Abstract
Divercin V41 (DvnV41) is a class IIa bacteriocin with potent antilisterial activity isolated from Carnobacterium divergens V41. Previously, we expressed from a synthetic gene, in Escherichia coli Origami, a recombinant DvnV41 designated DvnRV41, which possesses four additional amino acids (AMDP) in the N-terminal region that result from enzymatic cleavage and retains the initial DvnV41 activity. To unravel the relationship between the structure of DvnRV41 and its particularly elevated activity, we produced by site-directed mutagenesis eight variants in which a single amino acid replacement was specifically introduced into the sequence. The point mutations were designed to change either conserved residues in class IIa bacteriocins or residues specific to DvnV41 located mainly in the C-terminal region. The fusion proteins were purified from the cytosoluble fractions by immobilized affinity chromatography. DvnRV41 and its variants were released from the fusion proteins by enzymatic cleavage, using enterokinase. The purity of DvnRV41 and of the variants was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, high-performance liquid chromatography, and mass spectrometry. The antibacterial activity of DvnRV41 and its variants was assessed using different indicator strains, including Listeria monocytogenes EGDe and Enterococcus faecalis JH2-2. The activity of all of the variants appeared to be less than the activity of DvnRV41. The decrease in activity did not appear to be related to a global conformational change, as determined by circular dichroism. Overall, the variants of DvnRV41 produced in the present study provide interesting insights into structure-activity relationships of class IIa bacteriocins.
Lactic acid bacterium (LAB) bacteriocins are ribosomally synthesized antimicrobial peptides that could be useful as natural and nontoxic food preservatives. Most bacteriocins produced by LAB are known to be active only against gram-positive bacteria. A few exceptions to this general rule, including bacteriocins from Lactobacillus species (5, 31, 35, 49, 52), AS-48 from Enterococcus faecalis (1), enterocin E50-52 from Enterococcus faecium (50), and thermophylin produced by Streptococcus thermophilus (26), have been described; these bacteriocins exhibit various degrees of activity against gram-negative bacteria.
A classification of bacteriocins produced by LAB was first proposed by Klaenhammer (30) and then was modified by Nes and Holo (39) and further updated by Cotter et al. (9). Class I and class II bacteriocins are the most abundant and thoroughly studied bacteriocins. Class I bacteriocins, namely lantibiotics, have been widely studied, and one of them, nisin, is routinely used by the food industry as a preservative due to its high level of inhibitory activity against a wide range of bacterial pathogens. These bacteriocins are characterized by the presence in their primary structure of posttranslationally modified amino acid residues (lanthionine and methylanthionine). Bacteriocin class II is composed of three subclasses, designated classes IIa, IIb, and IIc, although two additional subclasses, classes IId and IIe, are expected. The most studied subclass is class IIa, also termed pediocin-like bacteriocins (11).
Class IIa bacteriocins are antilisterial, small (<10 kDa), heat-stable, nonlantibiotic peptides consisting of 37 to 48 amino acids containing the consensus sequence YGNGVXCX4C (where X is any amino acid) in the N-terminal region (11, 13). Class IIa bacteriocins are believed to exert their activity by forming pores in target cell membranes (6), but a recognition step involving the EIItMan mannose permease of the phosphotransferase system, which could act as a receptor for class IIa bacteriocins (10, 23), could take part in the mechanism of action in certain cases. Studies of the primary structure of class IIa bacteriocins have delineated two distinct domains: a highly conserved hydrophilic and cationic N-terminal domain stabilized by a disulfide bond and an amphiphilic or hydrophobic C-terminal domain, which is less conserved and is stabilized in some peptides, such as pediocin PA-1/AcH, by a second disulfide bond (11). The three-dimensional structures of several one-disulfide-bond class IIa bacteriocins have been determined (18, 21, 54, 56). Despite high sequence similarity, these molecules do not exhibit a common structure, but they share common elements, such as an amphipathic α-helix and most often a β-sheet domain. The N-terminal domain is thought to mediate the initial binding to target cell membranes through electrostatic interactions, while the C-terminal domain is thought penetrate into the hydrophobic part of the membrane bilayer target cell, causing membrane leakage (15, 36). Class IIa bacteriocins are secreted from their producing cells by dedicated ATP-biding cassettes (ABC transporters) and their accessory proteins, with cleavage of their N-terminal leaders (22). However, some class IIa bacteriocins are secreted by the general secretory pathway (sec) with cleavage of their N-terminal signal peptides (7, 29). Class IIa bacteriocins have been the focus of increased attention, which has been prompted by their potential use in food protection (8) and in medicine as antiviral agents (55) or antibiotic supplements (37). In particular, several structure-activity relationship studies have been performed (2, 14, 36, 38, 48, 53, 56).
Divercin V41 (DvnV41) is a class IIa bacteriocin and is naturally produced by Carnobacterium divergens V41, an LAB isolated from fish viscera (40). The broad spectrum of activity of DvnV41 against food-borne pathogenic bacteria, such as Listeria monocytogenes, in vitro and in vivo (43, 46) has attracted great interest and led to its heterologous production in Escherichia coli (43, 59) to enhance the peptide yield and simplify the purification procedure (45, 60). DvnV41 is synthesized as a prebacteriocin consisting of 66 amino acids. The 23-residue N-terminal extension is cleaved off, yielding the mature 43-amino-acid molecule DvnV41 with a molecular mass of 4.509 Da, which contains two disulfide bonds between Cys10 and Cys15 (N-terminal disulfide bond conserved in all class IIa bacteriocins) and between Cys25 and Cys43 (C-terminal disulfide bond specific to the class IIa bacteriocins closely related to DvnV41, such as pediocin PA-1/AcH [13]) (Fig. 1). In addition to the gene encoding the DvnV41 precursor, the genetic system also encodes putative proteins commonly encoded by bacteriocin operons, including an ATP-dependent transporter, two immunity-like proteins, and the two components of a lantibiotic-type signal-transducing system. The genetic organization of the nucleotide fragment suggests that there are gene rearrangements, which is consistent with the large repertoire of class IIa bacteriocins (11, 13).
FIG. 1.
(A) Sequence of DvnRV41 fusion protein. (B) Overview of the mutations created in the primary structure of DvnRV41 by site-directed mutagenesis. Cysteine residues are underlined. Compared to natural DvnV41, the DvnRV41 recombinant contains four additional amino acids at the N terminus (AMDP) due to specific cleavage by enterokinase. The highlighted residues correspond to amino acids inserted in the sequence in order to replace the original amino acids.
Taking advantage of our knowledge of the production of the DvnV41recombinant DvnRV41 in E. coli Origami from a synthetic gene, here we produced eight variants of DvnRV41 designed to assess the roles of individual residues that are either common to class IIa bacteriocins or essentially specific to the class IIa bacteriocins that include two disulfide bonds. All of these variants exhibited lower antibacterial activity than DvnRV41, but to different extents, reflecting the importance of their residues for the activity of DvnV41. This genetic approach, in combination with circular dichroism (CD) used to assess the conformational modifications induced by the mutations, allowed us to better understand the role of the selected individual amino acids in the activity of DvnRV41.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacterial growth conditions.
