Abstract
Since 1998, blooms of Alexandrium catenella associated with paralytic shellfish poisoning have been repeatedly reported for Thau Lagoon (French Mediterranean coast). Based on data obtained for rRNA gene markers, it has been suggested that the strains involved could be closely related to the Japanese temperate Asian ribotype of the temperate Asian clade. In order to gain more insight into the origin of these organisms, we carried out a genetic analysis of 61 Mediterranean and 23 Japanese strains using both ribosomal and microsatellite markers. Whereas the phylogeny based on ribosomal markers tended to confirm the previous findings, the analysis of microsatellite sequences revealed an unexpected distinction between the French and Japanese populations. This analysis also highlighted great intraspecific diversity that was not detected with the classical rRNA gene markers. The Japanese strains are divided into two differentiated A. catenella lineages: the Sea of Japan lineage and the east coast lineage, which includes populations from the Inland Sea and the Pacific Ocean. A. catenella strains isolated from Thau Lagoon belong to another lineage. These findings indicate that microsatellite markers are probably better suited to investigations of the population genetics of this species that is distributed worldwide. Finally, application of the population genetics concepts available for macroorganisms could support new paradigms for speciation and migration in phytoplankton assemblages.
Toxic phytoplankton blooms, also known as harmful algal blooms (HABs), have long been considered sporadic phenomena. These worldwide events occur particularly in coastal and confined waters when HAB species bloom or emerge from their “hidden flora status” (3). The increasing occurrence of these organisms is a significant and expanding threat to health and the fisheries and shellfish industries (21). Many toxic phytoplankton species involved in these toxic events belong to the class Dinophyceae (49). In French coastal waters, the genus Alexandrium Halim is represented by Alexandrium minutum on the North Brittany coast and by Alexandrium catenella (Whedon and Kofoid) Balech on the northwestern Mediterranean coast; both of these species produce neurotoxins responsible for paralytic shellfish poisoning (18). On the French Mediterranean coast, A. catenella was first observed in 1998 in Thau Lagoon, and it was responsible for the first toxic event at that time (P. Masselin, Z. Amzil, E. Abadie, E. Nézan, C. Le Bec, A. Carreras, C. Chiantella, and P. Truquet, presented at the IXth International Conference on Harmful Algal Blooms, Hobart, Tasmania, Australia, 2000). Since then, this species has repeatedly produced toxic blooms (in 2001, 2003, 2004, 2005, and 2007), particularly in the autumn and/or spring, during which shellfish removal, a significant economic activity in this area, was forbidden (25). Moreover, it is regularly detected along the neighboring Catalan coast (51).
Previous studies based on rRNA gene sequencing have highlighted the existence of a complex consisting of morphologically similar species named the “Alexandrium tamarense complex.” Within this complex, which included A. tamarense, A. catenella, and Alexandrium fundyense, the ribotypes were grouped into phylogeographic clusters closely related to geographic zones defined by the main oceanic coastal areas (18, 43, 45). These studies described the obvious spread of A. catenella by human activities, such as shipping or shellfish aquaculture (6). Lilly et al. (28) assumed that A. catenella could have recently been introduced into Thau Lagoon by ballast water from Japan. Indeed, paralytic shellfish poisoning events involving A. catenella have frequently caused damage in Japanese coastal waters over the last 25 years (54). This hypothesis was recently supported after a clear link was established between northwestern Mediterranean and Japanese A. catenella strains based on phylogenetic analyses of the 5.8S rRNA gene and internal transcribed spacer (ITS) regions (38).
Microsatellite markers, which are commonly used to analyze the population genetics of various terrestrial and aquatic macroorganisms (24), have recently been developed for phytoplankton species, including the diatoms Ditylum brightwellii (39-41) and Pseudo-nitzschia pungens (11) and the coccolithophore Emiliania huxleyi (23). These markers are recognized as valuable tools for understanding the genetic structure of phytoplankton populations at both temporal and spatial scales. For the Dinophyceae, many microsatellite markers have recently been developed in order to gain more insight into the development and dynamics of blooms of members of the genus Alexandrium, including A. tamarense, North American and Temperate Asian ribotypes (2, 30), A. minutum (32), and A. catenella (33). In the first study of the A. tamarense population, microsatellite markers revealed some correlation between genetic and geographic distances, as well as several segregated populations, in coastal waters around Japan (31).
In the present survey, we investigated the inter- and intraspecific diversity of French Mediterranean and Japanese strains by performing a comparative genetic analysis of 12 microsatellite markers developed for A. catenella (33) with population genetics tools. The phylogenetic relationships of these strains were established using the ribosomal marker spanning the region including ITS1, the 5.8S rRNA gene, ITS2, and the D1/D3 28S rRNA gene. In this survey we tried to measure the genetic diversity of French and Japanese A. catenella strains in order to explore the Japanese origin of the French Mediterranean strains; this study was also the first opportunity to check the ploidy level of the different phases of this dinoflagellate's life cycle.
MATERIALS AND METHODS
Sampling sites and isolate collection.
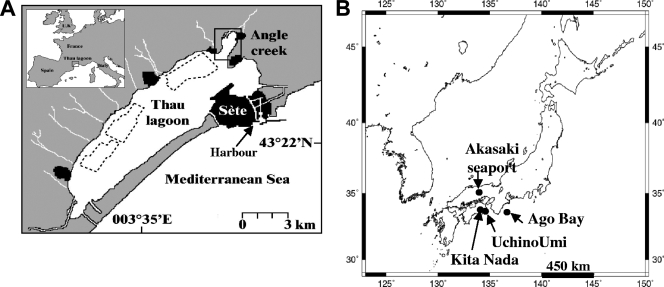
Thau Lagoon is the deepest marine lagoon located on the French Mediterranean coast (43°24′N, 3°36′E) and covers 75 km2, one-fifth of which is occupied by shellfish farming structures (Fig. 1). This lagoon is connected to the Mediterranean Sea by three channels, one of which opens into the Sète international harbor. Water and sediment samples were collected in Angle Creek, where A. catenella blooms are usually initiated. A. catenella cultures were developed from single vegetative cells (ACTEM) or single resting cysts (ACTKM) (Table 1). Resting cysts were isolated from the first 3 cm of sediment cores in April 2004. Isolation and germination of these cysts were performed as described by Genovesi-Giunti et al. (16). Planktonic vegetative cells were isolated from the bloom that occurred in November 2004. A. catenella cultures were grown in enriched natural seawater (22) at 20°C with a photocycle consisting of 12 h of light and 12 h of darkness, using a photon flux density of ∼100 μmol m−2 s−1. After centrifugation at 1,800 × g for 3 min, cultures were frozen at −20°C. Japanese strains were isolated from four locations, the Akasaki seaport in the Sea of Japan (35°51′11‴N, 133°65′69‴E), Uchino-Umi (34°22′03‴N, 134°60′57‴E) and Kita Nada (34°21′44‴N, 134°44′86‴E) in the Inland Sea, and Ago Bay (34°28′36‴N, 136°80′04‴E) on the Pacific coast (Fig. 1 and Table 1). A total of 61 monoclonal cultures from Thau Lagoon and 23 monoclonal cultures from Japanese waters and sediment were analyzed in this study (Table 1).
