Abstract
Francisella tularensis is a facultative intracellular pathogen and the causative agent of tularemia. We have shown that F. tularensis subspecies holarctica strain LVS prevents NADPH oxidase assembly and activation in human neutrophils, but how this is achieved is unclear. Herein, we used random transposon mutagenesis to identify LVS genes that affect neutrophil activation. Our initial screen identified carA, carB, and pyrB, which encode the small and large subunits of carbamoylphosphate synthase and aspartate carbamoyl transferase, respectively. These strains are uracil auxotrophs, and their growth was attenuated on cysteine heart agar augmented with sheep blood (CHAB) or in modified Mueller-Hinton broth. Phagocytosis of the uracil auxotrophic mutants triggered a respiratory burst in neutrophils, and ingested bacteria were killed and fragmented in phagosomes that contained superoxide. Conversely, phagocytosis did not trigger a respiratory burst in blood monocytes or monocyte-derived macrophages (MDM), and phagosomes containing wild-type or mutant bacteria lacked NADPH oxidase subunits. Nevertheless, the viability of mutant bacteria declined in MDM, and ultrastructural analysis revealed that phagosome egress was significantly inhibited despite synthesis of the virulence factor IglC. Other aspects of infection, such as interleukin-1β (IL-1β) and IL-8 secretion, were unaffected. The cultivation of carA, carB, or pyrB on uracil-supplemented CHAB was sufficient to prevent neutrophil activation and intramacrophage killing and supported escape from MDM phagosomes, but intracellular growth was not restored unless uracil was added to the tissue culture medium. Finally, all mutants tested grew normally in both HepG2 and J774A.1 cells. Collectively, our data demonstrate that uracil auxotrophy has cell type-specific effects on the fate of Francisella bacteria.
Francisella tularensis is a gram-negative facultative intracellular pathogen that is found throughout the Northern hemisphere. Tularemia was first described as a plaguelike illness of ground squirrels, and it is now clear that this organism infects a wide range of vertebrates and invertebrates and can persist in the environment for long periods of time (22, 56). Among F. tularensis subspecies, only F. tularensis subsp. tularensis (also called type A) and F. tularensis subsp. holarctica (type B) cause disease in healthy humans. Type A organisms are found predominantly in North America, whereas type B strains are more common in Europe and Asia (56). Natural reservoirs relevant to human infection include rabbits, beavers, mice, squirrels, deer, and Acanthamoeba castellani (56). In the United States, tularemia is most common in hunters who are exposed to the organism directly while skinning infected animals or indirectly via ticks that have fed on infected animals. Infection can also occur by inhalation of contaminated dust or ingestion of contaminated water, and recent outbreaks have occurred on Martha's Vineyard, MA (30). The clinical course of tularemia depends on the portal of entry and bacterial subspecies. Inhalation of as few as 10 type A organisms is sufficient to cause severe pneumonic tularemia, and at least 30% of untreated infections are fatal (55). In contrast, infections with type B strains can be severe but rarely result in death. An attenuated variant of F. tularensis subsp. holarctica was isolated in the 1960s (56). Because its mechanism of attenuation is unknown, this live vaccine strain (LVS) is not currently licensed for use in the United States. Nevertheless, LVS can cause fatal disease in mice and retains many features of virulent type A and type B organisms in vitro; for this reason LVS is widely studied, as is the related bacterium Francisella novicida (8, 28, 56, 59, 60).
The ability of macrophages and neutrophils (polymorphonuclear leukocytes [PMN]) to engulf and kill microbes is an essential component of innate defense, and the ability of F. tularensis to disrupt phagocyte function is a central aspect of virulence (3, 18, 56, 60). To gain entry into macrophages, F. tularensis binds complement receptor 3 (CR3) alone or together with the mannose receptor (MR) and scavenger receptor-A (SR-A) (7, 19, 62, 69). The fusion of nascent phagosomes with lysosomes is blocked, and bacteria escape this compartment to replicate in the cytosol (18, 20, 36, 56). Neutrophil phagocytosis of serum-opsonized F. tularensis is also mediated by CR3 (56), and our recent data demonstrate that LVS inhibits oxidative defense mechanisms by preventing NADPH oxidase assembly on forming phagosomes (3, 54). Consequently, LVS phagosomes are devoid of superoxide and other reactive oxygen species (ROS), and rapid blockade of NADPH oxidase activity is followed by phagosome escape and bacterial persistence in the cytosol (54). Several genes required for phagosome egress and intramacrophage growth have been defined, including the iglABCD operon in the Francisella pathogenicity island (FPI) (8, 13, 47, 59, 60), two identical copies of which are present in the genomes of F. tularensis subsp. tularensis and subsp. holarctica, including LVS (56). In contrast, the virulence factors that disrupt neutrophil NADPH oxidase activity are largely undefined. Previous studies indicate that in purified form, the F. tularensis acid phosphatase, AcpA, can prevent the activation of porcine neutrophils by phorbol esters and formyl peptides (64). However, the role of this enzyme in pathogenesis has been questioned, since transposon mutagenesis of F. novicida acpA does not diminish virulence in mice (10), and analysis of the F. tularensis proteome suggests that AcpA is nearly undetectable in LVS (40). These data may indicate that AcpA is sufficient but not necessary for inhibition of neutrophil NADPH oxidase activity. Additionally, the results of a recent study suggest that AcpA may affect other aspects of the Francisella life cycle, since deletion of acpA slows F. novicida's escape from macrophage phagosomes (58).
Herein we describe the use of random transposon mutagenesis to identify LVS genes that are required for inhibition of neutrophil NADPH oxidase activity. Our initial screen identified strains with mutations in carA, carB, and pyrB, and further characterization of these strains revealed that defects in pyrimidine biosynthesis have profound, cell type-specific effects on virulence and innate host defense, as indicated by the distinct fates of these mutants in human neutrophils, monocyte-derived macrophages (MDM), and epithelial cell lines.
MATERIALS AND METHODS
Materials.
Endotoxin-free Dulbecco's modified Eagle's medium (DMEM), HEPES-buffered RPMI 1640 medium, l-glutamine, phosphate-buffered saline (PBS), and Hank's buffered saline solution (HBSS) were from Cambrex (Walkersville, MD), and fetal bovine serum (FBS) was from HyClone (Logan, UT). Sodium pyruvate and nonessential amino acids were from Invitrogen (Carlsbad, CA). Bovine tendon collagen was obtained from Worthington Biochemicals (Lakewood, NJ). Defibrinated sheep blood was from Colorado Serum Co. (Denver, CO) or Remel (Lenexa, KS). Cysteine heart agar, Mueller-Hinton broth (MHB), Isovitalex, and rabbit anti-F. tularensis antiserum were purchased from Becton, Dickinson and Co. (Franklin Lakes, NJ). Murine anti-F. tularensis lipopolysaccharide (LPS) monoclonal antibodies (mAb) were from Biodesign International (Saco, ME). Ab specific for Francisella IglC were obtained from Karl Klose (University of Texas). Murine mAb specific for human gp91phox (54.1) and p22phox (44.1) were a gift from Algirdas Jesaitis and James Burritt (Montana State University, Bozeman, MT). Rabbit anti-GroEL polyclonal Ab (pAb) were from Sigma-Aldrich (St. Louis, MO). Fluorescein isothiocyanate and rhodamine-conjugated immunoglobulin G F(ab′)2 secondary Ab were from Jackson ImmunoResearch Laboratories (West Grove, PA). Paraformaldehyde and glutaraldehyde were from Electron Microscopy Sciences (Hatfield, PA). Restriction enzymes and other cloning reagents were from New England Biolabs (Beverly, MA). Interleukin-1β (IL-1β) and IL-8 enzyme-linked immunosorbent assay (ELISA) kits were from R&D Systems (Minneapolis, MN). Additional reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated.
Eukaryotic cell isolation and culture.
Heparinized venous blood was obtained from healthy volunteers using a protocol approved by the Institutional Review Board for Human Subjects at the University of Iowa, and all participants provided informed consent. Neutrophils and mononuclear cells were isolated using standard procedures (26, 54, 69). PMN (≥97% purity) were resuspended in HBSS without divalent cations, counted, and then diluted into appropriate media as indicated. Mononuclear cells were washed twice and then resuspended in HEPES-buffered RPMI supplemented with 20% fresh autologous serum (AS) and 2 mM l-glutamine at a concentration of 2 × 106/ml and differentiated into macrophages by incubation in Teflon jars for 5 to 7 days at 37°C (69).