E. coli K-12 strain DHα (Stratagene, Austin, TX) was used for standard cloning procedures, and E. coli K-12 strain Origami (DE3)(pLysS) (Novagen, Madison, WI) was used for DvnRV41 expression. E. coli transformants containing the recombinant plasmids (Table 1) were selected on Luria-Bertani (LB) agar containing ampicillin (100 μg/ml) (Sigma, St. Louis, MO).
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA endA1 gyr A96 thi-1 hsdR17 supE44 ΔlacU169(φ80 dlacZΔM15) deoR F− λ− | Invitrogen |
| E. coli Origami | Δ(ara-leu)7697 ΔlacX74 ΔphoA(PvuII) phoR araD139 ahpC galE galK rpsL F′ [lac+lacIqpro] | Novagen |
| L. innocua F | Indicator organism | DSVb |
| L. monocytogenes EGDe | Indicator organism | 19 |
| E. faecalis JH2-2 | Indicator organism | 59 |
| E. faecalis 35A1 | E. faecalis JH2-2 Δpde mutant, indicator organism | 4 |
| E. faecalis 36H4 | E. faecalis JH2-2 ΔglpQ mutant, indicator organism | 4 |
| E. faecalis 35H1 | E. faecalis JH2-2 ΔrpoN mutant, indicator organism | 4 |
| Plasmids | ||
| pCR03 | DvnRV41, Ampr | 44 |
| pCR04 | DvnRV41, Cmr Ampr | 44 |
| pET32b | Expression vector | Novagen |
| pCR11 | W19F substitution, Ampr | This study |
| pCR12 | Q21S substitution, Ampr | This study |
| pCR12 | A22G substitution, Ampr | This study |
| pCR13 | S23Y substitution, Ampr | This study |
| pCR14 | V30F substitution, Ampr | This study |
| pCR15 | L35F substitution, Ampr | This study |
| pCR16 | A38V substitution, Ampr | This study |
| pCR17 | P40V substitution, Ampr | This study |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
DSV, Direction des Services Vétérinaires de Nantes, Nantes, France.
L. monocytogenes EGDe (19), used as an indicator strain, was propagated in Elliker broth (Scharlau, Chemie S.A, Spain) and incubated for 24 h at 30°C. E. faecalis wild-type strain JH2-2 (59) and mutants of this strain resistant to DvnRV41 (4) were grown without shaking in M17 medium (Biokar Diagnostic, Beauvais, France) supplemented with glucose (0.5%, wt/vol) (51). For growth of enterococcal mutants, erythromycin (150 μg/ml) or tetracycline (15 μg/ml) was added to the growth medium.
Site-directed mutagenesis and synthesis of variants.
Point mutations were introduced into dvnRV41 through site-directed mutagenesis using the PCR described below. The pCR03 plasmid carrying the synthetic gene dvnRV41 (44) was used as a DNA template. Primers used for site-directed mutagenesis are shown in Table 2. PCR were performed using the Pfu polymerase (Sigma, Stenheim, Germany). The 50-μl reaction mixtures contained 40 ng of DNA template (plasmid pCR03), 0.25 μM of each primer, 0.25 mM of deoxynucleoside triphosphates, and 2.5 U of Pfu polymerase. After incubation for 4 min at 95°C, the PCR was initiated by incubation at 95°C for 1 min, which was followed by 19 cycles of denaturation at 95°C for 2 min, 55°C for 2 min, and 68°C for 10 min and then 68°C for 15 min. The PCR products were treated for 1 h at 37°C with the restriction enzyme DpnI (Fermentas, Biogen, Prague, Czech Republic) to eliminate the methylated DNA template. The size of the PCR product was determined by 1.5% agarose gel electrophoresis. Then the product was extracted by using a QIAquick gel extraction kit (Qiagen) and introduced into E. coli strain DH5α by heat shock (2 min, 42°C). The recombinant DH5α strain harboring the PCR product was grown in LB medium containing 100 μg/ml ampicillin, and the de novo-synthesized recombinant plasmid was extracted by using a Qiagen miniprep kit (Courtaboeuf, France) according to the manufacturer's recommendations. DNA fragments containing the mutated dvnRV41 genes were obtained by digestion with restriction enzymes NcoI and HindIII and ligated into the pET32b vector digested with the same restriction enzymes (Sigma), producing recombinant plasmids (Table 1). Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Beverly, MA) and from Promega (Charbonnieres, France), respectively. The cloning strategy used in this work allowed us to obtain a translational fusion with thioredoxin (TRX), TRX-His6-Asp4-Lys-DvnRV41 (DvnRV41-fP).
TABLE 2.
Primers used in this study for site-directed mutagenesis
| Primer | Sequence |
|---|---|
| W19F-forward | 5′-GCTGGGTGGATTTTGGCCAGGCGAGCGGC-3′ |
| W19F-reverse | 5′-GCCGCTCGCCTGGCCAAAATCCACCCAGC-3′ |
| Q21S-forward | 5′-GCTGGGTGGATTGGGGCTCGGCGAGCGGC-3′ |
| Q21S-reverse | 5′-GCCGCTCGCCGAGCCCCAATCCACCCAGC-3′ |
| A22G-forward | 5′-GGATTGGGGCCAGGGCAGCGGCTGCATTGGC-3′ |
| A22G-reverse | 5′-GCCAATGCAGCCGCTGCCCTGGCCCCAATCC-3′ |
| S23Y-forward | 5′-GGCCAGGCGTACGGCTGCATTGGCCAGACC-3′ |
| S23Y-reverse | 5′-GGTCTGGCCAATGCAGCCGTACGCCTGGCC-3′ |
| V30F-forward | 5′-GGCTGCATTGGCCAGACCAAGGTTGGCGGTTGGCTGGGC-3′ |
| V30F-reverse | 5′-GCCCAGCCAACCGCCAACCCTGGTCTGGCCAATGCAGCC-3′ |
| L35F-forward | 5′-CGTGGTTGGCGGTTGGTTTGGCGGTGCGATTCCGGGC-3′ |
| L35F-reverse | 5′-GCCCGGAATCGCACCGCCAAACCAACCGCCAACCACG-3′ |
| A38V-forward | 5′-GGCGGTTGGCTGGGCGGTGTGATTCCGGGCAAATGC-3′ |
| A38V-reverse | 5′-CGATTTGCCCGGAATCACACCGCCCAGCCAACCGCC-3′ |
| P40V-forward | 5′-GG CTG GGT GCG ATT GTG GGC AAA TGC-3′ |
| P40V-reverse | 5′-GCATTTGCCCACAATCGCACCCAGCC-3′ |
Plasmid pCR03 and recombinant plasmids pCR11 to pCR17 (Table 1) were isolated and sequenced to confirm the accuracy of the sequences (Genomic, Czech Republic). Once a sequence was in good agreement with the expected sequence, the plasmid was introduced into E. coli Origami (DE3)(pLysS) for further analysis.