FIG. 1.
Sampling locations in France (A) and in Japan (B). Panel A reprinted from reference 48 with permission of the publisher; panel B courtesy of Satoshi Nagai.
TABLE 1.
Designations and isolation locations for the French and Japanese A. catenella strains analyzed in this study
| Strain(s) | n | Sampling year | Locality | Prefecture | Source |
|---|---|---|---|---|---|
| France | |||||
| ATTL01 | 1 | 1998 | Thau Lagoon | Seawater | |
| ACT1, ACT2 | 2 | 2002 | Thau Lagoon | Sediment | |
| ACT3 | 1 | 2003 | Thau Lagoon | Seawater | |
| ACTEM1 to -47 | 47 | 2004 | Thau Lagoon | Seawater | |
| ACTKM1 to -10 | 10 | 2004 | Thau Lagoon | Sediment | |
| Japan | |||||
| AC0202-01 to -18 | 6 | 2001 | Uchino-Umi | Tokushima | Sediment |
| AC0206-06 to -20 | 7 | 2002 | Ago Bay | Mie | Seawater |
| AC0310-01 to -14 | 4 | 2003 | Kita Nada | Kagawa | Sediment |
| AC0409-02 to -13 | 6 | 2004 | Akasaki Seaport | Tottori | Sediment |
Molecular methods.
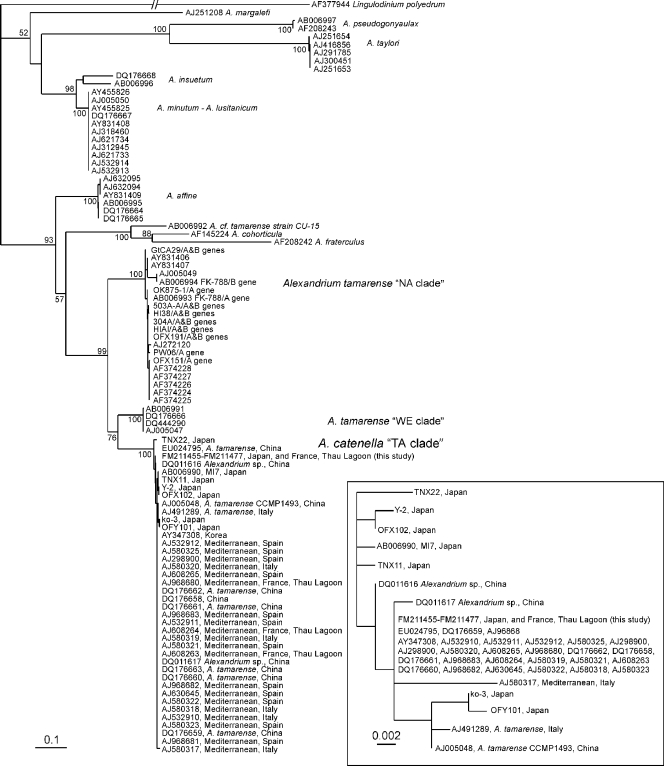
Total genomic DNA was extracted from monoclonal cultures using the classical phenol-chloroform method (42). rRNA gene sequencing was carried out for 23 selected A. catenella strains (11 strains from Thau Lagoon and 12 strains from Japan). An rRNA gene fragment spanning the region including ITS1, the 5.8S rRNA gene, ITS2, and the D1/D3 28S rRNA gene (∼1,500 bp) was PCR amplified with our novel forward primer 18S-ITS1-Ac-F (5′-CTTAGAGGAAGGAGAAGTCG-3′) specifically designed to cover the full length of the ITS1 region and reverse primer 28S-D3B-R (5′-TCGGAGGGAACCAGCTACTA-3′) (36). PCR amplification was performed using the proofreading DNA polymerase PrimeSTAR HS (Takara Bio Inc., Otsu, Shiga, Japan) and the following conditions: 95°C for 2 min, 35 cycles of 95°C for 30 s, 55°C for 15 s, and 72°C, 2 min, and a final extension at 72°C for 5 min. The PCR products checked on an agarose electrophoresis gel, and those containing a single band were purified using a MinElute PCR purification kit (Qiagen, Hilden, Germany). The purified PCR products were used as templates for direct sequencing using the two PCR primers and an additional internal primer. Sequencing was performed by Macrogen (Seoul, South Korea) using an ABI 3730XL genetic analyzer (Applied Biosystems). The sequence data were assembled using the software package Vector NTI (Invitrogen, Carlsbad, CA). Nucleotide sequence alignment, including alignment of data retrieved from GenBank, was performed using ClustalX (50). A maximum likelihood phylogenetic analysis was carried out using PHYML v2.4.4 (20) as follows: alignment spanning 515 A. catenella positions (ITS1, 5.8S rRNA gene, ITS2), 1,000 bootstraps, GTR substitution model, and an “invariant + Γ” among-site substitution rate distribution model approximated by eight category rates with all the parameters estimated by the program. The accession numbers of the deposited sequences used in this analysis are shown in the phylogenetic tree in Fig. 2.
FIG. 2.
Maximum likelihood phylogenetic tree of the genus Alexandrium, with Lingulodinium polyedrum as an outgroup, based on rRNA gene sequences (ITS1, 5.8S rRNA gene, ITS2) spanning 515 A. catenella nucleotide positions. The scale bar indicates 0.1 nucleotide substitution per site. Identical sequences were obtained for 23 selected A. catenella strains (12 strains from Japan and 11 strains from Thau Lagoon), and these sequences are also identical to most other sequences in the clade. (Inset) Relative genetic distances between the sequences belonging to the A. catenella clade. The scale bar indicates 0.002 nucleotide substitution per site, equivalent to one nucleotide difference for the sequences analyzed. The sequence data for A. catenella strains TNX22, TNY11, Y-2, OFX102, OFY101, ko-3, and MI7 (accession number AB006990) are from reference 1.
