. Proc. Natl. Acad. Sci. USA 968080-8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkins, C. C., K. Platt, J. Potempa, and J. Travis. 1998. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J. Biol. Chem. 2736611-6614. [DOI] [PubMed] [Google Scholar]
- 5.Fishburn, C. S., J. M. Slaney, R. J. Carman, and M. A. Curtis. 1991. Degradation of plasma proteins by the trypsin-like enzyme of Porphyromonas gingivalis and inhibition of protease activity by a serine protease inhibitor of human plasma. Oral Microbiol. Immunol. 6209-215. [DOI] [PubMed] [Google Scholar]
- 6.Gregory, R. L., D. E. Kim, J. C. Kindle, L. C. Hobbs, and D. R. Lloyd. 1992. Immunoglobulin-degrading enzymes in localized juvenile periodontitis. J. Periodontal Res. 27176-183. [DOI] [PubMed] [Google Scholar]
- 7.Houri-Haddad, Y., W. A. Soskolne, E. Shai, A. Palmon, and L. Shapira. 2002. Interferon-gamma deficiency attenuates local P. gingivalis-induced inflammation. J. Dent. Res. 81395-398. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura, Y., S. Matono, Y. Aida, T. Hirofuji, and K. Maeda. 2002. Gingipains in the culture supernatant of Porphyromonas gingivalis cleave CD4 and CD8 on human T cells. J. Periodontal Res. 37464-468. [DOI] [PubMed] [Google Scholar]
- 9.Kruse, N., B. J. Shen, S. Arnold, H. P. Tony, T. Muller, and W. Sebald. 1993. Two distinct functional sites of human interleukin 4 are identified by variants impaired in either receptor binding or receptor activation. EMBO J. 125121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruse, N., H. P. Tony, and W. Sebald. 1992. Conversion of human interleukin-4 into a high affinity antagonist by a single amino acid replacement. EMBO J. 113237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lantz, M. S., R. W. Rowland, L. M. Switalski, and M. Hook. 1986. Interactions of Bacteroides gingivalis with fibrinogen. Infect. Immun. 54654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larjava, H., V. J. Uitto, M. Haapasalo, J. Heino, and M. Vuento. 1987. Fibronectin fragmentation induced by dental plaque and Bacteroides gingivalis. Scand. J. Dent. Res. 95308-314. [DOI] [PubMed] [Google Scholar]
- 13.Matsuki, Y., T. Yamamoto, and K. Hara. 1992. Detection of inflammatory cytokine messenger-RNA (mRNA)-expressing cells in human inflamed gingiva by combined in situ hybridization and immunohistochemistry. Immunology 7642-47. [PMC free article] [PubMed] [Google Scholar]
- 14.McKenzie, A. N., B. Ely, and C. J. Sanderson. 1991. Mutated interleukin-5 monomers are biologically inactive. Mol. Immunol. 28155-158. [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczyk-Pawlinska, J., J. Travis, and J. Potempa. 1998. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 440282-286. [DOI] [PubMed] [Google Scholar]
- 16.Minamitake, Y., S. Kodama, T. Katayama, H. Adachi, S. Tanaka, and M. Tsujimoto. 1990. Structure of recombinant human interleukin 5 produced by Chinese hamster ovary cells. J. Biochem. (Tokyo) 107292-297. [DOI] [PubMed] [Google Scholar]
- 17.Norusis, M. J. 1993. SPSS for Windows base systems user's guide, release 6.0. SPSS, Inc., Chicago, IL.
- 18.Nusslein, H. G., and H. L. Spiegelberg. 1990. Interleukin-4 induces both Igg4 and Ige secretion by peripheral-blood B-cells. J. Clin. Lab. Anal. 4414-419. [DOI] [PubMed] [Google Scholar]
- 19.O'Brien-Simpson, N. M., C. L. Black, P. S. Bhogal, S. M. Cleal, N. Slakeski, T. J. Higgins, and E. C. Reynolds. 2000. Serum immunoglobulin G (IgG) and IgG subclass responses to the RgpA-Kgp proteinase-adhesin complex of Porphyromonas gingivalis in adult periodontitis. Infect. Immun. 682704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien-Simpson, N. M., R. D. Pathirana, R. A. Paolini, Y.-Y. Chen, P. D. Veith, V. Tam, N. Ally, R. N. Pike, and E. C. Reynolds. 2005. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 1753980-3989. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien-Simpson, N. M., R. D. Pathirana, G. Walker, and E. C. Reynolds. 29 December 2008. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect. Immun. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed]
- 22.O'Brien-Simpson, N. M., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4409-426. [DOI] [PubMed] [Google Scholar]
- 23.O'Garra, A., D. J. Warren, M. Holman, A. M. Popham, C. J. Sanderson, and G. G. Klaus. 1986. Interleukin 4 (B-cell growth factor II/eosinophil differentiation factor) is a mitogen and differentiation factor for preactivated murine B lymphocytes. Proc. Natl. Acad. Sci. USA 835228-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oido-Mori, M., R. Rezzonico, P.-L. Wang, Y. Kowashi, J.-M. Dayer, P. C. Baehni, and C. Chizzolini. 2001. Porphyromonas gingivalis gingipain-R enhances interleukin-8 but decreases gamma interferon-inducible protein 10 production by human gingival fibroblasts in response to T-cell contact. Infect. Immun. 694493-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pathirana, R. D., N. M. O'Brien-Simpson, P. D. Veith, P. F. Riley, and E. C. Reynolds. 2006. Characterization of proteinase-adhesin complexes of Porphyromonas gingivalis. Microbiology 1522381-2394. [DOI] [PubMed] [Google Scholar]