Expression of variants in E. coli Origami.
Overnight cultures of E. coli Origami (DE3)(pLysS) harboring plasmids pCR11 to pCR17 were grown aerobically at 37°C in 100 ml of LB medium containing 100 μg/ml ampicillin. When the optical density at 600 nm (measured with a spectrophotometer [Biotek Instruments, Winooski, VT]]) reached 0.6, gene expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) to a concentration of 1 mM. The cells were grown for an additional 3 h and then harvested by centrifugation (15 min, 6,000 × g, 4°C), and the cell pellet was kept at −20°C until it was used for extraction and purification of the fusion protein.
Purification of DvnRV41 and variants.
DvnRV41 and variants of DvnRV41 were purified as follows. Pellets from 50-ml samples of 3-h IPTG-induced cultures of E. coli containing plasmids pCR11 to pCR17 were resuspended in 5 ml of binding buffer containing 10 mM imidazole (pH 7.9) (Amersham Biosciences, Freiburg, Germany). The cells were disrupted by sonication (Aerosec Industrie, Fecamp, France) in ice-cold water (225 W, five times for 5 min each). The cytosoluble fraction (CSF) was separated from the cytoplasmic insoluble fraction and cell debris by centrifugation (15,000 × g, 15 min, 4°C). The CSF was incubated at 80°C for 15 min, filtered (0.45-μm-pore-size filter; Sartorius, Goettingen, Germany) and then loaded onto a 1-ml nickel His-Trap chelating column (His Trap FF column; Amersham Biosciences, Uppsala, Sweden). After loading, the column was eluted with increasing concentrations of imidazole (10 to 500 mM) at pH 7.9. The DvnRV41-fP fusion proteins were found in the 500 mM imidazole elution fraction (EF). This elution process was repeated four times, and the different EFs obtained were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to localize the fusion proteins. After this first immobilized metal affinity chromatography (IMAC) purification step, the EFs containing the fusion proteins were desalted using distilled water by gel filtration (G-25 PD-10 columns; Amersham Biosciences). The DvnRV41-fP fusion proteins were then cleaved with enterokinase EKMax used according to the manufacturer's recommendations (Invitrogen). DvnRV41 and its variants were separated from TRX-His6-Asp4-Lys by a second IMAC step.
Protein and immunoblot analysis.
Proteins were separated under reducing conditions by Tricine-SDS-PAGE (16.5% polyacrylamide [Sigma]) or glycine-SDS-PAGE (15% polyacrylamide), and the gels were stained with AgNO3 (Sigma) and Coomassie blue R-250 (Sigma), respectively. An ultra-low-range marker (Sigma) was used as a molecular mass marker (26.6, 17.0, 14.2, 6.5, 3.5, and 1.1 kDa). The protein concentration was determined by using a bicinchoninic acid protein assay reagent kit (Pierce, Rockford, IL) with bovine serum albumin as a standard. Immunoblotting using the proteins separated by Tricine-SDS-PAGE was performed with anti-DvnCt-keyhole limpet hemocyanin antibodies as previously described (45).
The EF containing DvnRV41 or the variants was loaded onto a C8 solid-phase extraction cartridge (Sep Pak; Waters) conditioned with acetonitrile (ACN) and equilibrated with 0.1% aqueous formic acid (FA). After sample loading, the cartridge was washed with 0.1% aqueous FA, and elution was performed stepwise with 20%, 30%, 40%, and 50% ACN in 0.1% aqueous FA. The 30% ACN fraction was dried under a vacuum and reconstituted in a mixture of ACN and 0.1% FA (50:50, vol/vol). The purity of DvnRV41 variants was analyzed by reversed-phase high-performance liquid chromatography using a C18 column (Capcell 5 μm; 4.6 by 250 mm; Interchim, France). Peptides were detected at 226 nm, and the peptide fractions were collected manually. Molecular masses and the purity of the peptides were analyzed by matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF MS) (Voyager-De-Pro; Applied Biosystems) with α-cyano-4-hydroxycinnamic acid as a matrix. The spectra were acquired in linear mode. Electrospray ionization mass spectrometry analyses were carried out with a quadrupole quadrupole time of flight mass spectrometer (Q-Star Pulsar; Applied Biosystems) to check for the presence of the two disulfide bonds expected for DvnRV41 and its variants. The concentrations of bacteriocins were determined by measurement of the UV absorption at 280 nm, which was converted to the protein concentration using the molecular extinction coefficients calculated from the contributions of individual amino acid residues.
Assessment of antilisterial activity.
The antibacterial activities of DvnRV41-fP and purified DvnRV41 and their variants were assayed by using spot-on-lawn inhibition tests with the sensitive strain L. monocytogenes EGDe (Scharlau, Chemie S.A, Spain) and were expressed in arbitrary activity units (40). We used this strain because of its virulence and the availability of the whole sequence. Ten microliters of each peptide was added to Elliker soft agar plates containing 107 CFU/ml of the indicator strain. The plates were incubated for 16 h at 30°C and then inspected to determine the formation of inhibition zones. The diameter of each inhibition zone was measured and integrated into the calculation for total activity (Table 3).
TABLE 3.
Total and specific antimicrobial activities of DvnRV41-fP and DvnRV41 and their variants with L. monocytogenes EGDea
| Peptide | Activity of fusion protein (AU) | Total activity of purified peptides (AU) | Sp act of purified peptides (AU/μg protein) |
|---|---|---|---|
| DvnRV41 | 3,200 | 76,800 | 958.5 |
| W19F | 0 | 400 | 3.2 |
| Q21S | 0 | 400 | 4.5 |
| A22G | 200 | 2,400 | 17.4 |
| S23Y | 3,200 | 51,200 | 312.6 |
| V30F | 0 | 3,200 | 24.5 |
| L35F | 0 | 12,800 | 93.4 |
| A38V | 0 | 6,400 | 53.8 |
| P40V | 200 | 8,000 | 65.7 |
Antibacterial activities are expressed in arbitrary activity units (AU).
MIC determination.
Two serial dilutions of DvnRV41 or its variants in Elliker broth were placed into wells of a microtiter plate (Nunc). Each well was inoculated with 50 μl of the indicator strain at a concentration of 105 CFU/ml. The microtiter plate cultures were incubated at 30°C for 18 h; the optical density at 600 nm was measured at regular intervals (1 h) using an UltraMicroplate reader (Bio-Tek Instruments). MICs were calculated using the highest dilution showing complete inhibition of the strain tested. MIC determination was repeated independently at least three times (Table 4).
TABLE 4.