The microsatellite analysis used the 12 loci described by Nagai et al. (33). The PCRs were carried out in 10-μl (total volume) mixtures containing 1 μl of 10× buffer, 2.5 mM MgCl2 (Promega), 0.2 mM of each deoxynucleoside triphosphate (Invitrogen), 0.5 μM of each primer labeled with either Cy5 or fluorescein (MWG-Biotech AG), 0.3 U of Taq polymerase (Sigma), 0.1 mg/ml of bovine serum albumin (Roche), 2 μl of DNA template (about 5 ng/μl), and DNA-free water (Sigma). The thermal cycling conditions consisted of initial denaturation at 94°C for 10 min, followed by 38 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min and then a final extension at 72°C for 5 min. Two microliters of the PCR mixture for each strain was electrophoresed onto an 8% denaturing polyacrylamide gel (Bio-Rad) along with a fluorescently labeled DNA ladder (100 to 600 bp; Promega). Labeled DNA fragments were visualized with an FMBIO II fluorescent imaging system (Hitachi), and the allele sizes were estimated using the FMBIOAnalysis 8.0 image analyzer program (Hitachi).
Statistical analyses of microsatellite markers.
The statistical analyses were performed using the program GENETIX (http://www.genetix.univ-montp2.fr/genetix/genetix.htm). A series of multidimensional analyses (factorial correspondence analysis [FCA] [4]) provided the overall genetic structure for the samples. The objective of this analysis was to represent each strain based on its complete genotype. For this, the genotypic data were first coded based on the presence of each allele as follows (47): 0, allele absent; 1, heterozygous for the allele; or 2, homozygous for the allele. The computation then aimed at finding composite axes which were a combination of the variables and optimized the differences between the individuals analyzed. The relationships among individuals can be visualized on two or three axes. In order to compare genotypes of different ploidies in the same analysis, we had to weight the haploid genotypes as 2, meaning that the allele of a haploid genotype was given the same weight as the two alleles of a diploid heterozygote genotype. The inertia values (i.e., the proportion of the total information contained by an axis, expressed as a percentage) along each axis were shown to be equivalent to linear combinations of the monolocus fixation index (Fst) values (19). The following standard parameters of population genetics were calculated for each group of strains: allele frequencies, P(0.95) and P(0.99) (proportions of polymorphic loci), and parameter A (mean number of alleles per locus). The observed heterozygosity (Ho) and unbiased expected heterozygosity (Hnb) (34) were estimated for the diploid samples (cyst cultures). For the F statistics of Wright, the Fis value (f parameter of Weir and Cockerham [53]) was used to test the departures from Hardy-Weinberg equilibrium of the cyst collection (expected to be diploid), and the Fst value (θ of Weir and Cockerham [53]) was used to measure the differentiation between samples using both cells and cysts. For these tests, the significance of departure from zero (null hypothesis) was estimated by performing 5,000 permutation tests, using Genetix options. The ploidy level of each cycle phase has seldom been tested, and microsatellite data can be used to estimate this level based on the heterozygote genotypes observed. However, apparent heterozygotes can also result from false monoclonal cultures. Indeed, in spite of thorough verification during the cell isolation procedure, two swimming cells could sometimes be isolated, which gave rise to a false monoclonal culture. Here, we compared the proportions of heterozygote genotypes in cultures of expected haploid cells and in cultures of expected diploid resting cysts.
Nucleotide sequence accession numbers.
Nucleotide sequence data have been deposited in the GenBank database under accession numbers FM211455 to FM211477.
RESULTS
Ribotype analysis.
Although direct sequencing of rRNA gene amplicons revealed some polymorphisms, as reported by Lilly et al. (27), the rRNA gene sequences spanning the region including ITS1, the 5.8S rRNA gene, ITS2, and the D1/D3 28S rRNA gene (∼1,500 bp) obtained from the 23 selected A. catenella strains from Thau Lagoon and Japan were identical. In the region including ITS1, the 5.8S rRNA gene, and ITS2 (515 bp), the consensus sequences were identical or nearly identical to most other A. catenella sequences deposited since 2000. This is shown in the phylogenetic tree of the genus Alexandrium that includes our new data and sequences deposited in GenBank in Fig. 2.
Microsatellite analyses.
The average proportion of missing genotypes per locus was 12.2%, and the proportions varied from 4.5% (loci Acat02 and Acat49) to 24.7% (locus Acat10). Missing genotypes represented 11.6% and 13.8% of the genotypes in the French and Japanese strains, respectively. The lack of amplification likely did not originate from cross-priming (the primers were designed using a Japanese strain DNA) but rather originated from sample preservation problems and/or null alleles. Microsatellite genotypes obtained for cultures originating from planktonic cells and germinated resting cyst cultures exhibited few heterozygote patterns. Although a chi-square test was not significant, the proportion of heterozygous genotypes in the French Mediterranean isolates was higher for cyst-derived cultures (5/114 genotypes, 4.4%) than for cultures obtained from planktonic cells (8/477, 1.7%). The number of Japanese strains investigated was not large enough to make the same comparison. In the following calculations, heterozygous cysts were considered true heterozygotes, whereas heterozygous planktonic or vegetative cells were assumed to be artifactual (e.g., possibly due to polyclonal cultures mistakenly produced during isolation or to occasional duplication and mutation of a microsatellite target region). In the French strains, heterozygotes were detected in loci Acat14 and Acat49 and in loci Acat14, Acat16, Acat20, Acat34, and Acat50, in cyst- and vegetative cell-derived cultures, respectively. In the Japanese samples, only one heterozygous genotype was observed at locus Acat14 in a vegetative cell-derived culture. For cyst samples only, expected to contain a diploid genetic reservoir, the Hardy-Weinberg equilibrium (genotype frequencies expected in panmixia) was tested by estimating the significance of Fis departure from zero. The Fis values were between 0.87 and 1.00, and the permutation tests (5,000 permutations) showed that the heterozygote deficit was highly significant (P < 0.001). The genetic diversity of the A. catenella samples was investigated using several methods. The classical descriptive parameters of population genetics showed that the genetic polymorphism was greater in Japan than in Thau Lagoon (Table 2). The Akasaki strains were considerably different from other Japanese and Mediterranean strains, having 10 private alleles (observed only in these samples) in seven loci (Acat02, Acat06, Acat14, Acat22, Acat37, Acat44, and Acat50). Parameter A (mean number of alleles per locus), which is dependent on the sample size, was 2.7 (42 strains) and 2.1 (10 strains) for vegetative cells and cysts, respectively, obtained in 2004 from Thau Lagoon, whereas for the main Japanese sample (the Akasaki seaport was not included) it was 3.7 for only 16 strains. The value obtained for the strains isolated in Akasaki seaport could not be considered because of the sample size was too small (six strains). This group of strains was then separated from the other Japanese strains because of its high level of differentiation (see FCA data below). In order to avoid strong distortions, only nearly complete genotypes were taken into account in the FCA (i.e., genotypes with at most 3 [25%] of the 12 loci missing due to either failed PCR or null alleles). As a result, the strains used in the analyses were as follows: 20 Japanese strains, 42 Thau Lagoon vegetative cell strains (but due to transformation of heterozygotes by creation of virtual haploid strains, this number increased 53), and 10 Thau Lagoon cyst strains (heterozygote genotypes were retained since they were considered true genotypes), plus ATTL01, ACT1 ACT2, and ACT3 (Table 1). The FCA of the whole data set showed that the Japanese strains from Akasaki were genetically very different from all the other strains (data not shown); the entire analysis was thus devoted to the differences between the Akasaki strains and the non-Akasaki strains. After the Akasaki strains were excluded, the FCA showed that there was a clear separation between the French Mediterranean and Japanese strains on the first FCA axis. The dispersion was much greater for the Japanese isolates, indicating greater polymorphism (Fig. 3). However, a cyst strain from Thau Lagoon seemed to be closer to the Japanese strains, and one strain from Ago Bay fell into the Thau Lagoon population (a second strain was discriminated along the third axis [data not shown]). When analyzed alone, the 61 Thau Lagoon strains showed no heterogeneity in the population structure (data not shown).