Cells of the murine macrophagelike cell line J774A.1 (hereinafter J774 cells) were maintained in DMEM containing 10% heat-inactivated FBS and 2 mM l-glutamine as we described previously (70). HepG2 cells were grown in MEM supplemented with 10% FBS, 100 μM nonessential amino acids, and 1 mM sodium pyruvate (46).
Cultivation of bacteria and opsonization.
F. tularensis subsp. holarctica strain LVS was grown on 9% sheep blood-cysteine heart agar (CHAB) plates in an atmosphere of 5% CO2 in air at 37°C (54, 69). Bacteria were collected, washed three times with HBSS, opsonized with 50% fresh AS (30 min, 37°C), washed with HBSS again, and then quantified by measurement of absorbance at 600 nm (54, 69). Where indicated, bacteria were left unopsonized or were killed by exposure to 5 to 10% formalin (1 h at 37°C) and then washed again prior to opsonization (54).
Tn5 library generation.
A detailed description of the LVS Tn5 library has been published (16). In brief, a system was developed based on the R6K pir-dependent plasmid pRL27 wherein the Francisella groES promoter was placed upstream of the hyperactive Tn5 transposase in pRL27 to drive expression in Francisella bacteria. Plasmid instability was alleviated by placing Tn5 under the control of the lac operator and cloning lacIq into the plasmid so that, in Escherichia coli bacteria, transposase expression is repressed. Expression of the kanamycin resistance cassette in the transposon is controlled by the Francisella omp26 promoter to ensure a high level of expression after chromosomal insertion. The resulting plasmid, pBDJ303, was then fused to a temperature-sensitive Francisella plasmid (51) to generate the E. coli-Francisella shuttle plasmid pBB107. Plasmid DNA was introduced into LVS by cryotransformation (42, 56), and transformants were selected at 30°C on plates containing spectinomycin. Transformants were then grown at 41°C on agar supplemented with 25 μg/ml kanamycin to cure the delivery plasmid and select for Tn5 insertion mutants. The transposition frequency was ∼10−3, and analysis of 15 randomly selected clones demonstrated that insertions were distributed throughout the LVS chromosome (data not shown).
Measurement of the respiratory burst.
To screen the Tn5 library, each clone was grown on CHAB plates containing 10 μg/ml kanamycin. After 48 h at 37°C, bacteria were harvested directly from the plate, washed, and then opsonized as described above. Bacteria were mixed with 1 × 106 neutrophils (multiplicity of infection [MOI], 50:1) in phenol red-free RPMI supplemented with 1% human serum albumin and 50 μM luminol, and ROS production was measured over 60 min at 37°C as luminol chemiluminescence (CL) using a BMG-Labtech NOVOstar (Durham, NC) as described previously (23, 54). PMN stimulated with wild-type LVS, formalin-killed LVS (MOI, 50:1), or 200 nM phorbol myristate acetate (PMA) were used as controls (2, 54). ROS generated by monocytes and MDM were measured in a similar manner using luminol CL and/or lucigenin CL assays (23, 45, 54). In this case, zymosan (MOI, 5:1) and 200 nM PMA were used as positive controls and bacteria were used at an MOI of 50:1. For both PMN and macrophages, the accumulation of superoxide inside phagosomes was detected as a blue-black formazan precipitate following nitroblue tetrazolium (NBT) staining as we described previously (2, 32, 54).
DNA sequencing and Southern blotting.
Disrupted genes were identified by rescue cloning and DNA sequencing. For LVS Tn5 plasmid recovery, single colonies picked from 48-h kanamycin-CHAB plates were grown overnight in 2 ml MHB at 37°C with shaking at 225 rpm and then processed for genomic DNA isolation using the Qiagen DNeasy tissue kit protocol (Valencia, CA). DNA was digested with MfeI, ethanol precipitated, and ligated overnight at 16°C. E. coli strain DH5α λ-pir was transformed by electroporation with ligation reactions. Plasmid minipreps were prepared from individual kanamycin-resistant transformants by using a QIAprep spin miniprep kit and digested with MfeI to confirm the size, and the remainder was sequenced at the University of Iowa DNA Facility.
For Southern blotting, Tn5 mutants were harvested from 48-h kanamycin-CHAB plates and grown overnight in 2 ml MHB at 37°C with shaking. Genomic DNA was isolated by using a Qiagen DNeasy tissue kit according to the manufacturer's instructions. Five micrograms of DNA was digested with MfeI, separated on a 0.8% agarose gel, and transferred to a nylon membrane. Digoxigenin-labeled kan probe preparations and hybridizations were performed using the Roche (Basel, Switzerland) DIG system according to the manufacturer's instructions.
Growth in liquid media.
Mutant and wild-type LVS (grown on CHAB with or without 10 μg/ml kanamycin, respectively) were used to inoculate 5-ml aliquots of morpholinepropanesulfonic acid (MOPS)-buffered MHB supplemented with 0.1% glucose, 0.025% ferric pyrophosphate, and 2% Isovitalex, and cultures were grown overnight at 37°C with shaking at 225 rpm. Liquid cultures were diluted to an optical density at 600 nm of 0.1 and then grown with shaking at 37°C for up to 48 h. Where indicated, the growth medium was supplemented with 200 μg/ml uracil and/or 200 μg/ml arginine. At each time point, aliquots were removed and quantified by measurement of absorbance at 600 nm.
Intracellular growth and survival.
Francisella survival in PMN was quantified as we described previously, with minor modifications (54). In brief, PMN (5 × 106/ml in RPMI containing 10% AS) in polypropylene tubes were infected with bacteria at an MOI of 20:1. After 15 min at 37°C, free bacteria were removed by washing cells with HBSS and PMN were processed immediately or returned to 37°C in complete medium. After a total of 15 min, 2 h, or 4 h at 37°C, aliquots of infected cells were washed with HBSS and then lysed by exposure to 1% saponin for 15 min. Each sample was serially diluted in PBS and plated on CHAB supplemented with ±200 μg/ml uracil, and live organisms were quantified by enumeration of CFU (54).
To quantify intramacrophage growth, J774 cells or MDM were plated in 24-well dishes (1 × 105 cells/well) in tissue culture medium and allowed to adhere for 2 h at 37°C. Thereafter, monolayers were washed twice with PBS and infected with wild-type or mutant LVS at an MOI of 20:1 in RPMI plus 2.5% AS (for MDM) or DMEM plus 10% heat-inactivated FBS (for J774 cells). After 1 h at 37°C, the medium was discarded and cells were washed extensively with PBS to remove uningested bacteria. Fresh medium was added, and samples in each well were either lysed immediately (0.5% saponin for 5 min) or incubated for an additional 3 to 47 h prior to saponin treatment. Viable bacteria in diluted samples were quantified by enumeration of CFU as described above.
Similarly, 2 × 105 HepG2 cells were plated in 24-well dishes precoated with bovine tendon collagen (Worthington Biochemical, Lakewood, NJ) and incubated overnight at 37°C. Unopsonized bacteria were dispersed in warm cell culture medium at 2 × 107 CFU/ml and added to HepG2 monolayers to achieve an MOI of 100:1. To facilitate binding and invasion, plates were centrifuged at 600 × g for 4 min (4, 46). After 4 h at 37°C, cells were washed with PBS and then incubated for 1 h in culture medium containing 10 μg/ml gentamicin as we described previously (46). Thereafter, cells were washed with PBS and transferred to antibiotic-free medium at 37°C. At 6 to 48 h postinfection (hpi), cells were lysed with 1% saponin, diluted in PBS, and plated for enumeration of CFU as described above.
Microscopy.
Immunofluorescence and confocal microscopy were performed by using our established methods (2, 54, 69). LVS was detected in fixed and permeabilized cells by using rabbit anti-F. tularensis antiserum or anti-LPS mAb and secondary F(ab′)2 Ab conjugated to rhodamine. NADPH oxidase components p22phox and gp91phox were detected as we described previously (2, 26, 54), using mAb 44.1 and 54.1 and secondary Ab conjugated to fluorescein isothiocyanate. Confocal imaging and differential interference contrast optics were used to assess the accumulation of superoxide inside phagosomes after NBT staining (2, 54). Images were obtained by using a Zeiss LSM-510 confocal microscope (Carl Zeiss, Inc., Thornwood, NY). All experiments were performed in triplicate on at least three occasions using cells from different donors.
Transmission electron microscopy (TEM) was used to assess phagosome escape (54). After 6 to 9 h at 37°C, infected MDM were fixed and processed for TEM as we described previously (1, 4, 54). Samples were analyzed by using a Jeol JEM-1230 TEM (Jeol Institute, Peabody, MA). Bacteria in sections from at least 30 independent cells were scored for each condition. Phagosome escape was defined as the loss of >50% of the surrounding membrane (54).