MICs of DvnRV41 variants following enterokinase cleavage and further purificationa
| Peptide | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| L. monocytogenes EGDe | E. faecalis JH2-2 | E. faecalis Δpde mutant | E. faecalis ΔglpQ mutant | E. faecalis ΔrpoN mutant | |
| DvnRV41 | 0.1 ± 0.01 | 0.6 ± 0.01 | 3.3 ± 1.11 | 2.1 ± 1.94 | 30.0 ± 10.01 |
| W19F | 21.1 ± 0.9 | 21.0 ± 10.5 | 128.0 ± 0.01 | 128.0 ± 0.01 | 128.0 ± 0.01 |
| Q21S | 18.3 ± 0.9 | 21.8 ± 0.01 | 116.0 ± 0.01 | 116.0 ± 0.01 | 116.0 ± 0.01 |
| A22G | 8.6 ± 0.01 | 17.2 ± 8.6 | 51.8 ± 17.3 | 21.8 ± 0.01 | 136.0 ± 0.01 |
| S23Y | 0.1 ± 0.01 | 1.0 ± 0.2 | 3.6 ± 0.01 | 5.5 ± 4.27 | 40.7 ± 0.01 |
| V30F | 1.3 ± 0.4 | 8.1 ± 0.01 | 12.2 ± 4.1 | 8.1 ± 8.13 | 65.0 ± 0.01 |
| L35F | 0.7 ± 0.2 | 3.2 ± 0.71 | 8.6 ± 0.01 | 4.3 ± 2.14 | 68.5 ± 0.01 |
| A38V | 1.5 ± 0.4 | 7.6 ± 0.01 | 11.4 ± 3.8 | 15.1 ± 0.01 | 60.5 ± 0.01 |
| P40V | 2.5 ± 0.9 | 12.6 ± 2.6 | 35.9 ± 17.1 | 38.4 ± 0.01 | 92.2 ± 30.7 |
The MICs were calculated using the highest dilution showing complete inhibition of the strain tested (the optical density at 600 nm was equal to the optical density at 600 nm of the blank) and were determined after 16 h of incubation at 30°C.
CD spectroscopy.
CD spectra were recorded using a Jasco J-810 instrument equipped with a Peltier effect-based thermostat-controlled cell holder. Samples were studied in quartz cells with path lengths of 0.5 mm at 25°C. They were prepared by dissolving the peptides to obtain a concentration of 50 μM either in 2,2,2-trifluoroethanol (TFE), in TFE-H2O (80:20, vol/vol), or in 10 mM sodium phosphate (pH 7) in the absence or presence of 50 mM dodecylphosphocholine (DPC) micelles. The spectral contribution from the background was subtracted, and the CD signals were normalized to the peptide concentration and expressed as the mean residue weight ellipticity values in degrees cm2 dmol−1. Samples were scanned over the 190- to 260-nm wavelength range by recording values every 1 nm with a scan rate of 50 nm min−1, a response time of 1 s, and a bandwidth of 1 nm. Each spectrum was the average of five scans. The percentages of the various secondary structures were calculated using the CD spectrum deconvolution software Dichroweb (58; http://www.cryst.bbk.ac.uk/cdweb/html/home.html). An estimate of the different structures theoretically present in DvnRV41 and its variants was obtained using the Psipred program (28, 34; http://bioinf.cs.ucl.ac.uk/psipred/).
RESULTS
Purification and characterization of DvnRV41 and its variants.
Eight amino acids were targeted for change in the DvnRV41 variants: Trp19, Gln21, Ala22, Ser23, Val30, Leu35, Ala38, and Pro40. These amino acids were selected because (i) they are common to class IIa bacteriocins (Trp19, Ala22) or (ii) they are essentially specific for the group of class IIa bacteriocins closely related to DvnV41, which possess two disulfide bonds (Fig. 1). They are located mainly in the region between the N- and C-terminal domains and in the C-terminal domain itself. In order to maintain similar local polarities, each amino acid selected was replaced by another amino acid with a similar polarity group, in agreement with the amino acid Venn diagram (51), which resulted in variants W19F, Q21S, A22G, S23Y, V30F, L35F, A38V, and P40V. The eight mutations selected were created by site-directed mutagenesis in vitro, resulting in eight DvnRV41 variants. Each mutation (one DNA triplet) was specifically introduced into the dvnRV41 gene, resulting in substitution of a single amino acid in the DvnRV41 sequence. Expression of the DvnRV41 fusion protein (DvnRV41-fP) and of its variants by the various E. coli Origami hosts harboring the recombinant plasmids listed in Table 1 was induced by addition of IPTG, as described in Materials and Methods. Induction of the T7 RNA polymerase promoter resulted in the expression of newly synthesized polypeptides, whose molecular masses were in agreement with the calculated theoretical molecular masses (data not shown). DvnRV41 and the variants were released enzymatically from the corresponding fusion proteins. The purification procedure allowed us to obtain pure peptides, as shown in Fig. 2 for variant W19F. Prior to further experiments, the presence of the two expected disulfide bonds and the substitutions generated in the DvnRV41 sequence were verified by electrospray ionization mass spectrometry and MALDI-TOF-MS (data not shown).
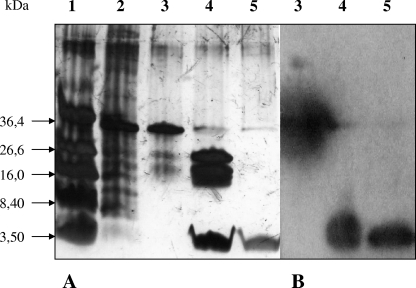
FIG. 2.
Expressed and purified W19F variant. (A) 16.5% Tricine-SDS-PAGE. Lane 1, 3 μl of molecular weight protein markers (Bio-Rad); lane 2, 2 μl of CSF before purification; lane 3, 2 μl of purified fusion protein DvnRV41-fP-W19F; lane 4, 5 μl of DvnRV41-fP-W19F digested with enterokinase EKMax (Invitrogen); lane 5, 5 μl of purified variant W19F. (B) Western blotting with DvnV41 polyclonal antibodies (45). Lane 1, 3 μl of fusion protein DvnRV41-fP-W19F after purification and desalting; lane 2, 3 μl of digested DvnRV41-fP-W19F; lane 3, 3 μl of purified variant W19F.
Determination of antibacterial activities.
The antibacterial activities of IMAC-immobilized eluted fractions (DvnRV41-fP and its variants) and of enterokinase cleavage products were determined by the agar diffusion assay, as previously reported (40, 43). As shown in Table 3, five fusion proteins were unable to inhibit the growth of L. monocytogenes EGDe. Remarkably, DvnRV41-fP-S23Y exhibited strong antilisterial activity, similar to that of DvnRV41-fP. The remaining fusion proteins, DvnRV41-fP-A22G and DvnRV41-fP-P40V, both exhibited low levels of activity under the conditions used.