TABLE 2.
Allele frequencies and classical parametersa
| Locus | Allele frequency in:
|
|||
|---|---|---|---|---|
| ACTEM | ACTKM | J06 | Akasaki | |
| n = 42 | n = 10 | n = 16 | n = 1 | |
| Acat02 | ||||
| 288 | 1 | 1 | 1 | 0 |
| 300 | 0 | 0 | 0 | 1 |
| n = 41 | n = 9 | n = 14 | n = 4 | |
| Acat06 | ||||
| 226 | 0 | 0 | 0.0714 | 0 |
| 230 | 0.4390 | 0.3333 | 0 | 0 |
| 232 | 0.5122 | 0.5556 | 0.5714 | 0 |
| 234 | 0 | 0.1111 | 0.1429 | 0 |
| 236 | 0 | 0 | 0.1429 | 0 |
| 238 | 0.0488 | 0 | 0.0714 | 0 |
| 240 | 0 | 0 | 0 | 0.7500 |
| 248 | 0 | 0 | 0 | 0.2500 |
| n = 39 | n = 7 | n = 13 | n = 0 | |
| Acat10 | ||||
| 264 | 0.0513 | 0 | 0 | |
| 266 | 0.8462 | 1 | 0.3846 | |
| 268 | 0.0513 | 0 | 0.0769 | |
| 270 | 0.0513 | 0 | 0.3077 | |
| 272 | 0 | 0 | 0.2308 | |
| n = 40 | n = 10 | n = 15 | n = 4 | |
| Acat14 | ||||
| 186 | 0.7625 | 0.3500 | 0.6333 | 0 |
| 188 | 0 | 0 | 0.3333 | 0.7500 |
| 190 | 0 | 0 | 0.0333 | 0 |
| 192 | 0.2375 | 0.6500 | 0 | 0 |
| 198 | 0 | 0 | 0 | 0.2500 |
| n = 41 | n = 9 | n = 15 | n = 4 | |
| Acat16 | ||||
| 250 | 0.0244 | 0 | 0.0667 | 0 |
| 252 | 0 | 0 | 0.0667 | 0 |
| 254 | 0.2439 | 0 | 0.2000 | 0 |
| 256 | 0.0366 | 0.4444 | 0.4000 | 0 |
| 258 | 0.6220 | 0.4444 | 0.0667 | 1 |
| 260 | 0.0732 | 0.1111 | 0.2000 | 0 |
| n = 38 | n = 9 | n = 13 | n = 0 | |
| Acat20 | ||||
| 277 | 0 | 0 | 0.0769 | |
| 279 | 0.9605 | 0.8889 | 0.7692 | |
| 281 | 0.0395 | 0.1111 | 0.1538 | |
| n = 40 | n = 10 | n = 16 | n = 4 | |
| Acat22 | ||||
| 212 | 0 | 0 | 0.0625 | 0.7500 |
| 214 | 0.9000 | 1 | 0.5000 | 0 |
| 216 | 0.1000 | 0 | 0.4375 | 0 |
| 238 | 0 | 0 | 0 | 0.2500 |
| n = 42 | n = 10 | n = 16 | n = 4 | |
| Acat34 | ||||
| 172 | 0.4881 | 0.7000 | 0 | 1 |
| 174 | 0 | 0 | 0.0625 | 0 |
| 176 | 0 | 0 | 0.0625 | 0 |
| 178 | 0 | 0 | 0.2500 | 0 |
| 180 | 0.4881 | 0.3000 | 0.5625 | 0 |
| 182 | 0.0238 | 0 | 0.0625 | 0 |
| n = 42 | n = 10 | n = 16 | n = 4 | |
| Acat37 | ||||
| 219 | 0 | 0 | 0 | 0.7500 |
| 223 | 1 | 1 | 1 | 0 |
| 229 | 0 | 0 | 0 | 0.2500 |
| n = 37 | n = 9 | n = 12 | n = 4 | |
| Acat44 | ||||
| 208 | 0 | 0 | 0.0833 | 0 |
| 212 | 0.2973 | 0.2222 | 0.3333 | 0 |
| 214 | 0.4865 | 0.7778 | 0.3333 | 0 |
| 216 | 0.1351 | 0 | 0.1667 | 0 |
| 218 | 0 | 0 | 0.0833 | 0 |
| 220 | 0.0811 | 0 | 0 | 0.7500 |
| 228 | 0 | 0 | 0 | 0.2500 |
| n = 41 | n = 10 | n = 16 | n = 4 | |
| Acat49 | ||||
| 232 | 0.0244 | 0 | 0.3125 | 0.7500 |
| 234 | 0.4390 | 0.4500 | 0.5625 | 0.2500 |
| 236 | 0 | 0.0500 | 0.1250 | 0 |
| 238 | 0.5366 | 0.5000 | 0 | 0 |
| n = 42 | n = 10 | n = 16 | n = 4 | |
| Acat50 | ||||
| 164 | 0 | 0 | 0.0625 | 0.2500 |
| 166 | 0 | 0.0500 | 0.1250 | 0 |
| 168 | 0.2500 | 0.1000 | 0.0625 | 0 |
| 170 | 0.5357 | 0.7000 | 0.6250 | 0 |
| 172 | 0.2143 | 0.1500 | 0.0625 | 0 |
| 174 | 0 | 0 | 0.0625 | 0 |
| 176 | 0 | 0 | 0 | 0.7500 |
| n = 42 | n = 10 | n = 16 | n = 4 | |
| P(0.95) | 0.7500 | 0.6667 | 0.8333 | 0.7000 |
| P(0.99) | 0.8333 | 0.6667 | 0.8333 | 0.7000 |
| A | 2.7500 | 2.0833 | 3.7500 | 1.7000 |
ACTEM, French cultures obtained from planktonic cells, ACTKM, French cultures obtained from cysts; J06, Japanese cultures obtained from planktonic cells or cysts; Akasaki, cultures obtained from cysts in Akasaki harbor. P(0.95) and P(0.99) are the proportions of polymorphic loci determined using the 0.95 and 0.99 criteria, respectively, and A is the mean number of alleles per locus.