Immunoblotting.
Bacteria harvested from agar plates were washed with PBS, normalized by measurement of absorbance at 600 nM, and lysed in radioimmunoprecipitation assay buffer containing protease inhibitors (1, 78). Total protein was determined by using a Pierce bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL). Proteins in normalized lysates were boiled in sample buffer, resolved by 10% or 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene fluoride membranes (78). Blocked membranes (1) were probed with rabbit anti-IglC pAb (from Karl Klose, University of Texas) or anti-GroEL pAb (Sigma) to ensure equal loading. Secondary Ab conjugated to horseradish peroxidase were obtained from Amersham (Fairfield, CT), and bands were detected by using Pierce SuperSignal West Pico substrate (Thermo Scientific, Rockford IL).
IL-1β ELISA.
MDM or monocytes attached to 24-well dishes (5 × 105 cells/well) were left untreated or infected with bacteria at an MOI of 100:1 in duplicate. After 1 h at 37°C, samples were washed with PBS to removed uningested organisms and then incubated for another 23 h at 37°C in fresh medium. Thereafter, supernatants were collected, and released IL-1β and IL-8 were measured by ELISA according to the manufacturer's instructions (R&D Systems, Minneapolis, MN).
Statistical analysis.
Statistical significance (P < 0.05) was assessed by using the f test, unpaired Student's t test, and one-way analysis of variance (54).
RESULTS
Identification of LVS Tn5 mutants that no longer prevent neutrophil activation.
We have shown previously that LVS inhibits the respiratory burst of human neutrophils (54). How this is achieved is not well defined, and our published data suggest that one or more bacterial factors prevent NADPH oxidase assembly on the LVS phagosome and also impair overall cell responsiveness (54). To identify bacterial factors involved in inhibition of PMN NADPH oxidase activity, a library of random Tn5 mutants generated in LVS (16; also see Materials and Methods) was screened by using the luminol CL assay to identify mutants that no longer prevented neutrophil activation. An initial screen of 480 clones identified three mutants that, like killed wild-type LVS (54), triggered a respiratory burst in PMN (Fig. 1A). Rescue cloning and DNA sequencing demonstrated that mutants 3E10, 5H9, and 1C9 contained transposon insertions in carB, carA, and pyrB (Table 1), which encode the large and small subunits of carbamoylphosphate synthetase and aspartate carbamoyl transferase, respectively. These enzymes catalyze the first two steps in the pyrimidine nucleotide biosynthetic pathway in many bacteria, including Francisella (63), and are required for the virulence of several pathogens, including Salmonella and E. coli (38, 50).
FIG. 1.
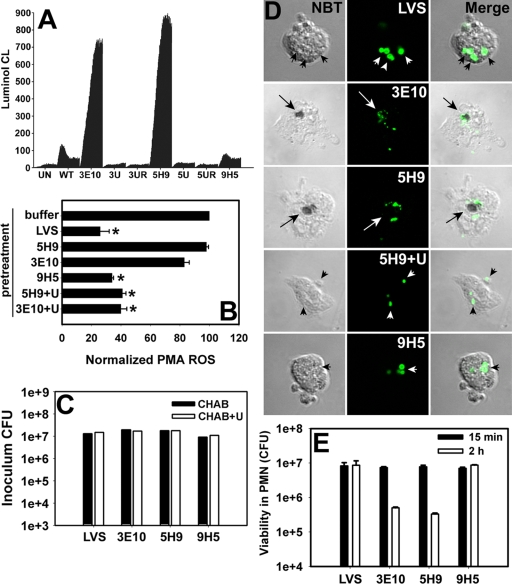
Identification of LVS mutants defective for inhibition of neutrophil NADPH oxidase activity. (A) ROS production by untreated PMN (UN) or cells infected with formalin-killed (FK) or live LVS (WT) or Tn5 mutant strain 1C9, 3E10, or 5H9 at an MOI of 50:1. Data indicate luminol CL in counts per second (cps) generated over 60 min at 37°C and are the means ± standard errors of the means of the results for triplicate samples from a representative experiment. (B) Southern blot of MfeI-digested genomic DNA probed to detect the kanamycin resistance cassette of the transposon, ahpA3. Lanes indicate molecular size standards (23.1, 9.4, 6.1, 4.3, 2.3, and 2.0 kbp; lane 1), LVS (lane 2), and mutants 3E10 (lane 3), 5H9 (lane 4), and 9H5 (lane 5). (C) Growth curves for LVS (WT), 3E10, and 5H9 in MHB or MHB supplemented with 200 μg/ml uracil (+Ura), 200 μg/ml arginine (+Arg) or both uracil and arginine (+/+). Data indicate the means ± standard errors of the means of the results for triplicate samples from one experiment that is representative of six. Where not visible, error bars are smaller than symbols. WT, wild type.
TABLE 1.
LVS Tn5 mutants
| Mutant | LVS gene | Tn5 insertion site | Protein encoded (gene name) |
|---|---|---|---|
| 1C9 | FTL0028 | 26,958 | Aspartate carbamoyltransferase (pyrB) |
| 3E10 | FTL0029 | 28,100 | Carbamoylphosphate synthase, large subunit (carB) |
| 5H9 | FTL0030 | 31,367 | Carbamoylphosphate synthase, small subunit (carA) |
| 9H5a | FTL0113 | 105,442 | Intracellular growth locus C (iglC) |
9H5 contains one normal copy and one disrupted copy of iglC and is used throughout this study as a control for the presence of Tn5 in the LVS genome.
Of note, strains with transposon insertion mutations in carA, carB, and pyrB of virulent F. tularensis subsp. tularensis SchuS4 (63) and an LVS carA mutant (52) have been identified by other research groups, and all these strains grow normally in J774 macrophages (52, 63). Conversely, replication of SchuS4 pyrB in HepG2 epithelial cells is somewhat impaired (63). We therefore studied the interactions of our mutants with primary human neutrophils and mononuclear phagocytes, as well as macrophage and hepatic cell lines, to better understand the roles these genes play in F. tularensis virulence.
Other studies of this transposon library have shown that one copy of iglC is disrupted in mutant 9H5 (Table 1 and Fig. 1B, lane 5). Because Tn5 insertion into one of the two identical copies of iglC in the LVS genome had no effect on the amount of IglC in bacterial lysates (see Fig. 5C below), we used 9H5 throughout this study as a control for the presence of Tn5 in the chromosome. Our data demonstrate that this single-iglC mutant was indistinguishable from wild-type LVS with respect to inhibition of neutrophil ROS production, serum resistance, and evasion of intracellular killing (see Fig. 2 below).
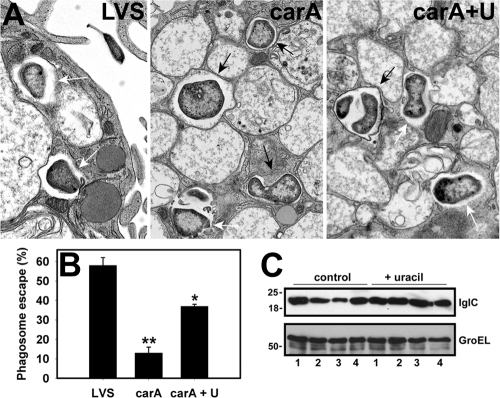
FIG. 5.
Quantitation of phagosome escape and IglC. (A) MDM were infected with LVS, the carA mutant, or the carA mutant that had been grown on uracil-CHAB (carA+U). After 9 h at 37°C, samples were fixed and processed for electron microscopy. Black arrows and white arrows indicate intraphagosomal and cytosolic bacteria, respectively. (B) Pooled data indicate the extent of bacterial escape from phagosomes in MDM. Error bars indicate means ± standard errors of the means. *, P < 0.05 versus results for LVS; **, P < 0.01 versus results for LVS and carA+U. (C) Bacteria were grown on CHAB plates (control) or CHAB supplemented with uracil (+ uracil) at 37°C. Immunoblots show IglC and the GroEL loading control in normalized bacterial lysates. Lanes 1 to 4 show LVS, 3E10 (carB), 5H9 (carA), and 9H5 (single-iglC mutant), respectively. Data shown are representative of two independent determinations.
FIG. 2.