It was apparent, however, that following enterokinase cleavage and purification, DvnRV41 and its variants all displayed antilisterial activity (Table 3). When both L. monocytogenes EGDe (19) and E. faecalis JH2-2 (4) were used as sensitive indicator strains, MIC measurements (Table 4) showed that the S23Y variant exhibited the highest level of activity, similar to that of unmodified DvnRV41, while the W19F and Q21S variants were the variants most affected by the amino acid replacements. Finally, the other variants exhibited intermediate results, with moderately reduced activities in the following order: L35F > V30F = A38V > P40V > A22G > W19F = Q21S. Overall, after replacement of the selected amino acids, the DvnRV41 MICs increased by factors 10 to 200 and 5 to 30 for L. monocytogenes EGDe and E. faecalis JH2-2, respectively.
The activities of DvnRV41 and its variants were also examined using three E. faecalis JH2-2 mutants (ΔrpoN, Δpde, and ΔglpQ mutants) (Table 4) recently isolated and shown to exhibit intermediate resistance to DvnRV41 (4). All three E. faecalis mutants exhibited higher levels of resistance to DvnRV41 than the wild-type strain, and the MICs were higher by factors of 5 (Δpde mutant), 3 (ΔglpQ mutant), and 50 (ΔrpoN mutant). While the wild-type E. faecalis JH2-2 strain was sensitive to the DvnRV41 variants, the resistant mutants showed considerable levels of resistance to the variants, and the MICs were ≥100 μg/ml for the W19F and Q21S variants (Table 4). When the three E. faecalis JH2-2 mutants resistant to DvnRV41 were examined, the order of activity was the same as that previously determined for the DvnRV41 variants with the wild-type strain. Of the mutants tested, it was notable that the ΔrpoN mutant was the mutant that was least affected by the amino acid replacements; the MICs of the variants increased only by factors of 2 to 4, while the MICs increased by factors of 4 to 40 and 2 to 60 for the Δpde and ΔglpQ mutants, respectively.
CD analysis of DvnRV41 and its variants.
Since they exhibited the greatest loss of antibacterial activity, the W19F, Q21S, A22G, and P40V variants were chosen to examine by CD spectroscopy the impact of the mutations on the global conformation of DvnRV41. The purity and molecular masses of these variants were checked by high-performance liquid chromatography and MALDI-TOF MS (data not shown). CD measurements were obtained either in phosphate buffer at pH 7 or in the presence of DPC micelles and in 80% TFE to mimic membrane environments. The CD spectrum of DvnRV41 in aqueous buffer had a negative band at 200 nm and a positive band at 230 nm characteristic of random coil conformation. Upon addition of DPC or in the presence of 80% TFE, the spectrum showed both a β-sheet contribution, as indicated by the minimum around 215 nm, and the characteristics typical of a right-handed helical structure, with negative bands at 207 and 220 nm accompanied by a positive band around 190 nm. Thus, DvnRV41 appeared to be unstructured under aqueous conditions and to acquire a mixed α-helix-β-sheet structure in the presence of DPC micelles or in 80% TFE. None of the amino acid replacements appeared to result in a complete loss or modification of the structure. The variants exhibited very similar behaviors, with a mainly random coil structure in aqueous medium and a mixed α-helix-β-sheet structure in membrane-mimicking media, except for the W19F variant, which differed in that its CD spectrum was similar under all of the conditions used. An estimate of the different secondary structure contributions obtained using the Dichroweb program (http://www.cryst.bbk.ac.uk/cdweb/html/home.html) (Table 5) allowed comparison of the global folds of the variants. A slight but significant decrease in the helix content, accompanied by a slight increase in the β-sheet content, was observed for all of the variants, particularly for A22G, in both the DPC and TFE environments. Prediction of the three-dimensional structure of DvnV41 using the program Psipred (http://bioinf.cs.ucl.ac.uk/psipred/) indicated that it is potentially organized into 11% helix, 50% β-sheet, and 39% random coil structures, which does not contradict our CD data. The same prediction analysis applied to the variants did not reveal major changes in the structure.
TABLE 5.
CD analysis of the secondary structure of DvnRV41 and its variants in phosphate buffer, DPC micelles, and 80% TFE, determined using the CD spectra deconvolution software Dichroweb
| Prepn | Peptide | % of total
|
||
|---|---|---|---|---|
| α-Helix | β-Sheet | Unordered | ||
| Phosphate buffer | DvnRV41 | 2 | 34 | 64 |
| W19F | 7 | 32 | 61 | |
| Q21S | 5 | 27 | 68 | |
| A22G | 3 | 34 | 63 | |
| P40V | 4 | 36 | 60 | |
| DPC micelles | DvnRV41 | 15 | 24 | 61 |
| W19F | 11 | 27 | 62 | |
| Q21S | 7 | 31 | 62 | |
| A22G | 5 | 31 | 64 | |
| P40V | 8 | 29 | 63 | |
| 80% TFE | DvnRV41 | 14 | 28 | 58 |
| W19F | 9 | 31 | 60 | |
| Q21S | 6 | 32 | 62 | |
| A22G | 4 | 34 | 62 | |
| P40V | 6 | 36 | 58 | |
DISCUSSION
The amino acid content is important for the folding and bioactivity of bacteriocins. Recently, more than 123 bacteriocins produced by gram-positive and gram-negative bacteria have been classified on the basis of their primary structures, and a database has been created (20). This original database has revealed that 93.5% of bacteriocins contain at least one glycine, 25% do not contain cysteine, 32% contain only one pair of cysteines, and the proline content is surprisingly low. Until the industrial use of class IIa bacteriocins is authorized, it is important to unravel the mode of action of these molecules, as well as the structure-function relationships, in order to be able to optimize their use as food additives or antiinfectives. Clearly, the presence of two cysteine residues in a disulfide bond appears to be a prerequisite for the antibacterial activity of class IIa bacteriocins (11). However, the bacteriocins which contain two disulfide bonds exhibit much higher activity than their one-disulfide-bond counterparts (12, 43); this was hypothesized to be due to better structure of the C-terminal domain. Indeed, the C-terminal domain and the second disulfide bridge located in this region have been proposed to be important for determining the target cell specificity of class IIa bacteriocins (15, 16).