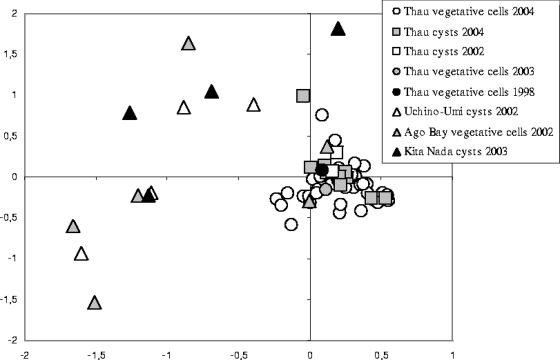
FIG. 3.
Diagram of the first plan of the CA (axis 1 and axis 2) including all samples except those from Akasaki, which were considered outliers due to their high levels of genetic difference. The inertia for the two axes was 16.83% for a total of 59 axes.
Fst estimates were calculated, and a statistical evaluation of the significance of the departure from zero was performed by using 5,000 permutations (Table 3). The values indicated that (i) there was no differentiation between vegetative cells and cysts obtained from Thau Lagoon in 2004 (θ = 0.04, not significant), (ii) the differentiation between France and the Pacific coast of Japan was significant to highly significant (0.15 < θ < 0.29), (iii) the differentiation between France and the Sea of Japan was greater (0.55 < θ < 0.58), and (iv) the differentiation between the two Japanese coasts was not significant (0.48 < θ 0.54). The fact that the significant values were similar to the nonsignificant ones for Akasaki and the other stations indicated that this distortion was probably due to very small sample sizes. Finally, it was clear that there was no differentiation among Pacific stations in Japan.
TABLE 3.
Fst values calculated using the Weir-Cockerham estimator (θ)a
| Culture | Fst value
|
||||
|---|---|---|---|---|---|
| ACTKM | Uchino-Umi | Ago Bay | Kita Nada | Akasaki | |
| ACTEM | 0.04326 NS | 0.26852** | 0.14961** | 0.26801** | 0.55077** |
| ACTKM | 0.27977** | 0.17752** | 0.29159* | 0.58133* | |
| Uchino-Umi | −0.01687 NS | 0.05485 NS | 0.53721 NS | ||
| Ago Bay | 0.02373 NS | 0.48041 NS | |||
| Kita Nada | 0.52256 NS | ||||
Using GENETIX software, 5,000 permutations were used to calculate the P values. ACTEM, French cultures obtained from planktonic cells; ACTKM, French cultures obtained from cysts. *, significant at the 0.05 level; **, significant at the 0.01 level; NS, not significant.
DISCUSSION
Phylogenetic relationships among A. catenella ribotypes.
Until now, differentiation among Alexandrium lineages has been investigated mainly by using ribosomal sequences (18, 37, 38, 43-46). These analyses showed that the Alexandrium lineages are notably related to their geographic origins, and the lineages are defined as the North American, Western European, Mediterranean, Temperate Asian, Tropical Asian, and Tasmanian ribotypes. Since 1998, when a toxic event caused by A. catenella was observed for the first time in Thau Lagoon, many studies have been carried out in order to understand the ecophysiology and growth strategies of this organism (7-9, 25). Among the causation theories advanced by Hallegraeff (21) concerning HAB expansion, the appearance of A. catenella in Thau Lagoon through dispersal of resting cysts in ballast water (28) and in transplanted shellfish stocks from aquaculture (25) has been considered a putative source of the new and recurrent outbreaks. Lilly et al. (28) first described the strains isolated in 1998 as members of the Japanese ribotype of the Temperate Asian clade and recently renamed them group IV in the A. tamarense complex phylogeny (27). According to the phylogenetic analysis, isolates obtained in November 2004 are similar to the isolates obtained in 1998, 2002, and 2003. The phylogenetic tree (Fig. 2), which includes most of the published ITS1-5.8S rRNA gene-ITS2 sequences from the A. tamarense-A. catenella-A. fundyense species complex, including sequences of the 23 new French and Japanese A. catenella strains, indicated that the ribosomal operon could not elucidate the intraspecific variability within and between A. catenella populations having a common geographic origin. Furthermore, the A. catenella cluster contains some A. tamarense morphotypes from China. The phylogenic grouping of isolates in this complex is consistent with the ribotype clades but not with the morphospecies that form the complex (37). They are in accordance with the recently developed trends (27, 37, 52) which question the phylogenetic, proteomic, biological, and morphological species definition for the A. tamarense complex. Because ribosomal genes are ubiquitous and orthologous (14) and display little polymorphism due to a limited mutation rate (26), they should still be useful and valuable markers at the species level. However, they may not be the most judicious and accurate molecular markers for analyzing the intraspecific diversity essential for understanding migration and speciation phenomena within A. catenella assemblages.
Genetic diversity within the Temperate Asian A. catenella clade as shown by microsatellite markers.
In contrast, it appears that microsatellite markers may be appropriate genetic markers for investigating the intraspecific diversity of the A. catenella ribotype. For the first time, Japanese and Mediterranean A. catenella isolates belonging to the Japanese Temperate Asian ribotype of the Temperate Asian clade were compared using these highly polymorphic genetic markers designed by using Japanese A. catenella strains. Thus, the analysis distinctly showed that the Japanese samples studied were divided into two differentiated “A. catenella” lineages: the Sea of Japan lineage (including the differentiated sample from Akasaki seaport) and the east coast lineage, which included populations from the Inland Sea (Uchino-Umi, Kita Nada) and the Pacific Ocean (Ago Bay) (Fig. 1). This is the first time that such heterogeneity within Japanese A. catenella populations has been described. It also appeared that A. catenella strains isolated from Thau Lagoon could belong to another lineage. Several discordances (private alleles) were in agreement with Fst values, which were always significant between France and Japan (Table 3).