Uracil auxotrophs activate PMN, and intracellular survival is impaired. (A) PMN were left untreated (UN) or infected with wild-type LVS (WT) or mutant 3E10 (carB), 5H9 (carA), or 9H5 (single-iglC mutant). Where indicated, mutants 3E10 and 5H9 were grown on CHAB supplemented with uracil (3U and 5U, respectively) or uracil and arginine (3UR and 5UR, respectively) prior to use. Data indicate luminol CL in counts per second and are the averages ± standard errors of the means (SEM) of the results for triplicate samples from one experiment that is representative of four. (B) Same as described for panel A except that neutrophils were stimulated with 200 nM PMA 10 min after infection with the indicated strain of bacteria. “+U” indicates 3E10 and 5H9 that were grown on uracil-CHAB. Data are normalized to the signal obtained for neutrophils stimulated with 200 nM PMA after pretreatment with buffer alone and are the averages ± SEM from three determinations. *, P < 0.05 versus results for PMA control. (C) Viability of opsonized bacteria prior to infection of neutrophils. Opsonized bacteria were washed, diluted, and then plated on CHAB or CHAB supplemented with uracil (CHAB+U) for enumeration of CFU. Data from a representative experiment are shown. (D) PMN were infected with the indicated strain of bacteria (shown in green), and intraphagosomal superoxide was detected by NBT staining. Arrows and arrowheads indicate NBT-positive and NBT-negative phagosomes, respectively. Note that bacteria in NBT-positive phagosomes appear fragmented. (E) Viability of bacteria inside PMN was quantified 15 min and 2 h after uptake by measurement of CFU. Data are the averages ± SEM of the results for triplicate samples from one experiment that is representative of four.
LVS carA, carB, and pyrB mutants are uracil auxotrophs.
Southern blotting of genomic DNA was used to quantify the number of transposition events that occurred in each mutant. Our data indicate that Tn5 was not present in LVS and inserted only once into the bacterial chromosome of carA (5H9), carB (3E10) (Fig. 1B), and pyrB (1C9) (not illustrated) mutants. These findings, together with the fact that carbamoylphosphate is an intermediate in the uracil and arginine biosynthetic pathways, suggested that these mutants may be uracil and/or arginine auxotrophs (14, 52, 63). Indeed, we found that our carA, carB, and pyrB mutants were unable to grow in Chamberlain's defined medium and grew more slowly than wild-type LVS on CHAB (data not shown). In modified MHB, carA and carB mutants exhibited no apparent growth defect during the first 8 h at 37°C (Fig. 1C). Thereafter, the growth of mutant cultures slowed markedly relative to the growth of wild-type LVS and ceased at ∼24 h (Fig. 1C). The addition of uracil to modified MHB (Fig. 1C) or CHAB (not shown) reversed this growth defect. On the other hand, arginine supplementation was without effect, and data obtained using medium supplemented with uracil and arginine were identical to those obtained for uracil alone (Fig. 1C). Similar data were also obtained for the pyrB mutant (not illustrated). Thus, the LVS mutants identified in our screen are uracil auxotrophs.
Uracil supplementation restores inhibition of the neutrophil respiratory burst.
Next, we determined the extent to which growth in uracil-supplemented medium would confer functional complementation of carA and carB with respect to inhibition of the neutrophil respiratory burst. To this end, PMN were infected with wild-type LVS, the carA or carB mutant, or mutant organisms that had been grown on CHAB supplemented with uracil and/or arginine, and ROS production was measured using the luminol assay. The data shown in Fig. 2A indicate that growth on medium supplemented with uracil (or uracil and arginine) restored the ability of carA and carB mutants to inhibit PMN ROS production.
A key feature of LVS is the fact that this organism not only prevents NADPH oxidase assembly at its own phagosome, but also inhibits PMN activation by heterologous stimuli (54). Therefore, we also determined the extent to which carA or carB mutants affected ROS production by neutrophils stimulated with 200 nM PMA. Concordant with our previously published data (54), the infection of neutrophils with LVS 10 min prior to stimulation with PMA inhibited phorbol ester-triggered ROS production by ∼74% compared with the PMA control. In marked contrast, carA and carB mutants were unable to retard cell activation by PMA unless bacteria had been grown on uracil-supplemented medium (Fig. 2B) (P < 0.05 versus results for PMA-only control, n = 3).
We have shown that LVS remains viable inside PMN for at least 12 h (54). The ability of uracil auxotrophs to trigger oxidant production suggested that their survival inside PMN might be compromised. To test this hypothesis, we quantified bacterial viability before and after infection. The data in Fig. 2C indicate that, like LVS, the carA and carB mutants were viable after complement opsonization (Fig. 2C) and as such were serum resistant (56). NBT staining and confocal microscopy of infected PMN confirmed the absence of superoxide inside LVS phagosomes (Fig. 2D) (54). In contrast, superoxide accumulated inside phagosomes of neutrophils infected with the carA and carB mutants, and bacteria appeared fragmented and degraded (Fig. 2D). Quantitation of viable intracellular bacteria confirmed that a majority of the carA and carB mutants were killed by 2 h after uptake by PMN (Fig. 2E), and similar data were obtained at 4 hpi (not shown). The accumulation of intraphagosomal superoxide and bacterial degradation and killing were ablated by growth of these mutants on uracil-supplemented CHAB prior to infection of PMN (Fig. 2D and data not shown). Collectively, these data suggest that inhibition of NADPH oxidase activity is essential for F. tularensis survival in neutrophils (54).
Mutants replicate normally in J774 macrophages and HepG2 cells.
It is established that Francisella bacteria replicate in the cytosol of primary macrophages and macrophagelike cell lines (18, 56, 66), and recent data indicate that these organisms also parasitize primary alveolar epithelial cells and epithelial cell lines (21, 35, 46, 57, 61). In this regard, it is noteworthy that disruption of pyrB in SchuS4 slows bacterial growth in HepG2 epithelial cells (63). In contrast, uracil auxotrophy does not impair the growth of SchuS4 or LVS in the J774 murine macrophagelike cell line (52, 63). We now show that the carA and carB mutants generated in this study replicated normally in both HepG2 (Fig. 3A) and J774 cells (Fig. 3B) during the first 48 hpi. Similar data were obtained for mutant 1C9 (pyrB mutant) (data not shown) and the single-iglC mutant, 9H5 (Fig. 3B).
FIG. 3.
Differential growth and survival of uracil auxotrophs in MDM, HepG2, and J774 cells. Bacteria were grown on CHAB (A to C) or CHAB plus uracil (D) prior to infection of host cells. (A) Intracellular growth and survival of unopsonized LVS, 5H9 (carA), and 3E10 (carB) in HepG2 cells over 48 h at 37°C. (B) Similar to experiment described for panel A except that J774 cells were infected with unopsonized LVS, 5H9, 3E10, or 9H5 (single-iglC mutant). (C, D) Extent of opsonized LVS, 5H9, 3E10, and 9H5 growth and survival in MDM over 48 h at 37°C. In panel C, “+UM” indicates the addition of uracil to RPMI at the time of infection with 5H9. In the experiment whose results are shown in panel D, all bacteria were cultivated on uracil-CHAB prior to infection of MDM in normal medium. For panels A to D, data indicate the means ± standard errors of the means of the results for triplicate samples from one experiment that is representative of three. Where not visible, error bars are smaller than symbols.
Growth and survival of uracil auxotrophs in primary human macrophages are impaired.
Whether carA, carB, or pyrB is required for LVS replication in primary human macrophages is unknown. To address this question, MDM were infected with opsonized bacteria in RPMI containing fresh AS, and live intracellular organisms were quantified after 1 to 48 h at 37°C. The data shown in Fig. 3C demonstrate that, in sharp contrast to the results with J774 cells, neither the carA nor the carB mutant replicated in MDM. Moreover, the number of viable, intracellular carA and carB mutants declined by 85.2% ± 12.5% (mean ± standard deviation) and 71.0% ± 23.9%, respectively, over this time course, whereas LVS bacteria (and the 9H5 control) increased by two orders of magnitude (P ≤ 0.002, n = 4). Similar data were obtained for MDM infected with the pyrB mutants and monocytes infected with the carB mutants (data not shown).