- 26.Paul, W. E. 1991. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood 771859-1870. [PubMed] [Google Scholar]
- 27.Paul, W. E. 1987. Interleukin 4/B-cell stimulatory factor-I: one lymphokine, many functions. FASEB J. 1456-461. [DOI] [PubMed] [Google Scholar]
- 28.Paul, W. E., and J. Ohara. 1987. B-cell stimulatory factor-1/interleukin-4. Annu. Rev. Immunol. 5429-459. [DOI] [PubMed] [Google Scholar]
- 29.Pike, R. N., J. Potempa, W. McGraw, T. H. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J. Bacteriol. 1782876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabhu, A., B. S. Michalowicz, and A. Mathur. 1996. Detection of local and systemic cytokines in adult periodontitis. J. Periodont. 67515-522. [DOI] [PubMed] [Google Scholar]
- 31.Preiss, D. S., and J. Meyle. 1994. Interleukin-1-beta concentration of gingival crevicular fluid. J. Periodont. 65423-428. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson, C. J. 1992. Interleukin-5, eosinophils, and disease. Blood 793101-3109. [PubMed] [Google Scholar]
- 33.Sanderson, C. J., M. L. De Boer, M. S. Salerno, G. T. F. Schwenger, M. Mordvinov, V. A. Thomas, and S. Karlen. 1999. Regulation of interleukin-5 gene expression. Interleukin-5: from molecule to drug target for asthma, p. 267-287. In C. J. Sanderson (ed.), Lung biology in health and disease. Marcel Dekker, Inc., New York, NY.
- 34.Sanderson, C. J., D. J. Warren, and M. Strath. 1985. Identification of a lymphokine that stimulates eosinophil differentiation in vitro. Its relationship to interleukin 3, and functional properties of eosinophils produced in cultures. J. Exp. Med. 16260-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, C. F., E. J. Whitaker, B. F. Hammond, and R. W. Colman. 1993. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J. Biol. Chem. 2687935-7942. [PubMed] [Google Scholar]
- 36.Stashenko, P., R. B. Goncalves, B. Lipkin, A. Ficarelli, H. Sasaki, and A. Campos-Neto. 2007. Th1 immune response promotes severe bone resorption caused by Porphyromonas gingivalis. Am. J. Pathol. 170203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundqvist, G., A. Bengtson, and J. Carlsson. 1988. Generation and degradation of the complement fragment C5a in human serum by Bacteroides gingivalis. Oral Microbiol. Immunol. 3103-107. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, K., T. Azuma, H. Motohira, D. F. Kinane, and S. Kitetsu. 2005. The potential role of interleukin-17 in the immunopathology of periodontal disease. J. Clin. Periodontol. 32369-374. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, T., N. Yamaguchi, S. Mita, Y. Yamaguchi, T. Suda, A. Tominaga, Y. Kikuchi, Y. Miura, and K. Takatsu. 1990. Structural comparison of murine T-cell (B151K12)-derived T-cell-replacing factor (IL-5) with RIL-5: dimer formation is essential for the expression of biological activity. Mol. Immunol. 27911-920. [DOI] [PubMed] [Google Scholar]
- 40.Takatsu, K., A. Tominaga, N. Harada, S. Mita, M. Matsumoto, T. Takahashi, Y. Kikuchi, and N. Yamaguchi. 1988. T-cell-replacing factor (Trf) interleukin-5 (Il-5): molecular and functional properties. Immunol. Rev. 102107-135. [DOI] [PubMed] [Google Scholar]
- 41.Tony, H. P., B. J. Shen, P. Reusch, and W. Sebald. 1994. Design of human interleukin-4 antagonists inhibiting interleukin-4-dependent and interleukin-13-dependent responses in T-cells and B-cells with high efficiency. Eur. J. Biochem. 225659-665. [DOI] [PubMed] [Google Scholar]
- 42.Uitto, V. J., M. Haapasalo, T. Laakso, and T. Salo. 1988. Degradation of basement membrane collagen by proteases from some anaerobic oral micro-organisms. Oral Microbiol. Immunol. 397-102. [DOI] [PubMed] [Google Scholar]
- 43.Walker, C., J. C. Virchow, P. L. B. Bruijnzeel, and K. Blaser. 1991. T-cell subsets and their soluble products regulate eosinophilia in allergic and nonallergic asthma. J. Immunol. 1461829-1835. [PubMed] [Google Scholar]
- 44.Yun, P. L., A. A. Decarlo, C. Collyer, and N. Hunter. 2001. Hydrolysis of interleukin-12 by Porphyromonas gingivalis major cysteine proteinases may affect local gamma interferon accumulation and the Th1 or Th2 T-cell phenotype in periodontitis. Infect. Immun. 695650-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun, P. L., A. A. DeCarlo, and N. Hunter. 1999. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect. Immun. 672986-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]