Genetic diversity within the A. catenella lineage from Thau Lagoon.
Microsatellite markers revealed relatively low genetic diversity in all the Mediterranean isolates from Thau Lagoon (ATTL01, ACT1, ACT2, ACT3, ACTEM, ACTKM) compared to Japanese populations. Based on these markers, these Thau Lagoon strains could belong to the same lineage. This could be explained in several ways: (i) the introduction of a small exogenous population which found its ecological niche or (ii) the specific environmental characteristics of Thau Lagoon (i.e., high temperatures [up to 26°C] and high salinities [up to 39 to 40 practical salinity units in the summer]) could have selected adapted ecotypes. Despite the low recorded diversity with microsatellites, the A. catenella individuals within the population would not homogeneously participate in formation of the following generation because no panmixia was apparent according to the Fis values. When the following generation is not the product of random pooling in the initial gametic pool (15), prezygotic selection should be considered. The low polymorphism and absence of panmixia in cysts could be explained by differential reproductive success, as follows. (i) If very few cells produce resting cysts during a bloom, a founding effect would be generated for each reproduction. (ii) The original settlement of the Thau Lagoon population could stem from very few cells of unknown origin. (iii) Resting cysts sampled in 2004, which could settle for several years in the sediment, could not represent a population at equilibrium. Thus, more accurate genetic markers are required for describing the diversity within the A. catenella population and consequently for understanding its adaptive strategy and the success of its recurrent dominance.
Ploidy level and artifacts.
Many dinoflagellates, including the genus Alexandrium, have a complex life cycle with a predominant vegetative haploid (n) phase with asexual replication and a short sexual diploid (2n) phase (5, 17, 29). Haploid cells divide mitotically in the water column, and some cells differentiate into a gametic stage. Compatible gametes (n) fuse in the water column to form a short-lived diploid zygote called a planozygote (2n), which then transforms into a resting cyst (2n). After a dormant period, the cyst germinates and regenerates a planomeiocyte (2n), which undergoes meiotic division, resulting in new haploid cells (n) that develop as vegetative cells (n). The distinction between haploid and diploid cells in population genetics can be made by examining the genotypes of polymorphic nuclear markers. A polymorphic nuclear marker shows heterozygous genotypes in a diploid individual but not in a haploid individual. In our study, the genotypes were obtained from a monoclonal culture of a “swimming cell” or a resting cyst. Isolating a single cell or cyst is a critical step, in which false heterozygotes can be obtained if two (or more) cells are inadvertently isolated together or a planomeiocyte (2n) or planozygote (2n) rather than a haploid vegetative cell is isolated.
In the present study, the proportion of heterozygote genotypes was higher in cyst cultures (4.4%) than in planktonic cell cultures (1.7%), where the number of vegetative cell genotypes obtained was more than four times higher than the number of cyst genotypes obtained. Despite the fact that this difference in heterozygote genotypes is not significant, we could attribute the 1.7% vegetative cell heterozygote genotypes to mishandling in the long process of monoclonal culture and/or to a percentage of planomeiocytes and planozygotes isolated in situ among the “swimming cells.” We also considered the possibility that within the estimated 4.4% cyst heterozygote genotypes, false heterozygotes also occurred in the same proportion that they occurred in cell samples. This could show that the global diploid phase analyzed has low heterozygosity, which has been overestimated (Ho, <0.04). This preliminary result is encouraging, but it is obvious that new highly polymorphic microsatellites are necessary. This is why new markers were designed by the same team (35). When analyzing the expected Hardy-Weinberg equilibrium in the three cyst samples large enough to allow the test, we observed an extreme Fis disequilibrium between 0.9 and 1 (P < 0.001), meaning that only a few expected heterozygous genotypes were detected. Panmixia (free sexuality) was not observed, highlighting the sexual crossing (i.e., heterothallism) which occurred in the A. catenella populations. The fusion of compatible gametes from the same clone (selfing, homothallism) results in a lack of effective recombination, whereas the fusion of gametes from different clones (outcrossing, heterothallism) may result in recombination. Therefore, A. catenella from Thau Lagoon may exhibit heterothallic behavior like that described by Yoshimatsu (55) for Japanese strains. The life cycle of A. catenella must be studied in more detail because sexual stages remain unclear, in contrast to Alexandrium excavatum (10), Alexandrium tamutum, A. minutum (13), and Alexandrium peruvianum (12), whose sexual stages have been well described. Such knowledge is essential for understanding the origin and occurrence of toxic events due to A. catenella.
Is the recent Mediterranean invasion of Japanese origin?
We are far from having a complete description of Japanese A. catenella polymorphism. The greater polymorphism and the structure of the Japanese isolates confirm that east Asia is the natural origin of distribution of A. catenella. In contrast, the low genetic diversity among the Mediterranean strains from Thau Lagoon could be related to recent introduction of genotypes whose exact origin remains to be determined. These preliminary results suggest that the hypothesis that the A. catenella blooming in Thau Lagoon (French Mediterranean) had a Japanese origin should be precisely explored. Based on the observed and potential differentiation of the species from Asian coasts, more strains from both regions must be analyzed before a conclusion can be drawn. This study urges workers to compare other A. catenella isolates from the Mediterranean area using the same markers in order to (i) determine if these populations belong to the same lineage and (ii) understand the genetic distribution of the species in the Mediterranean basin. A genetic analysis will require the use of more microsatellite markers (35) and more samples, not only samples from Japan. This genetic study must be extended to other Asiatic sites and geographic areas in the world in order to clarify the origin of A. catenella established in Thau Lagoon. We consider the present survey the first step of a multiyear project examining A. catenella all around the world in order to understand the long-distance exchanges related to human activity.
These findings should provide insight into other environmental factors affecting the niche adaptations of these microorganisms in situ. Large variations in single-cell characteristics and physiological capabilities (adaptation to temperature, salinity, nutrient assimilation, allelopathic activity) can be expected within and between lineages, which could have ecological consequences, such as HABs. A genetic approach could underpin a new paradigm for speciation and migration in phytoplankton assemblages.
Acknowledgments
We thank Pierre-Alexandre Gagnaire for his help with statistics, Annie Pastoureaud (Ifremer LER/LR-Sète) for her helpful collaboration, Patrick Gentien (Ifremer DYNECO-Brest) for his continuous support, and the Ifremer staff (LER/LR-Sète) for technical assistance. We also thank the anonymous reviewers for their helpful comments and suggestions.
This research was supported by grants from the French National Programme “Ecosphère Continentale et Côtière” (EC2CO-PNEC) and the Agence Nationale de la Recherche (ANR −05-BLAN-0219 XPressFloral) and by the ALCAT program of Ifremer.