What accounts for this cell type-specific effect of uracil auxotrophy on bacterial growth and survival is unclear. The results of control experiments indicate that these divergent outcomes were not due to differences in the tissue culture medium, since bacterial replication was neither diminished by infection of J774 cells in RPMI nor restored by infection of MDM in DMEM (not shown). Thus, we hypothesized that essential nutrients may be more limited in quantity or less available to bacteria in MDM than in J774 cells. To test this hypothesis, we added uracil to RPMI at the time of infection or grew bacteria on uracil-CHAB prior to infection of MDM in normal medium. The data shown in Fig. 3C indicate that the addition of uracil to RPMI restored carA mutant replication in MDM. On the other hand, cultivation of carA and carB mutants on uracil-CHAB enhanced intra-MDM survival of mutant organisms during the first 24 hpi but was not sufficient to confer intracellular growth (Fig. 3D). The results of control experiments indicated that neither mode of uracil supplementation altered the intra-MDM growth of wild-type LVS or strain 9H5 (compare Fig. 3C and D). Altogether, our data confirm previous results for J774 cells (52, 63) and demonstrate for the first time that disruption of carA, carB, or pyrB ablates LVS replication in primary human macrophages.
Neither wild-type nor mutant bacteria trigger an oxidative burst in human monocytes and macrophages.
Like PMN, primary human macrophages and monocytes contain the phagocyte NADPH oxidase and have the capacity to generate large amounts of toxic ROS. On the other hand, most macrophagelike cell lines (including the J774 cells we use) have lost or downregulated this key element of host defense (24). Therefore, we considered the possibility that our carA, carB, and pyrB mutants may activate MDM and that exposure to intraphagosomal oxidants might account for their inability to replicate in this cell type.
Because effects of Francisella tularensis on MDM NADPH oxidase assembly and activation have not been described, we first determined whether phagocytosis of wild-type bacteria triggered the synthesis of superoxide. As expected, treatment of MDM with well-characterized NADPH oxidase agonists, such as unopsonized zymosan particles or PMA (33, 73, 74, 77), triggered a rapid and transient respiratory burst (Fig. 4A). NBT staining demonstrated specific accumulation of superoxide anions inside zymosan phagosomes (Fig. 4B), and confocal microscopy revealed profound enrichment of the NADPH oxidase subunits gp91phox and p22phox in the zymosan phagosome membrane (Fig. 4C). In contrast, MDM were not activated by LVS (Fig. 4A) (lucigenin CL, 93% ± 5% of baseline; n = 3), and phagosomes containing this organism lacked both NADPH oxidase subunits (Fig. 4C) and ROS (Fig. 4B). Here it is important to note that the basal lucigenin CL generated by uninfected macrophages (Fig. 4A) is not due to the NADPH oxidase but rather reflects the fact that superoxide is generated during normal mitochondrial metabolism in this cell type (29, 45, 65). Taken together, these data demonstrate for the first time that phagocytosis of LVS by MDM does not trigger NADPH oxidase assembly and activation at the F. tularensis phagosome.
FIG. 4.
Neither wild-type nor mutant bacteria trigger a respiratory burst in primary human monocytes and MDM. (A) Superoxide production by resting MDM (Un) or cells stimulated with 200 nM PMA, zymosan (Zymo., MOI of 5:1), LVS, the carB mutant, or formalin-killed LVS (FK-LVS) (each at an MOI of 50:1) was quantified at 30-s intervals over 60 min at 37°C as lucigenin CL. Data indicate the means of the results for triplicate samples from one experiment that is representative of four. (B) NBT staining of MDM demonstrates accumulation of superoxide inside zymosan phagosomes (Zym, black arrows) but not compartments containing LVS or the carB mutant (white arrowheads). Data shown are representative of three independent experiments. (C) Confocal sections of MDM stained to show gp91phox/p22phox heterodimers in green and LVS in red. Left panel, distribution of gp91phox/p22phox in uninfected MDM. Arrows indicate the plasma membrane, and the arrowhead indicates the biosynthetic-secretion pathway. Middle panel, MDM infected with zymosan for 15 min. Arrows indicate phagosomes. Right panel, MDM infected with LVS (red) for 15 min. Arrows indicate LVS phagosomes. (D) Superoxide production by resting monocytes (Un) or cells stimulated with zymosan (Zymo.; MOI, 5:1), LVS, the carA or carB mutant, or the single-iglC mutant (each at an MOI of 50:1) was quantified over 60 min at 37°C as lucigenin CL. Data indicate the means of the results for triplicate samples from one experiment that is representative of four. (E) Same as described for panel D except that monocyte ROS were detected by using luminol. Un, untreated.
Although phagocytosis of our carA, carB, and pyrB mutants and formalin-killed LVS all triggered a respiratory burst in PMN (Fig. 1A), we found that this was not the case in macrophages, as indicated by the results of lucigenin CL assays and NBT staining (Fig. 4A and B and data not shown). Similar data were obtained for peripheral blood monocytes using either the lucigenin or luminol CL assay to detect ROS (Fig. 4D and E). Thus, neither live nor formalin-killed LVS nor any of the mutant bacteria tested (including 9H5) triggered NADPH oxidase activation in primary human mononuclear phagocytes. Our findings identify differences in phagocyte responses to this organism and also suggest that in marked contrast to PMN, MDM and monocytes use oxidant-independent mechanisms for intracellular killing of carA, carB, and pyrB mutants.
Effects of uracil auxotrophy on IglC synthesis and phagosome escape.
A distinguishing feature of F. tularensis is its ability to escape from phagosomes in both macrophages and PMN (20, 36, 54, 56). Previous studies have shown that phagosome escape and intramacrophage growth require FPI genes, including iglC (8, 47, 56, 59). Of note in this regard is the fact that the FPI is duplicated in the genomes of F. tularensis subsp. tularensis and subsp. holarctica (including LVS) (56), and it has been suggested that changes in the expression of iglABCD can profoundly affect virulence (25, 59). At the same time, the fate of carA and carB in human macrophages is unknown, and whether uracil auxotrophy retards phagosome egress or the expression of virulence factors such as IglC has not been determined.
To begin to address these questions, we infected MDM with wild-type LVS or the carA mutant, and samples were fixed and processed for TEM at 6 to 9 hpi. In each case, phagosome escape was defined as the loss of at least 50% of the phagosome membrane (20, 54). By this assay, 58% ± 4% of wild-type bacteria reached the MDM cytosol by 9 hpi (Fig. 5A and B), confirming published data (20). Conversely, the ability of the carA mutant to breach the phagosome membrane was inhibited by 77%, and only 13% ± 3% of these organisms were free in the cytosol (P = 0.01 versus results for LVS) (Fig. 5A and B). These data are concordant with the 71% decline in the carA mutant's viability during the first 9 hpi that we described above (Fig. 3C), as well as the results of previous studies which indicate that killed LVS is retained in macrophage phagolysosomes (20, 56). Of note, the cultivation of the carA mutant on uracil-CHAB prior to infection supported bacterial survival in MDM (Fig. 3D) and increased phagosome escape (Fig. 5A and B); nevertheless, the fraction of mutant organisms in the cytosol did not reach wild-type levels.
As IglC is required for phagosome escape (47), we used immunoblotting to assess the amount of this protein in bacterial lysates. The data shown in Fig. 5C indicate that the carA and carB mutants contained ∼50% less IglC than LVS after growth on normal medium yet were indistinguishable from wild-type bacteria after cultivation on uracil-CHAB. Conversely, all bacteria tested contained similar amounts of GroEL regardless of the culture medium employed (Fig. 5C).
As noted above, other studies of this transposon library have shown that one copy of iglC is disrupted in mutant 9H5 (Table 1 and Fig. 1B, lane 5). Unlike the results for the carA and carB mutants, Tn5 insertion into one of the two copies of iglC in the LVS genome had no effect on the amount of IglC in bacterial lysates (Fig. 5C). For this reason, we used the single-iglC mutant 9H5 throughout this study as a control for the presence of Tn5 in the chromosome. As noted above, phagocytosis of 9H5 did not trigger a respiratory burst in monocytes (Fig. 4D and E), MDM (Fig. 4A), or neutrophils (Fig. 2A and B), and 9H5 survived and/or grew normally in all eukaryotic cell types tested (Fig. 2D and 3B to D). Thus, the phenotype of our uracil auxotrophs is not due to nonspecific effects of Tn5 insertion into the LVS genome.
Phagosome egress is sufficient to trigger IL-1β secretion.