DvnV41 has strong antilisterial activity, making it a potential agent in the fight against the food-borne pathogen L. monocytogenes, which continues to cause major illnesses and industrial damage all over the globe. Pediocin PA-1/AcH may be considered an archetype of the class IIa bacteriocins that contain two disulfide bonds and have the highest levels of activity (11). Its antibacterial activity could be related to perturbation of membrane bilayers, and chemical modifications of specific amino acids have been used previously to delineate this process (2). A recognition step involving components of the mannose permease is also probably involved in the mechanism of action (23). Taking into account the previous structure-activity relationship studies of class IIa bacteriocins (2, 14, 36, 38, 48, 53, 56) and the study of Marion and colleagues (2), who specifically examined the roles of the N-terminal region and of the two disulfide bonds in bacteriocin activity, we designed eight variants in order to gain more insight into the structure-activity relationships of DvnV41. As the roles of the disulfide bonds, of the consensus sequence YGNGVXCX4C, and of the positively charged residues mainly located in the N-terminal domain have been studied previously, here we designed variants with single amino acid replacements located in the central and C-terminal parts of the molecule, using a genetic approach which previously has been proven to be efficient (47). We also took advantage of our knowledge of the production of DvnRV41, a recombinant form of this two-disulfide-bond class IIa bacteriocin which retained the antibacterial activity of the natural bacteriocin (44, 60), in order to develop an efficient production system for the desired variants. In order to determine the precise role of the amino acid side chains, each selected amino acid was replaced by another amino acid belonging to a similar polarity group, in agreement with the amino acid Venn diagram (51). To investigate the roles of individual amino acids that are either common to class IIa bacteriocins (Trp19, Ala22) or essentially specific to two-disulfide-bond class IIa bacteriocins, we compared the antibacterial activities of W19F, Q21S, A22G, S23Y, V30F, L35F, A38V, and P40V variants of DvnRV41 and studied the global conformational features of the most affected variants.
A CD analysis of the secondary structure elements of DvnRV41 and its variants in phosphate buffer at pH 7, in 80% TFE (3), and in DPC micelles at pH 7 showed that DvnRV41 and its variants have mainly an unordered structure in an aqueous environment, while they have a mixed conformation with both regions organized in β-sheets and α-helices in an anisotropic solvent or in the presence of DPC micelles, as previously observed for other class IIa bacteriocins (17, 18, 56, 57). Such a mixed α-helix-β-sheet conformation has also been determined by nuclear magnetic resonance and molecular modeling for other class IIa bacteriocins, curvacin A (21), leucocin A (18), and sakacin P (54), while carnobacteriocin B2 is organized mainly in an α-helix conformation (56). The CD data and calculated percentages of different structures (Table 5) showed that the global conformation is not substantially modified by the single amino acid replacements, indicating that no major perturbation of the conformation occurred upon amino acid replacement and that the local side chain modification is responsible by itself for the decreases in activity observed for the variants. Even if the three-dimensional structures available for one-disulfide-bond class IIa bacteriocins (18, 21, 54, 56) have some differences, which are presumably due to the peptides themselves and also to different environments, they are apportioned mainly into mixed α-helix-β-sheet structures, with the helix located at the center and/or in the C-terminal region. However, the solution structure of the class IIa bacteriocins that contain two disulfide bonds has not been determined until now. At the very most, a scheme for the pediocin PA-1/AcH structure, resulting from CD measurements in micelle and liposome environments, has been proposed (57). This structure, which is in agreement with the structure predictions resulting from the pediocin PA-1/AcH amino acid sequence, proposes that the second disulfide bond linking the two cysteines located at the central and very C-terminal regions folds the C-terminal domain back on the central helix. In the case of DvnRV41, both structure prediction and estimation of the percentages for the secondary structure by CD spectra deconvolution do not contradict this hypothesis and indicate that this bacteriocin should include an N-terminal β-sheet domain, as also observed for one-disulfide-bond bacteriocins, and a short helix located at the center of the peptide. The activities of our variants may be analyzed in light of this hypothetical structure.
Compared to the DvnRV41 activities, the activities against L. monocytogenes and E. faecium were altered for all of the variants except S23Y. All of the selected amino acids except Ser23 thus appeared to be important for bacteriocin activity. Notably, class IIa bacteriocins contain one to four tryptophan residues, one of which is highly conserved (11). The three Trp residues in DvnV41 have been shown previously to be crucial for activity, since modification of any one of them resulted in inactivation of the bacteriocin activity (2). An essential role for this residue was also described previously for sakacin P (14), mesentericin Y105 (38), and pediocin PA-1/AcH (36), which is structurally more similar to DvnV41. In this study, replacement of Trp19 in DvnRV41 had, as expected, a deleterious effect on the antibacterial activity, thus confirming the major role of this residue in the mechanism of action of DvnV41. It is worth noting that even when another aromatic residue, such as phenylalanine in the W19F variant, is used to replace Trp, the molecule is not active. The irreplaceable role of tryptophan in DvnV41 activity is presumably related to its determinative role in the adsorption and orientation of proteins in membranes due to its capacity to form hydrogen and hydrophobic bonds with the polar and nonpolar groups of polar lipids (24, 25). Other aromatic residues have been shown to play important roles in the antibacterial activity of class IIa bacteriocins (41). Indeed, substitution of Phe33 with serine eliminated the antibacterial activity of carnobacteriocin B2 (42). In our case, introducing a supplementary phenylalanine residue at position 30 or 35 (variants V30F and L35F) did not increase the activity. DvnV41 has three tyrosine residues, and modification by nitration of each of them results in a molecule that has activity, while modification of all of them results in a loss of the antilisterial activity (2). In this study, we added a supplementary Tyr residue to the DvnRV41 sequence through Tyr substitution at Ser23, but the resulting variant exhibited the initial activity of DvnRV41 (Table 4). Thus, introducing a supplementary Tyr residue into the DvnV41 sequence, at least at position 23 located in the putative helix domain, is not beneficial for activity.
The modified residue in the Q21S variant results in a drastic decrease in activity (the activity with L. monocytogenes decreases by a factor 200). Shortening the side chain and exchanging an amide function for a hydroxyl function in the putative helix thus have a dramatic effect on the activity. Variant A22G is also much affected by the amino acid replacement (the activity with L. monocytogenes decreases by a factor 100). The putatively helical central region thus appears to play a pivotal role in the activity. It is interesting to point out that the P40V variant also shows a significant decrease in antibacterial activity (by a factor 20). The importance of proline in the structure and biological properties of peptides and proteins has been extensively documented. The P40 residue is considered a helix and β-sheet breaker (32) due to the inability of this amino acid to establish a stabilizing hydrogen bond and its ability to induce bends or hinges in helices. Studies of the structure of bacteriorhodopsin have revealed its major role in transmembrane α-helices (33). In diphtheria toxin, proline 345 located in the TH8 helix undergoes a conformational change, which is a prerequisite for toxin translocation (27). Introduction of proline into the mesentericin Y105 sequence significantly altered the activity of this pediocin-like bacteriocin (38) through perturbation of the helix. It is clear from our CD data that the decrease in the activity of variant P40V, observed with both L. monocytogenes and E. faecalis, was not related to a profound and global conformational change. This is not really surprising, since in our case the proline is located at the extreme C terminus of the sequence. As the C terminus of class IIa bacteriocins has been proposed to be essential for activity and target specificity, the presence of a proline at this position would be hypothesized to be more important for recognition by the mannose permease EIItMan, which is thought to act as a receptor for class IIa bacteriocins (23), than for interaction with membrane bilayers. Position 22 also appears to be critical, since the activity of variant A22G is among the activities most affected. Taken together, these data clearly show that (i) the central region (amino acids 19 to 23), which is part of the putative helix, and (ii) the extreme C terminus are pivotal for activity, as shown by the results for variants W19F, Q21S, A22G, S23Y, and P40V, while the region including amino acids 30 to 38, exemplified by variants V30F, L35F, A38V, appears to be less important. Thus, it could be hypothesized that the central region is responsible for the correct spatial positioning of DvnV41 side chains required for its interaction with membrane bilayers and/or recognition by the receptor.