Footnotes
Published ahead of print on 5 February 2009.
REFERENCES
- 1.Adachi, M., Y. Sako, and Y. Ishida. 1996. Analysis of Alexandrium (Dinophyceae) species using sequences of the 5.8S ribosomal DNA and internal transcribed spacer regions. J. Phycol. 32:424-432. [Google Scholar]
- 2.Alpermann, T. J., U. E. John, L. K. Medlin, K. J. Edwards, P. K. Hayes, and K. M. Evans. 2006. Six new microsatellite markers for the toxic marine dinoflagellate Alexandrium tamarense. Mol. Ecol. Notes 6:1057-1059. [Google Scholar]
- 3.Anderson, D. M. 1989. Toxic algal blooms and red tides: a global perspective, p. 11-16. In T. Okaichi, D. M. Anderson, and T. Nemoto (ed.), Red tides: biology, environmental science and toxicology. Elsevier, New York, NY.
- 4.Benzecri, J. P. 1982. L'analyse des données. II. L'analyse des correspondances. Dunod, Paris, France.
- 5.Blackburn, S. I., and N. Parker. 2005. Microalgal life cycles: encystment and excystment, p. 399-417. In R. A. Anderson (ed.), Algal culturing techniques. Elsevier Academic Press, Amsterdam, The Netherlands.
- 6.Bolch, C. J. S., and M. F. de Salas. 2007. A review of the molecular evidence for ballast water introduction of the toxic dinoflagellates Gymnodinium catenatum and the Alexandrium “tamarensis complex” to Australasia. Harmful Algae 6:465-485. [Google Scholar]
- 7.Collos, Y., C. Gagne, M. Laabir, A. Vaquer, P. Cecchi, and P. Souchu. 2004. Nitrogenous nutrition of Alexandrium catenella (Dinophyceae) in cultures and in Thau lagoon, southern France. J. Phycol. 40:96-103. [Google Scholar]
- 8.Collos, Y., M. Lespilette, A. Vaquer, M. Laabir, and A. Pastoureaud. 2006. Uptake and accumulation of ammonium by Alexandrium catenella during nutrient pulses. Afr. J. Mar. Sci. 28:313-318. [Google Scholar]
- 9.Collos, Y., A. Vaquer, M. Laabir, E. Abadie, T. Laugier, A. Pastoureaud, and P. Souchu. 2007. Contribution of several nitrogen sources to growth of Alexandrium catenella during blooms in Thau lagoon, southern France. Harmful Algae 6:781-789. [Google Scholar]
- 10.Destombe, C., and A. Cembella. 1990. Mating-type determination, gametic recognition and reproductive success in Alexandrium excavatum (Gonyaulacales, Dinophyta), a toxic red-tide dinoflagellate. Phycologia 29:316-325. [Google Scholar]
- 11.Evans, K. M., S. F. Kuhn, and P. K. Hayes. 2005. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J. Phycol. 41:506-514. [Google Scholar]
- 12.Figueroa, R. I., I. Bravo, and E. Garces. 2008. The significance of sexual versus asexual cyst formation in the life cycle of the noxious dinoflagellate Alexandrium peruvianum. Harmful Algae 7:653-663. [Google Scholar]
- 13.Figueroa, R. I., E. Garces, and I. Bravo. 2007. Comparative study of the life cycles of Alexandrium tamutum and Alexandrium minutum (Gonyaulacales, Dinophyceae) in culture. J. Phycol. 43:1039-1053. [Google Scholar]
- 14.Fitch, W. M. 1970. Toward defining the tree of maximum parsimony, p. 160-178. In G. F. Estabrook (ed.), Eighth International Conference on Numerical Taxonomy. Freeman, San Francisco, CA.
- 15.Genovesi-Giunti, B. 2006. Initiation, maintien et récurrences des efflorescences toxiques d'Alexandrium catenella (Dinophyceae) dans une lagune méditerranéenne (Thau, France): rôle du kyste dormant. Ph.D. thesis. Université Montpellier II, Montpellier, France.
- 16.Genovesi-Giunti, B., M. Laabir, and A. Vaquer. 2006. The benthic resting cyst: a key actor in harmful dinoflagellate blooms—a review. Vie Milieu Life Environ. 56:327-337. [Google Scholar]
- 17.Giacobbe, M. G., and X. M. Yang. 1999. The life history of Alexandrium taylori (Dinophyceae). J. Phycol. 35:331-338. [Google Scholar]
- 18.Guillou, L., E. Nezan, V. Cueff, E. E. L. Denn, M. A. Cambon-Bonavita, P. Gentien, and G. Barbier. 2002. Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis, and Karenia) from French coasts. Protist 153:223-238. [DOI] [PubMed] [Google Scholar]
- 19.Guinand, B. 1996. Use of a multivariate model using allele frequency distributions to analyse patterns of genetic differentiation among populations. Biol. J. Linn. Soc. 58:173-195. [Google Scholar]
- 20.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 21.Hallegraeff, G. M. 1993. A review of harmful algal blooms and their apparent global increase. Phycologia 32:79-99. [Google Scholar]
- 22.Harrison, P. J., R. E. Waters, and F. J. R. Taylor. 1980. A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J. Phycol. 16:28-35. [Google Scholar]
- 23.Iglesias-Rodriguez, M. D., O. M. Schofield, J. Batley, L. K. Medlin, and P. K. Hayes. 2006. Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): the use of microsatellite analysis in marine phytoplankton population studies. J. Phycol. 42:526-536. [Google Scholar]
- 24.Jarne, P., and P. J. L. Lagoda. 1996. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 11:424-429. [DOI] [PubMed] [Google Scholar]
- 25.Laabir, M., Z. Amzil, P. Lassus, E. Masseret, Y. Tapilatu, R. De Vargas, and D. Grzebyk. 2007. Viability, growth and toxicity of Alexandrium catenella and Alexandrium minutum (Dinophyceae) following ingestion and gut passage in the oyster Crassostrea gigas. Aquat. Living Resour. 20:51-57. [Google Scholar]
- 26.Lecointre, G., H. Philippe, H. L. Van Le, and H. Le Guyader. 1993. Species sampling has a major impact on phylogenetic inference. Mol. Phylogenet. Evol. 2:205-224. [DOI] [PubMed] [Google Scholar]
- 27.Lilly, E. L., K. M. Halanych, and D. M. Anderson. 2007. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae). J. Phycol. 43:1329-1338. [Google Scholar]
- 28.Lilly, E. L., D. M. Kulis, P. Gentien, and D. M. Anderson. 2002. Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: evidence from DNA and toxin analysis. J. Plankton Res. 24:443-452. [Google Scholar]