Detection of whole bacteria or bacterial products by pattern recognition receptors in the cytosol of macrophages triggers activation of the inflammasome, which in turn leads to caspase-1-dependent processing and release of IL-1β and IL-18 (43, 71). Several research groups have shown that infection of human monocytes and dendritic cells or murine inflammatory macrophages with LVS or F. novicida causes caspase-1 activation and IL-1β secretion (12, 34, 44, 53). While it is clear that access to the cytosol is essential for IL-1β secretion triggered by Francisella bacteria, whether phagosome egress is sufficient in the absence of replication has not, to our knowledge, been tested directly (34, 39, 44, 53). We therefore infected MDM with wild-type or mutant bacteria and quantified the amount of mature IL-1β released into the tissue culture medium after 24 h at 37°C. The results of a representative experiment are shown in Fig. 6A. These data indicate that LVS-infected MDM secreted 60-fold more IL-1β than uninfected cells. Uracil auxotrophy had only a modest effect on IL-1β processing and release (19.8% ± 5.4% reduction) that was not statistically significant, despite the fact that intracellular replication of mutant bacteria was ablated (Fig. 3C) and phagosome egress was significantly impaired (Fig. 5A and B). To assess specificity, we also quantified IL-8 release. The data in Fig. 6B show that MDM infected with either wild-type or mutant bacteria secreted four- to fivefold more IL-8 than resting cells.
FIG. 6.

Both wild-type and mutant bacteria trigger secretion of IL-1β and IL-8 from mononuclear phagocytes. MDM (A and B) and monocytes (C) were infected with LVS, the carA mutant (5H9), the carB mutant (3E10), or the single-iglC mutant (9H5) as described in Materials and Methods, and at 24 hpi, the amount of mature IL-1β (A and C) or IL-8 (B) present in the tissue culture medium was quantified by ELISA. In each case, error bars indicate the means ± standard errors of the means of the results for triplicate samples from one experiment that is representative of three. Uninf, uninfected.
Previous studies have shown that the differentiation of human monocytes into macrophages compromises their ability to secrete IL-1β after stimulation with LPS (34, 75). Therefore, we also quantified the amount of IL-1β released by Francisella-infected monocytes. In this case, the tissue culture medium contained ∼2.0 ng/ml IL-1β, a 200-fold increase over the baseline amount and a >10-fold increase relative to the amount released by infected MDM, regardless of whether cells were infected with the carA or carB mutant, LVS, or the single-iglC mutant, 9H5 (Fig. 6C). These data indicate that monocytes secrete more IL-1β than MDM after infection with F. tularensis.
DISCUSSION
A distinguishing feature of F. tularensis is its ability to inhibit neutrophil NADPH oxidase assembly and activation (3, 54), but how this is achieved is unclear. The results of this study demonstrate that random transposon mutagenesis, together with the luminol CL assay, can be used to identify LVS genes that affect neutrophil function. We characterized carA, carB, and pyrB mutants and show that these uracil auxotrophs activate human neutrophils and are killed and fragmented inside phagosomes that contain superoxide. More importantly, we demonstrate that carA and carB mutants exhibit selective and cell type-specific virulence defects, as indicated by the fact that they grow normally in HepG2 and J774 cells and retain serum resistance, yet are killed not only by neutrophils but also by human monocytes and MDM. Furthermore, our data demonstrate that mononuclear phagocytes and neutrophils use distinct mechanisms to control the uracil auxotrophic mutants, since neither wild-type nor mutant bacteria triggered NADPH oxidase activation in MDM or monocytes. Rather, mutant bacteria were eliminated by oxidant-independent mechanisms that act in part via effects on phagosome escape.
How F. tularensis prevents NADPH oxidase activation and assembly at its own phagosome and elsewhere in infected cells is unclear, and the results of this study demonstrate that random transposon mutagenesis can be used to identify LVS genes that are required for the inhibition of the neutrophil respiratory burst. Our initial screen identified mutants with Tn5 insertions in carA, carB, and pyrB. We show that these organisms are uracil auxotrophs, phagocytosis of which triggers NADPH oxidase activation in PMN. Intracellular survival is diminished, and microscopy analysis revealed that mutant bacteria are digested in phagosomes that contain superoxide. Because the ability of LVS to prevent neutrophil activation by other stimuli is specific for live bacteria (54), it is likely that rapid elimination of mutant organisms accounts at least in part for the fact that neutrophils infected with mutants lacking functional carA, carB, or pyrB respond normally to other NADPH oxidase agonists, such as PMA. We also show that cultivation of the mutants on uracil-supplemented medium is sufficient to confer functional complementation with respect to the in vitro growth rate and blockade of neutrophil activation. Thus, our data are in agreement with the results of other studies which have shown that defects in nucleotide biosynthesis compromise the virulence of many intracellular pathogens, including various strains of Francisella (38, 50, 52, 61, 63, 72). Nevertheless, additional studies will be required to define F. tularensis virulence factors that directly inhibit NADPH oxidase. In future studies, it will also be of interest to determine whether the uracil auxotrophic carA, carB, or pyrB mutants are more sensitive than wild-type LVS to the toxic effects of oxidants, perhaps due to diminished levels of superoxide dismutase or KatG (6, 48).
Although the ability to produce toxic ROS is essential to the killing arsenal of macrophages and monocytes, as well as neutrophils, there are important differences in these phagocyte types. Pertinent here are the fact that macrophages lack myeloperoxidase, which catalyzes the conversion of hydrogen peroxide into HOCl and is required for the amplification of luminol CL (41), and the fact that CR3-mediated phagocytosis is not coupled to NADPH oxidase activation in mononuclear phagocytes as it is neutrophils (76). Ligation of MR and SR-A also fail to trigger a respiratory burst (5, 74). For this reason, CR3, MR, and SR-A have been described as “safe portals of entry,” and it is noteworthy that all of these receptors have been implicated in macrophage uptake of F. tularensis (7, 19, 62, 69) and are also exploited by other intracellular pathogens, such as Mycobacterium tuberculosis and Leishmania promastigotes (5, 11, 68, 74, 79).
At the same time, the results of this study provide the first direct evidence that phagocytosis of F. tularensis is not coupled to NADPH oxidase activation in monocytes or MDM. Our data demonstrate that uptake of LVS does not trigger a respiratory burst, as indicated by the results of the luminol and lucigenin CL assays and NBT staining, and our confocal data show that this is due, at least in part, to the fact that forming phagosomes exclude flavocytochrome b558 (gp91phox/p22phox heterodimers), which contains the redox center of the NADPH oxidase and is the docking site for cytosolic subunits p47phox, p67phox, p40phox, and Rac (3, 27). Because phagocytosis of carA, carB, and pyrB mutants and killed LVS also failed to activate monocytes and MDM for production of ROS, our findings also define potentially important differences in the responses of human mononuclear phagocytes and neutrophils to F. tularensis. Thus, when considered together, the data suggest that instead of actively inhibiting the respiratory burst, as occurs in PMN, wild-type and mutant LVS strains, as well as formalin-killed bacteria, enter monocytes and MDM silently, utilizing receptors that are not coupled to NADPH oxidase assembly and activation. In keeping with this model, we have shown that forming LVS phagosomes accumulate CR3 alone or together with MR (69), and previous studies of activated macrophages from p47phox- and/or inducible nitric oxide synthase-deficient mice indicate that an inability to produce NO profoundly diminishes Francisella killing, whereas the deletion of genes that encode NAPDH oxidase subunits is of little or no consequence (49).
Although the mechanisms used by MDM to control carA, carB, and pyrB mutants have not been precisely defined, our data indicate that a majority of mutant bacteria are killed, phagosome escape is significantly impaired, and organisms that reach the cytosol are unable to replicate despite their ability to trigger secretion of IL-8 and IL-1β. Among the genes required for phagosome egress and intracellular growth is the FPI gene iglC (13, 47, 56, 67), and we show here that IglC levels are reduced approximately ∼50% in carA and carB mutants relative to those in wild-type bacteria. Because cultivation of carA and carB mutants on uracil-CHAB restored normal IglC synthesis, enhanced phagosome escape, and ablated intracellular killing, our data lend further support to the notion that small changes in FPI gene expression can significantly alter virulence (25, 59). At the same time, our data also show that IglC is not sufficient for intracellular growth, since replication of carA and carB mutants in MDM was not achieved unless uracil was added to the tissue culture medium at the time of infection or shortly thereafter. Because we also found that IglC levels are normal in mutant 9H5, which contains one intact and one disrupted copy of iglC, and this strain exhibits no apparent virulence defects, it is likely that uracil deficiency also alters the expression of other, as-yet-unidentified virulence genes outside the iglABCD operon or the FPI, perhaps via effects on key regulatory factors, such as MglA, SspA, or FevR (9, 13, 15, 17, 37, 47). Our data also suggest that either sustained access to uracil or higher concentrations of this nutrient are required for intracellular replication, reminiscent of a study by Fortier et al. which demonstrated that the elevation of phagosome pH by using NH4Cl or related agents ablates LVS growth in murine peritoneal macrophages and enhances phagocytic killing by preventing bacterial access to transferrin iron (31). In this system, intramacrophage growth was restored by the addition of ferric pyrophosphate to the tissue culture medium.