Resistance of E. faecalis to DvnV41 is associated with the rpoN, glpQ, and pde genes, which encode the σ54 factor that is involved in expression of the docking molecule that participates in the recognition mechanism, a putative glycerophosphoryl diester phosphodiesterase (GlpQ) that participates in degradation of phospholipids and fatty acids, and a protein with a putative phosphodiesterase function, respectively (4). Compared to the DvnRV41 activity, the activities of the different variants with the E. faecalis strains with intermediate resistance (ΔglpQ and Δpde mutants) are globally similarly or less affected than the activities with the wild-type strain. The highest level of resistance, which is conferred by rpoN, is associated with reduced sensitivity to the amino acid replacements, as the MICs are decreased only by factors of 2 to 4 for the ΔrpoN mutant, compared to factors of 4 to 60 for the Δpde and ΔglpQ mutants.
Convincingly, the present study underlines the important and different roles of individual amino acids located in the center of the DvnV41 sequence, such as Trp19, Gln21, or Ser23, and at the C terminus in the antibacterial activity of DvnV41. Indeed, CD used to examine the effects of single substitutions on the global structure has not revealed any major differences at the structural level. Furthermore, none of the single amino acid replacements is able to increase the activity of the bacteriocin or to change the specificity of the bacteria targeted by the bacteriocin. This study paves the way for further detailed understanding of the DvnV41 mechanism of action, using randomized mutagenesis for the production of a great number of variants and determination of the three-dimensional structure, which will be our next focus.
Acknowledgments
We thank Paul Cotter for critical reading of the manuscript and improvement of the English.
Research at ENITIAA was supported in part by la Région des Pays de la Loire through the VANAM II program. J.R. was a recipient of a Ph.D. scholarship awarded by the French Government.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Abriouel, H., E. Valdivia, A. Galvez, and M. Maqueda. 2001. Influence of physico-chemical factors on the oligomerization and biological activity of bacteriocin AS-48. Curr. Microbiol. 42:89-95. [DOI] [PubMed] [Google Scholar]
- 2.Bhugaloo-Vial, P., J. P. Douliez, D. Moll, X. Dousset, P. Boyaval, and D. Marion. 1999. Delineation of key amino acid side chains and peptide domains for antimicrobial properties of divercin V41, a pediocin-like bacteriocin secreted by Carnobacterium divergens V41. Appl. Environ. Microbiol. 65:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, M. 1998. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31:297-355. [DOI] [PubMed] [Google Scholar]
- 4.Calvez, S., A. Rincé, Y. Auffray, H. Prévost, and D. Drider. 2007. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 153:1609-1618. [DOI] [PubMed] [Google Scholar]
- 5.Caridi, A. 2002. Selection of Escherichia coli-inhibiting strains of Lactobacillus paracasei subsp. paracasei. J. Ind. Microbiol. Biotechnol. 29:303-308. [DOI] [PubMed] [Google Scholar]
- 6.Chikindas, M. L., M. J. García-Garcerá, A. J. Driessen, A. M. Ledeboer, J. Nissen-Meyer, I. F. Nes, T. Abee, W. N. W. N. Konings, and G. Venema. 1993. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 59:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cintas, L. M., P. Casaus, L. S. Håvarstein, P. E. Hernández, and I. F. Nes. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 10.Dalet, K., Y. Cenatiempo, P. Cossart, Y. Héchard, and The European Listeria Genome Consortium. 2001. A σ54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263-3269. [DOI] [PubMed] [Google Scholar]
- 11.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eijsink, V. G., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 14.Fimland, G., V. G. Eijsink, and J. Nissen-Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 15.Fimland, G., O. R. Blingsmo, K. Sletten, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1996. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 62:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fimland, G., L. Johnsen, L. Axelsson, M. B. Brurberg, I. F. Nes, V. G. H. Eijsink, and J. Nissen-Meyer. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J. Bacteriol. 182:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleury, Y., M. A. Dayem, J. J. Montagne, E. Chaboisseau, J. P. Le Caer, P. Nicolas, P., and A. Delfour. 1996. Covalent structure, synthesis, and structure-function studies of mesentericin Y105, a defensive peptide from gram-positive bacteria Leuconostoc mesenteroides. J. Biol. Chem. 271:14421-14429. [DOI] [PubMed] [Google Scholar]
- 18.Fregeau Gallagher, N. L., M. Sailer, W. P. Niemczura, T. T. Nakashima, M. E. Stiles, and J. C. Vederas. 1997. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062-15072. [DOI] [PubMed] [Google Scholar]
- 19.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, F. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. García-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, F. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 20.Hammami, R., A. Zouhir, J. Benhamida, and I. Fliss. 2007. BACTIBASE: a new web-accessible data base for bacteriocin characterization. BMC Microbiol. 7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haugen, H. E., G. Fimland, J. Nissen-Meyer, and P. E. Kristiansen. 2005. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A. Biochemistry 44:16149-16157. [DOI] [PubMed] [Google Scholar]
- 22.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 23.Héchard, Y., C. Pelletier, Y. Cenatiempo, and J. Frère. 2001. Analysis of σ54-dependent genes in Enterococcus faecalis: a mannose PTS permease (EIIMan) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575-1580. [DOI] [PubMed] [Google Scholar]
- 24.Hu, W., and T. A. Cross. 1995. Tryptophan hydrogen bonding and electric dipole moments: functional roles in the gramicidin channel and implications for membrane proteins. Biochemistry 34:14147-14155. [DOI] [PubMed] [Google Scholar]
- 25.Hu, W., N. D. Lazo, and T. A. Cross. 1995. Tryptophan dynamics and structural refinement in a lipid bilayer environment: solid state NMR of the gramicidin channel. Biochemistry 34:14138-14146. [DOI] [PubMed] [Google Scholar]
- 26.Ivanova, I., V. Miteva, T. Stefanova, A. Pantev, I. Budakov, S. Danova, P. Moncheva, I. Nikolova, X. Dousset, and P. Boyaval. 1998. Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 42:147-158. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, V. G., P. J. Nicholls, W. H. Habig, and R. J. Youle. 1993. The role of proline 345 in diphtheria toxin translocation. J. Biol. Chem. 268:3514-3519. [PubMed] [Google Scholar]