- 29.Montresor, M. 1995. The life history of Alexandrium pseudogonyaulax (Gonyaulacales, Dinophyceae). Phycologia 34:444-448. [Google Scholar]
- 30.Nagai, S., C. Lian, M. Hamaguchi, Y. Matsuyama, S. Itakura, and T. Hogetsu. 2004. Development of microsatellite markers in the toxic dinoflagellate Alexandrium tamarense (Dinophyceae). Mol. Ecol. Notes 4:83-85. [Google Scholar]
- 31.Nagai, S., C. Lian, S. Yamaguchi, M. Hamaguchi, Y. Matsuyama, S. Itakura, H. Shimada, S. Kaga, H. Yamauchi, Y. Sonda, T. Nishikawa, C. H. Kim, and T. Hogetsu. 2007. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J. Phycol. 43:43-54. [Google Scholar]
- 32.Nagai, S., L. McCauley, N. Yasuda, D. L. Erdner, D. M. Kulis, Y. Matsuyama, S. Itakura, and D. M. Anderson. 2006. Development of microsatellite markers in the toxic dinoflagellate Alexandrium minutum (Dinophyceae). Mol. Ecol. Notes 6:756-758. [Google Scholar]
- 33.Nagai, S., M. Sekino, Y. Matsuyama, and S. Itakura. 2006. Development of microsatellite markers in the toxic dinoflagellate Alexandrium catenella (Dinophyceae). Mol. Ecol. Notes 6:120-122. [Google Scholar]
- 34.Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishitani, G., S. Nagai, E. Masseret, C. Lian, S. Yamaguchi, N. Yasuda, S. Itakura, D. Grzebyk, P. Berrebi, and M. Sekino. 2007. Development of compound microsatellite markers in the toxic dinoflagellate Alexandrium catenella (Dinophyceae). Plankton Benthos Res. 2:128-133. [Google Scholar]
- 36.Nunn, G., B. Theisen, B. Christensen, and P. Arctander. 1996. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. J. Mol. Evol. 42:211-223. [DOI] [PubMed] [Google Scholar]
- 37.Penna, A., S. Fraga, M. Maso, M. G. Giacobbe, I. Bravo, E. Garces, M. Vila, E. Bertozzini, F. Andreoni, A. Luglie, and C. Vernesi. 2008. Phylogenetic relationships among the Mediterranean Alexandrium (Dinophyceae) species based on sequences of 5.8S gene and internal transcript spacers of the rRNA operon. Eur. J. Phycol. 43:163-178. [Google Scholar]
- 38.Penna, A., E. Garces, M. Vila, M. G. Giacobbe, S. Fraga, A. Luglie, I. Bravo, E. Bertozzini, and C. Vernesi. 2005. Alexandrium catenella (Dinophyceae), a toxic ribotype expanding in the NW Mediterranean Sea. Mar. Biol. 148:13-23. [Google Scholar]
- 39.Rynearson, T. A., and E. V. Armbrust. 2000. DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol. Oceanogr. 45:1329-1340. [Google Scholar]
- 40.Rynearson, T. A., and E. V. Armbrust. 2004. Genetic differentiation among populations of the planktonic marine diatom Ditylum brightwellii (Bacillariophyceae). J. Phycol. 40:34-43. [Google Scholar]
- 41.Rynearson, T. A., and E. V. Armbrust. 2005. Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol. Ecol. 14:1631-1640. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Scholin, C. A., and D. M. Anderson. 1994. Identification of group-specific and strain-specific genetic-markers for globally distributed Alexandrium (Dinophyceae). 1. RFLP analysis of SSU ribosomal-RNA genes. J. Phycol. 30:744-754. [Google Scholar]
- 44.Scholin, C. A., and D. M. Anderson. 1996. LSU rDNA-based RFLP assays for discriminating species and strains of Alexandrium (Dinophyceae). J. Phycol. 32:1022-1035. [Google Scholar]
- 45.Scholin, C. A., M. Herzog, M. Sogin, and D. M. Anderson. 1994. Identification of group-specific and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). 2. Sequence analysis of a fragment of the LSU ribosomal-RNA gene. J. Phycol. 30:999-1011. [Google Scholar]
- 46.Sebastian, C. R., S. M. Etheridge, P. A. Cook, C. O'Ryan, and G. C. Pitcher. 2005. Phylogenetic analysis of toxic Alexandrium (Dinophyceae) isolates from South Africa: implications for the global phylogeography of the Alexandrium tamarense species complex. Phycologia 44:49-60. [Google Scholar]
- 47.She, J. X., M. Autem, G. Kotoulas, N. Pasteur, and F. Bonhomme. 1987. Multivariate analysis of genetic exchanges between Solea aegyptiaca and Solea senegalensis (Teleosts, Soleidae). Biol. J. Linn. Soc. 32:357-371. [Google Scholar]
- 48.Souchu, P., A. Vaquer, Y. Collos, S. Landrein, J. M. Deslous-Paoli, and B. Bibent. 2001. Influence of shellfish farming activities on the biogeochemical composition of the water column in Thau lagoon. Mar. Ecol. Prog. Ser. 218:141-152. [Google Scholar]
- 49.Sournia, A. 1995. Red tide and toxic marine phytoplankton of the world ocean: an inquiry into biodiversity, p. 103-112. In P. Lassus, G. Arzul, E. Erard-Le Denn, P. Gentien, and C. Marcaillou-Le Baut (ed.), Harmful marine algal blooms. Lavoisier, Intercept Ltd., Paris, France.
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vila, M., E. Garces, M. Maso, and J. Camp. 2001. Is the distribution of the toxic dinoflagellate Alexandrium catenella expanding along the NW Mediterranean coast. Mar. Ecol. Prog. Ser. 222:73-83. [Google Scholar]
- 52.Wang, D. Z., L. Lin, H. F. Gu, L. L. Chan, and H. S. Hong. 2008. Comparative studies on morphology, ITS sequence and protein profile of Alexandrium tamarense and A. catenella isolated from the China Sea. Harmful Algae 7:106-113. [Google Scholar]
- 53.Weir, B. S., and C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358-1370. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, T., Y. Sako, and A. Uchida. 2001. Geographic differences in paralytic shellfish poisoning toxin profiles among Japanese populations of Alexandrium tamarense and Alexandrium catenella (Dinophyceae). Phycol. Res. 29:13-21. [Google Scholar]
- 55.Yoshimatsu, S. 1981. Sexual reproduction of Protogonyaulax catenella in culture. I. Heterothallism. Bull. Plankton Soc. Jpn. 28:131-139. [Google Scholar]