In marked contrast to their growth in MDM and neutrophils, the carA and carB mutants grew normally in J774 and HepG2 cells and in this manner were indistinguishable from wild-type LVS. What accounts for this cell type-specific virulence defect is unclear, and our data indicate that differences in fitness are not due to the use of specific tissue culture media, bacterial uptake by phagocytosis versus invasion, host cell capacity for ROS production, or eukaryotic cell species. On the other hand, J774 and HepG2 cells are rapidly growing, transformed cell lines, whereas MDM, monocytes, and neutrophils are terminally differentiated and do not divide. Thus, it is attractive to predict that pyrimidine nucleotides may be more abundant or more available to bacteria in J774 and HepG2 cells, perhaps because of metabolic changes associated with their transformed state. In support of this notion, other groups have shown that similar mutants generated in SchuS4 or LVS exhibit no apparent virulence defects in J774 cells (52, 63), and Tempel et al. have shown that both wild-type F. novicida and a carB mutant grow much more readily in J774 and RAW264.7 macrophage cell lines than in primary murine bone marrow-derived macrophages (72). Whether an inability to synthesize uracil impairs LVS infection of primary human or murine epithelial cells remains to be determined.
In summary, we have shown that random transposon mutagenesis of LVS can be used to identify genes that affect neutrophil function and are required for survival in primary human phagocytes. Our initial screen identified carA, carB, and pyrB, and we show that defects in the pyrimidine biosynthetic pathway have pleiotropic effects that diminish LVS virulence in a cell type-specific manner. Additional studies of these organisms, in vivo and in vitro, may lead to the identification of virulence factors that specifically affect NADPH oxidase activity and phagosome escape.
Acknowledgments
We thank Randy Nessler of the University of Iowa Central Microscopy Research Facility for assistance with electron microscopy and Karl Klose (University of Texas, San Antonio) for the generous gift of antibodies to IglC.
This study was supported by NIH P01-AI44642 and R01-AI073835 (to L.-A.H.A.). R.L.M. is a postdoctoral fellow who was supported in part by NIH T32-AI07343 and is the recent recipient of a VA career development award. S.R.L. is supported by a U.S. Department of Homeland Security (DHS) graduate student fellowship and performed this research while on appointment as a DHS Fellow under the DHS Scholarship and Fellowship Program administered by the Oak Ridge Institute for Science and Education (ORISE) for DHS through an interagency agreement with the U.S. Department of Energy (DOE). ORISE is managed by Oak Ridge Associated Universities under DOE contract number DE-AC05-06OR23100.
All opinions expressed in this paper are the authors' and do not reflect the policies or views of DHS, DOE, or ORISE.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 9 February 2009.
REFERENCES
- 1.Allen, L.-A. H., and A. Aderem. 1995. Protein kinase C regulates MARCKS cycling between the plasma membrane and lysosomes in fibroblasts. EMBO J. 141109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, L.-A. H., B. R. Beecher, J. T. Lynch, O. V. Rohner, and L. M. Wittine. 2005. Helicobacter pylori disrupts NADPH oxidase targeting in human neutrophils to induce extracellular superoxide release. J. Immunol. 1743658-3667. [DOI] [PubMed] [Google Scholar]
- 3.Allen, L.-A. H., and R. L. McCaffrey. 2007. To activate or not to activate: distinct strategies used by Helicobacter pylori and Francisella tularensis to modulate the NADPH oxidase and survive in human neutrophils. Immunol. Rev. 219103-117. [DOI] [PubMed] [Google Scholar]
- 4.Allen, L.-A. H., L. S. Schlesinger, and B. Kang. 2000. Virulent strains of Helicobacter pylori demonstrate delayed phagocytosis and stimulate homotypic phagosome fusion in macrophages. J. Exp. Med. 191115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astarie-Dequeker, C., E. N. N′Diaye, V. Le Cabec, M. G. Rittig, J. Prandi, and I. Maridonneau-Parini. 1999. The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect. Immun. 67469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakshi, C. S., M. Malik, K. Regan, J. A. Melendez, D. W. Metzger, V. M. Pavlov, and T. J. Sellati. 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 1886443-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balagopal, A., A. S. MacFarlane, N. Mohapatra, S. Soni, J. S. Gunn, and L. S. Schlesinger. 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 745114-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker, J. R., and K. E. Klose. 2007. Molecular and genetic basis of pathogenesis in Francisella tularensis. Ann. N. Y. Acad. Sci. 1105138-159. [DOI] [PubMed] [Google Scholar]
- 9.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29247-259. [DOI] [PubMed] [Google Scholar]
- 10.Baron, G. S., T. J. Reilly, and F. E. Nano. 1999. The respiratory burst-inhibiting acid phosphatase AcpA is not essential for the intramacrophage growth or virulence of Francisella novicida. FEMS Microbiol. Lett. 17685-90. [DOI] [PubMed] [Google Scholar]
- 11.Blackwell, J. M. 1985. Role of macrophage complement and lectin-like receptors in binding Leishmania parasites to host macrophages. Immunol. Lett. 11227. [DOI] [PubMed] [Google Scholar]
- 12.Bolger, C. E., C. A. Forestal, J. K. Italo, J. L. Benach, and M. B. Furie. 2005. The live vaccine strain of Francisella tularensis replicates in human and murine macrophages but induces only the human cells to secrete proinflammatory cytokines. J. Leukoc. Biol. 77893-897. [DOI] [PubMed] [Google Scholar]
- 13.Bonquist, L., H. Lindgren, I. Golovliov, T. Guina, and A. Sjostedt. 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 763502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bringel, F., and J.-C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2-dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 692674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brotcke, A., and D. M. Monack. 2008. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 763473-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchan, B. W., M. K. McLendon, and B. D. Jones. 2008. Identification of differentially regulated Francisella tularensis genes using a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 742637-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charity, J. C., M. M. Costante-Hamm, E. L. Balon, D. H. Boyd, E. J. Rubin, and S. L. Dove. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemens, D. L., and M. A. Horwitz. 2007. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105160-186. [DOI] [PubMed] [Google Scholar]
- 19.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 735892-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craven, R. R., J. D. Hall, J. R. Fuller, S. Taft-Benz, and T. H. Kawula. 2008. Francisella tularensis invasion of lung epithelial cells. Infect. Immun. 762833-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross, J. T., Jr., and R. L. Penn. 2000. Francisella tularensis (tularemia), p. 2393-2402. In G. F. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, PA.