- 28.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 29.Kalmokoff, M. L., S. K. Banerjee, T. Cyr, M. A. Hefford, and T. Gleeson. 2001. Identification of a new plasmid-encoded sec-dependent bacteriocin produced by Listeria innocua 743. Appl. Environ. Microbiol. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 31.Ko, S.-H., and C. Ahn. 2000. Bacteriocin production by Lactococcus lactis KCA2386 isolated from white kimachi. Food Sci. Biotechnol. 9:263-269. [Google Scholar]
- 32.Li, S. C., N. K. Goto, K. A. Williams, and C. M. Deber. 1996. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. USA 93:6676-66781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, H., T. Marti, and P. J. Booth. 2001. Proline residues in transmembrane alpha helices affect the folding of bacteriorhodopsin. J. Mol. Biol. 308:437-446. [DOI] [PubMed] [Google Scholar]
- 34.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 35.Messi, P., M. Bondi, C. Sabia, R. Battini, and G. Manicardi. 2001. Detection and preliminary characterization of a bacteriocin (plantaricin 35d) produced by a Lactobacillus plantarum strain. Int. J. Food Microbiol. 64:193-198. [DOI] [PubMed] [Google Scholar]
- 36.Miller, K. W., R. Schamber, O. Osmanagaoglu, and B. Ray. 1998. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl. Environ. Microbiol. 64:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minahk, C. J., F. Dupuy, and R. D. Morero. 2004. Enhancement of antibiotic activity by sub-lethal concentrations of enterocin CRL35. J. Antimicrob. Chemother. 53:240-246. [DOI] [PubMed] [Google Scholar]
- 38.Morisset, D., J. M. Berjeaud, D. Marion, C. Lacombe, and J. Frère. 2004. Mutational analysis of mesentericin Y105, an anti-Listeria bacteriocin, for determination of impact on bactericidal activity, in vitro secondary structure, and membrane interaction. Appl. Environ. Microbiol. 70:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50-61. [DOI] [PubMed] [Google Scholar]
- 40.Pilet, M. F., X. Dousset, R. Barré, G. Novel, M. Desmazeaud, and J. C. Piard. 1995. Evidence for two bacteriocins produced by Carnobacterium divergens V41 and Carnobacterium piscicola V1 isolated from fish and active against Listeria monocytogenes. J. Food Prot. 3:256-262. [DOI] [PubMed] [Google Scholar]
- 41.Porcelli, F., R. Verardi, L. Shi, K. A. Henzler-Widman, A. Ramamoorthy, and G. Veglia. 2008. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry 47:5565-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quadri, L. E., L. Z. Yan, M. E. Stiles, and J. C. Vederas. 1997. Effect of amino acid substitutions on the activity of carnobacteriocin B2. Overproduction of the antimicrobial peptide, its engineered variants, and its precursor in Escherichia coli. J. Biol. Chem. 272:3384-3388. [DOI] [PubMed] [Google Scholar]
- 43.Richard, C., R. Cañon, K. Naghmouchi, D. Bertrand, H. Prévost, and D. Drider. 2006. Evidence on correlation between number of disulfide bridge and toxicity of class IIa bacteriocins. Food Microbiol. 23:175-183. [DOI] [PubMed] [Google Scholar]
- 44.Richard, C., D. Drider, K. Elmorjani, D. Marion, and H. Prévost. 2004. Heterologous expression and purification of active divercin V41, a class IIa bacteriocin encoded by a synthetic gene in Escherichia coli. J. Bacteriol. 186:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richard, C., D. Drider, I. Fliss, S. Denery, and H. Prévost. 2004. Generation and utilization of polyclonal antibodies to a synthetic C-terminal amino acid fragment of divercin V41, a class IIa bacteriocin. Appl. Environ. Microbiol. 70:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard, C., A. Brillet, M. F. Pilet, H. Prévost, and D. Drider. 2003. Evidence on inhibition of Listeria monocytogenes by divercin V41 action. Lett. Appl. Microbiol. 36:288-292. [DOI] [PubMed] [Google Scholar]
- 47.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 48.Simon, L., C. Fremaux, Y. Cenatiempo, and J. M. Berjeaud. 2002. Sakacin g, a new type of antilisterial bacteriocin. Appl. Environ. Microbiol. 68:6416-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. P. Levchuk, O. E. Svetoch, and B. S. Seal. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svetoch, E. A., B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. N. Borzenkov, V. P. Levchuk, O. E. Svetoch, Y. N. Kovalev, Y. G. Stepanshin, G. R. Siragusa, B. S. Seal, and N. J. Stern. 2008. Diverse antimicrobial killing by Enterococcus faecium E 50-52 bacteriocin. J. Agric. Food Chem. 56:1942-1948. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, W. R. 1986. The classification of amino acid conservation. J. Theor. Biol. 119:205-218. [DOI] [PubMed] [Google Scholar]
- 52.Todorov, S. D., and L. M. T. Dicks. 2004. Effect of medium components on bacteriocin production by Lactobacillus pentosus ST151BR, a strain isolated from beer produced by the fermentation of maize, barley and soy flour. World J. Microbiol. Biotechnol. 20:643-650. [Google Scholar]
- 53.Tominaga, T., and Y. Hatakeyama. 2006. Determination of essential and variable residues in pediocin PA-1 by NNK scanning. Appl. Environ. Microbiol. 72:1141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uteng, M., H. H. Hauge, P. R. L. Markwick, G. Fimland, D. Mantzilas, J. Nissen-Meyer, and C. Muhle-Goll. 2003. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42:11417-11426. [DOI] [PubMed] [Google Scholar]
- 55.Wachsman, M. B., V. Castilla, A. P. de Ruiz Holgado, R. A. de Torres, F. Sesma, and C. E. Coto. 2003. Enterocin CRL35 inhibits late stages of HSV-1 and HSV-2 replication in vitro. Antiviral Res. 58:17-24. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., M. E. Henz, N. L. Gallagher, S. Chai, A. C. Gibbs, L. Z. Yan, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 1999. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438-15447. [DOI] [PubMed] [Google Scholar]
- 57.Watson, R. M., R. W. Woody, R. V. Lewis, D. S. Bohle, A. H. Andreotti, B. Ray, and K. W. Miller. 2001. Conformational changes in pediocin AcH upon vesicle binding and approximation of the membrane-bound structure in detergent micelles. Biochemistry 40:14037-14046. [DOI] [PubMed] [Google Scholar]
- 58.Whitmore, L., and B. A. Wallace. 2004. Dichroweb, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668-W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yildirim, S., D. Konrad, S. Calvez, D. Drider, H. Prévost, and C. Lacroix. 2007. Production of recombinant bacteriocin divercin V41 by high cell density Escherichia coli batch and fed-batch cultures. Appl. Microbiol. Biotechnol. 77:525-531. [DOI] [PubMed] [Google Scholar]