- 23.Dahlgren, C., and A. Karlsson. 1999. Respiratory burst in human neutrophils. J. Immmunol. Methods 2323-14. [DOI] [PubMed] [Google Scholar]
- 24.Damiani, G., C. Kiyotaki, W. Soeller, M. Sasada, J. Peisach, and B. R. Bloom. 1980. Macrophage variants in oxygen metabolism. J. Exp. Med. 152808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bruin, O., J. Ludu, and F. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLeo, F. R., L.-A. H. Allen, M. Apicella, and W. M. Nauseef. 1999. NADPH oxidase activation and assembly during phagocytosis. J. Immunol. 1636732-6740. [PubMed] [Google Scholar]
- 27.DeLeo, F. R., and M. T. Quinn. 1996. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J. Leukoc. Biol. 60677-691. [DOI] [PubMed] [Google Scholar]
- 28.Elkins, K. L., S. C. Cowley, and C. M. Bosio. 2007. Innate and adaptive immunity to Francisella. Ann. N. Y. Acad. Sci. 1105284-324. [DOI] [PubMed] [Google Scholar]
- 29.Esterline, R. L., and M. A. Trush. 1989. Lucigenin chemiluminescence and its relationship to mitochondrial respiration in phagocytic cells. Biochem. Biophys. Res. Commun. 159584. [DOI] [PubMed] [Google Scholar]
- 30.Feldman, K. A., R. E. Enscore, S. L. Lathrop, B. T. Matyas, M. McGuill, M. E. Schriefer, D. Stiles-Enos, D. T. Dennis, L. R. Petersen, and E. B. Hayes. 2001. An outbreak of primary pneumonic tularemia on Martha's Vineyard. N. Engl. J. Med. 3451601-1606. [DOI] [PubMed] [Google Scholar]
- 31.Fortier, A. H., D. A. Leiby, R. B. Narayanan, E. Asafoadjei, R. M. Crawford, C. A. Nacy, and M. S. Meltzer. 1995. Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect. Immun. 631478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallois, A., J. R. Klein, L.-A. H. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 1665741-5748. [DOI] [PubMed] [Google Scholar]
- 33.Gantner, B. N., R. M. Simmons, S. J. Canavera, S. Akira, and D. M. Underhill. 2003. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 1971107-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrilin, M. A., I. J. Bouakl, N. L. Knatz, M. D. Duncan, M. W. Hall, J. S. Gunn, and M. D. Wewers. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1β processing and release. Proc. Natl. Acad. Sci. USA 103141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentry, M., J. Taormina, R. B. Pyles, L. Yeager, M. Kirtley, V. L. Popov, G. Klimpel, and T. Eaves-Pyles. 2007. Role of primary human alveolar epithelial cells in host defense against Francisella tularensis infection. Infect. Immun. 753969-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guina, T., D. Radulovic, A. J. Bahrami, D. L. Bolton, L. Rohmer, K. A. Jones-Isaac, J. Chen, L. A. Gallagher, B. Gallis, S. Ryu, G. K. Taylor, M. J. Brittnacher, C. Manoil, and D. R. Goodlett. 2007. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J. Bacteriol. 1896580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 371417-1435. [DOI] [PubMed] [Google Scholar]
- 39.Henry, T., A. Brotcke, D. S. Weiss, L. J. Thompson, and D. M. Monack. 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernychova, L., P. Halada, A. Macela, M. Kroca, T. Johansson, and M. Malina. 2001. Construction of a Francisella tularensis two dimensional electrophoresis database. Proteomics 1508-515. [DOI] [PubMed] [Google Scholar]
- 41.Johansson, A., and C. Dahlgren. 1992. Differentiation of human peripheral blood monocytes to macrophages is associated with changes in the cellular respiratory burst activity. Cell Biochem. Funct. 1087-93. [DOI] [PubMed] [Google Scholar]
- 42.Lai, X. H., I. Golovliov, and A. Sjostedt. 2004. Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb. Pathog. 37225-230. [DOI] [PubMed] [Google Scholar]
- 43.Lamkanfi, M., T.-D. Kanneganti, L. Franchi, and G. Nunez. 2007. Caspase-1 inflammasomes in infection and inflammation. J. Leukoc. Biol. 82220-225. [DOI] [PubMed] [Google Scholar]
- 44.Li, H., S. Nookala, X. R. Bina, J. E. Bina, and F. Re. 2006. Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J. Leukoc. Biol. 80766-773. [DOI] [PubMed] [Google Scholar]
- 45.Li, Y., H. Zhu, P. Kuppusamy, V. Roubaud, J. L. Zweier, and M. A. Trush. 1998. Validation of lucigenin (Bis-N-methylacridinium) as a chemilumigenic probe for detecting superoxide anion radical production by enzymatic and cellular systems. J. Biol. Chem. 2732015-2023. [DOI] [PubMed] [Google Scholar]
- 46.Lindemann, S. R., M. K. McLendon, M. A. Apicella, and B. D. Jones. 2007. An in vitro model system used to study adherence and invasion of Francisella tularensis live vaccine strain in nonphagocytic cells. Infect. Immun. 753178-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindgren, H., I. Golovliov, V. Baranov, R. K. Ernst, M. Telepnev, and A. Sjostedt. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53953-958. [DOI] [PubMed] [Google Scholar]
- 48.Lindgren, H., H. Shen, C. Zingmark, I. Golovliov, W. Conlan, and A. Sjostedt. 2007. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 751303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindgren, H., L. Stenman, A. Tarnvik, and A. Sjostedt. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7467-475. [DOI] [PubMed] [Google Scholar]
- 50.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 707511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier, T. M., R. Pechous, M. Casey, T. C. Zahrt, and D. W. Frank. 2006. In vivo himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 721878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariathasan, S., D. S. Weiss, V. M. Dixit, and D. M. Monack. 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 2021043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCaffrey, R. L., and L.-A. H. Allen. 2006. Pivotal advance: Francisella tularensis evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 801224-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCrumb, F. R. 1961. Aerosol infection of man with Pasturella tularensis. Bacteriol. Rev. 25262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLendon, M. K., M. Apicella, and L.-A. H. Allen. 2006. Francisella tularensis: taxonomy, genetics and immunopathogenesis of a potenial agent of biowarfare. Annu. Rev. Microbiol. 60167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melillo, A., D. D. Sledjeski, S. Lipski, R. M. Wooten, V. Basrur, and E. R. Lafontaine. 2006. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263102-108. [DOI] [PubMed] [Google Scholar]
- 58.Mohapatra, N. P., A. Balagopal, S. Soni, L. S. Schlesinger, and J. S. Gunn. 2007. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect. Immun. 75390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nano, F., and C. Schmerk. 2007. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105122-137. [DOI] [PubMed] [Google Scholar]
- 60.Oyston, P. C. F. 2008. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J. Med. Microbiol. 57921-930. [DOI] [PubMed] [Google Scholar]
- 61.Pechous, R. D., T. R. McCarthy, N. P. Mohapatra, S. Soni, R. M. Penoske, N. H. Salzman, D. W. Frank, J. S. Gunn, and T. C. Zahrt. 2008. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE 3e2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pierini, L. M. 2006. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell. Microbiol. 81361-1370. [DOI] [PubMed] [Google Scholar]
- 63.Qin, A., and B. Mann. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiology 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reilly, T. J., G. S. Baron, F. E. Nano, and M. S. Kuhlenschmidt. 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 27110973-10983. [DOI] [PubMed] [Google Scholar]
- 65.Rembish, S. J., and M. A. Trush. 1994. Further evidence that lucigenin-derived chemiluminescence monitors mitochondrial superoxide generation in rat alveolar macrophages. Free Radic. Biol. Med. 17117-126. [DOI] [PubMed] [Google Scholar]
- 66.Santic, M., M. Molmeret, K. E. Klose, and Y. Abu Kwaik. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 1437-44. [DOI] [PubMed] [Google Scholar]
- 67.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7969-979. [DOI] [PubMed] [Google Scholar]
- 68.Schlesinger, L. S. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 1502920-2930. [PubMed] [Google Scholar]
- 69.Schulert, G. S., and L. A. H. Allen. 2006. Differential infection of mononuclear phagoctyes by Francisella tularensis: role of the macrophage mannose receptor. J. Leukoc. Biol. 80563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz, J. T., and L.-A. H. Allen. 2006. Role of urease in megasome formation and Helicobacter pylori survival in macrophages. J. Leukoc. Biol. 791214-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sutterwala, F. S., Y. Ogura, and R. A. Flavell. 2007. The inflammasome in pathogen recognition and inflammation. J. Leukoc. Biol. 82259-264. [DOI] [PubMed] [Google Scholar]
- 72.Tempel, R., X.-H. Lai, L. Crosa, B. Kozlowicz, and F. Heffron. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 745095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Underhill, D. M. 2003. Toll-like receptors: networking for success. Eur. J. Immunol. 331767-1775. [DOI] [PubMed] [Google Scholar]
- 74.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20825-852. [DOI] [PubMed] [Google Scholar]
- 75.Wewers, M. D., and D. J. Herzyk. 1989. Alveolar macrophages differ from blood monocytes in human IL-1 beta release. Quantitation by enzyme-linked immunoassay. J. Immunol. 1431635-1641. [PubMed] [Google Scholar]
- 76.Wright, S. D., and S. C. Silverstein. 1983. Receptors for C3b and C3bi promote phagocytosis but not the release of toxic oxygen from human phagocytes. J. Exp. Med. 1582016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yagisawa, M., A. Yuo, M. Yonemaru, S. Imajoh-Ohmi, S. Kanegasaki, Y. Yazaki, and F. Takaku. 1996. Superoxide release and NADPH oxidase components in mature human phagocytes: correlation between functional capacity and amount of functional proteins. Biochem. Biophys. Res. Commun. 228510-516. [DOI] [PubMed] [Google Scholar]
- 78.Yang, C., S. Mora, J. W. Ryder, K. J. Coker, P. Hansen, L.-A. H. Allen, and J. E. Pessin. 2001. VAMP3 null mice display normal constitutive, insulin-, and exercise-regulated vesicle trafficking. Mol. Cell. Biol. 211573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zimmerli, S., S. Edwards, and J. D. Ernst. 1996. Selective receptor blockade during phagocytosis does not alter the survival and growth of Mycobacterium tuberculosis in human macrophages. Am. J. Respir. Cell Mol. Biol. 15760-770. [DOI] [PubMed] [Google Scholar]